Abstract
Pentachlorophenol (PCP) bioremediation by the fungal strains amongst the cork-colonising community has not yet been analysed. In this paper, the co- and direct metabolism of PCP by each of the 17 fungal species selected from this community were studied. Using hierarchical data analysis, the isolates were ranked by their PCP bioremediation potential. Fifteen isolates were able to degrade PCP under co-metabolic conditions, and surprisingly Chrysonilia sitophila, Trichoderma longibrachiatum, Mucor plumbeus, Penicillium janczewskii and P. glandicola were able to directly metabolise PCP, leading to its complete depletion from media. PCP degradation intermediates are preliminarily discussed. Data emphasise the significance of these fungi to have an interesting potential to be used in PCP bioremediation processes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Pentachlorophenol (PCP) was first produced in the 1930s as a wood preservative. Subsequently, it has been used worldwide as a herbicide, biocide and pesticide, resulting in significant contamination of terrestrial and aquatic ecosystems [13]. Due to health and environmental concerns, PCP use is now widely restricted, but it is still authorised, e.g., as a wood preservative. Importantly, man-made PCP has the potential for long-range atmospheric transport, representing an indirect source for the contamination of human foods and drinking-water (for revision see, e.g., [7, 15]). PCP is considered a priority hazardous substance and is believed to be an endocrine disruptor, reinforcing the concerns of the consequences of long-term exposure to humans and wildlife [7]. Recently, Taylor et al. [33] evaluated PCP for its ability to affect natural human killer cell function and demonstrated that 10 μM PCP for 1 h caused a progressive loss (>80%) of lytic function within 6 days of exposure.
Currently available approaches to pollution mitigation, such as incineration and storage, are expensive, inefficient and lead to additional environmental problems through the introduction of hazardous by-products. The most promising approach is bioremediation exploiting the catabolic capability of microorganisms. Numerous reports have shown that PCP is degraded in the environment by microorganisms (e.g., [25, 31, 35]), further emphasising the potential of bioremediation for its efficient elimination. Filamentous fungi are unique microorganisms, ensuring the degradation of the lignocellulose plant polymers: the most important degradation event in the carbon cycle of earth. There are about 72,000 named species, and new species are being added at the rate of about 700–1,500 each year [4].
PCP bioremediation by basidiomycetes and ascomycetes (and zygomycetes) fungi has not been fully assessed, as the latter are less studied. The best-studied basidiomycete, the white-rot fungus Phanerochaete chrysosporium, is known to mineralize several chlorinated phenols, including PCP [20]. The pathway for the degradation of PCP has been elucidated, identifying as the main metabolites: tetrachlorobenzoquinone (TeCBQ) and tetrachlorodihydroquinone (TeCHQ). The pathways, presumably, lead to CO2 mainly through reductive dechlorination reactions. However, PCP degradation is not an exclusive feature of white rot fungi; neither can it be correlated solely with ligninolytic strains [19]. For example, the brown rot species Gloeophyllum striatum and G. trabeum efficiently degraded PCP in the absence of ligninolytic activities [9]. Additionally, PCP disappearance is also efficiently mediated by non-producing and producing phenoloxidases ascomycetes [19]. That is the case for the nonligninolytic ascomycete Trichoderma harzianum that can actively degrade PCP [36].
Cork, a material of vegetable origin, is the bark of the cork oak (Quercus suber), whose main component is suberin, a complex aromatic biopolymer structurally related to lignin and, accordingly, highly resistant to degradation. Several soft rot species, amongst the cork-colonising consortia, were able to completely perforate the cork cell wall, suggesting their biodegradation ability [29]. Cork-colonising fungi isolates were also observed to degrade some chlorophenols, leading to the formation of chloroanisoles as a minor [28] or major intermediate [1].
In this study, the co- and direct metabolism of PCP by each of the 17 fungal strains selected from the cork-colonising community were studied. Data ranked the isolates by their PCP bioremediation potential, and surprisingly 15 of the fungal strains degraded PCP up to a certain level.
Materials and methods
Biological
All fungal strains used were environmental isolates previously isolated from cork samples purchased from several Portuguese cork industries [8, 17, 30] and are part of the Instituto de Biologia Experimental e Tecnológica (IBET) culture collection. They were: Chrysonilia sitophila (DSM 16514), Mucor plumbeus Bonord (DSM 16513), Trichoderma longibrachiatum Rifai (DSM 16517), Cladosporium herbarum, Penicillium glabrum (Wehmer) Westling (DSM 16516), P. olsonii Bainier and Sartory (DSM 16515), Eupenicillium hirayamae D.B. Scott and Stolk (anamorph state of P. hirayamae), P. brevicompactum Dierckx, P. glandicola (Oudem) Seifert and Samson, P. variabile Sopp, P. diversum Raper and Fennell, P. decumbens Thom, P. cf. janczewskii K.M. Zalessky, P. corylophilum Dierckx, P. adametzii Zaleski, P. fennelliae Stolk and P. restrictum J.C. Gilman and E.V. Abbot.
Fungal cultures
Each fungal strain was conserved as a suspension of spores in glycerol (10%, v/v) at –80°C. The fungal spore suspensions were prepared as previously described [28]. Briefly, the spores were harvested from fungal colonies after ageing for 15 days at 27°C on malt extract agar (Merck) or dichloranglycerol agar (Oxoid) with a saline solution (0.085 g/l NaCl), filtered through glass wool and washed by repeated centrifugation at 4°C (20,000g, 20 min). The spore suspension was allowed to thaw slowly at 4°C and washed in minimal medium (see below) before being used. The optical density (scatter) of the suspension was measured at 600 nm and calibrated by counting the number of spores in a haemocytometer. The initial fungal spores’ density in the cultures was standardised to 105 spores/ml.
Fungal cultures were grown in media containing sodium nitrate as the sole source of nitrogen. The minimal culture media contained, per liter of water, 1 g K2HPO4; 3 g NaNO3; 10 mg ZnSO4 · 7H2O; 5 mg CuSO4 · 5H2O; 0.5 g MgSO4 · 7H2O; 10 mg FeSO4 · 7H2O; 0.5 g KCl and glucose: 0 and 10 g for minimal media (MM) and glucose minimal media (GMM), respectively. For the PCP degradation experiments the toxic was added to the culture media from a concentrated stock solution (7.5 g/l) in ethanol. The final PCP concentrations in the media ranged from 0 to 20 mg/l, thus guaranteeing that ethanol was not affecting fungal growth (data not shown).
PCP degradation experiments
The inhibitory effect of PCP on fungal spore germination was inferred by measuring the optical density of the culture media at 600 nm during the incubation period. The method used was adapted from a method for the determination of the broth microdilution minimal inhibitory concentration (MIC) [10]. Briefly, 250 μl of the culture media (MM or GMM) containing a final PCP concentration in the range 1-20 mg/l was spread onto 96-well microtitre plates. The fungal spore suspensions were used to inoculate the culture media, and the plates were incubated in an incubation chamber (Aralab Clima Plus) at 27°C and 70% relative humidity, in the dark, for a maximum period of 60 days. The experiments were performed in quadruplicate, including controls without PCP and controls for PCP abiotic decay.
After incubation a chloramphenicol solution (Sigma) in ethanol (for volume normalisation) was added to the wells. The culture media were centrifuged at 4°C for removal of cell debris and solids (20,000g, 20 min). Samples were stored at –20°C until further analysis.
PCP quantification by HPLC
PCP in samples was analysed by high performance liquid chromatography (HPLC), consisting of a Waters 510 Pump, Waters 715 plus Autosampler, Waters Temperature Control Module and Waters 486 Absorbance detector (Waters Chromatography, Milford, MA). Data acquisition was accomplished with the Millenium32 version 3.05.01, 1998 system (Waters Chromatography). PCP and other chlorinated compounds were detected at 230 nm. The chromatographic separation was carried out using a Symmetry C18 column (4.6 × 250 cm), 5-μm particle size (Waters Chromatography) and set at 35°C. The mobile phase, at a flow rate of 0.8 ml/min, consisted of a solution of 0.1% phosphoric acid (solvent A) and acetonitrile (solvent B). The gradient program was implemented as follows: 90–1.0% of A in 30 min, 2 min to return to the initial conditions and 10 min to re-equilibrate the column to the initial conditions. The external standard method was used for PCP quantification (retention time: 30.3), within the limits of 0.1–15 mg/l.
Pure standards of the following chlorinated compounds were also analysed: hydroquinone (HQ) (Aldrich), chlorohydroquinone (CHQ) (Aldrich), tetrachlorohydroquinone (TeCHQ) (Sigma), 2,6-dichloro-1,4-benzoquinone (DCBQ) (Aldrich), tetrachloro-1,4-benzoquinone (TeCBQ) (Riedel-de Haën) and pentachloroanisole (PCA) (Sigma) (retention times: 6.3, 12.6, 21.4, 19.9, 22.1 and 35.6; respectively). All HPLC solvents used were of the highest analytical grade, and water was obtained from a Milli-Q system (Millipore).
Glucose quantification
Glucose analysis was undertaken using the HPLC system described above, connected to a LKB 2142 Differential Refractometer (Bromma) detector. Data acquisition was as previously described and chromatographic separation undertaken using an Aminex HPX-87H (300 × 7.8 mm), 9-μm particle size (Bio-Rad) and set at 60°C. Elution was carried out isocratically, at a flow rate of 0.5 ml/min, with 0.005 N of H2SO4. For the quantification the external standard method was also used (quantification limits: 0.125–50 g/l).
Detection of PCP degradation intermediates
The HPLC software (Millenium32 version 3.05.01, 1998, Waters Chromatography) was used to retrieve the full peak list of each 230-nm chromatographic analysis. Integration parameters (equal to those used to integrate the PCP peak) were set to: peak width = 15.0, threshold = 30.0, minimum area = 0 and minimum height = 300. Only the peaks detected between 6.5 and 35 min were considered. PCP and chloramphenicol peaks were used to align the chromatograms. The peak lists were initially compared using Microsoft Excel tools (pivot tables) to select data containing solely the peaks that were PCP dependent. The retention times of the peaks in the resulting matrix were compared to those of the pure standards, which provided preliminary identity.
Computational interpretation of data
The numerical data for PCP decay—PCPd (calculated as the percentage of PCP removed from the media relatively to its initial concentration) and residual glucose—Gr (calculated as the percentage of glucose in the media relatively to its initial concentration) were analysed by hierarchical cluster analysis using the complete linkage between the groups calculated by the squared Euclidian distance method (SPSS v. 16.0). The distance between two clusters was calculated as the greatest distance between the members of the relevant clusters.
Results
Fungal growth behaviour in the presence of PCP
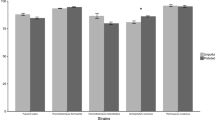
The inhibitory effect of PCP on germination ability of the fungal spores under co-metabolic conditions (GMM) was inferred measuring the optical density of the submerged culture media at 600 nm during the incubation period. Increase of absorbance was taken as an indication of growth. Spore formation (gauged by eye) and/or the absorbance approaching a constant value indicated that the culture had entered a stationary growth phase. The fungal isolates can be divided into four main groups according to their PCP tolerance level (mg/l): 5 ≤ [C. sitophila, P. diversum; P. fenneliae, P. restrictum] < 10; 10 ≤ [C. herbarum, M. plumbeus, P. variabile] < 15; 15 ≤ [P. adametzii, P. corylophilum, P. decumbens, P. glandicola] < 20; 20 ≤ [P. brevicompactum, E. hirayamae, P. janczewskii, P. olsonii, P. glabrum, T. longibrachiatum] (Table 1). For each fungal strain, the growth curve in GMM containing PCP can provide additional information on its toxicity (example shown in Fig. 1). In fact, for example in the growing cultures of C. sitophila, the lag phase of growth increases (5 mg/l PCP), but in M. plumbeus the growth curves are either rather similar (5 mg/l PCP) or suggest a lower production of fungal biomass (10 mg/l PCP) (see Fig. 1).
Analysis of fungal growth profiles in GMM containing PCP. The growth curves of C. sitophila (a) and M. plumbeus (b) strains are shown as an example. Fungal growth was monitored along the incubation period measuring the media scatter at 600 nm. Initial PCP concentration in media was in mg/l: 0 (filled circle); 5 (open square); 10 (open triangle) and 15 (multiplication)
PCP abiotic degradation
Analyses of the abiotic degradation of PCP in the non-inoculated media (MM and GMM) were made to evaluate the contribution of physical-chemical events in the overall decay. Abiotic decay of PCP in the GMM and in the MM were 33 ± 10% and 7.5 ± 1.5%, respectively. Independent tests on PCP absorption to the plates, the fungal spores and the mycelium showed an insignificant contribution to the overall abiotic decay (data not shown).
PCP biotic degradation in co-metabolic conditions (GMM)
PCPd and the percentage of Gr measured in the 17 fungal cultures in co-metabolic conditions (GMM) are presented in Table 1. The fungus micro-cultures were treated as a single sample, selecting only the individual wells that reported the same growth behaviour. Independent runs indicated the cultures behaviour to be consistent (data not shown), and replicates of the same fungus showed a consistent standard deviation (<10%) for PCP decay. The chloramphenicol concentration measured chromatographically was use to infer the cultures' final volume, allowing to calculate accurately PCP decay in media (thus eliminating errors introduced by evaporation).
PCP decay observed in the cultures fully inhibited by the toxic was 29 ± 10%, agreeing with the abiotic decay determined in the non-inoculated GMM controls. In these cultures glucose was not consumed, or only minor consumption was detected (≤20%), confirming the growth inhibition data. Active fungal growth results in glucose exhaustion from the media (Table 1). In view of that, overall decay of PCP in the active growing cultures has only a minor contribution from abiotic decay of PCP (≥7.5 ± 1.5%). Glucose quantification for different replicates showed a consistently low standard deviation (≤2%). Amongst the isolates tested, those able to degrade PCP in all concentrations in order of efficiency were: P. brevicompactum and P. olsonii > P. janczewskii > P. glabrum > E. hirayamae ≫ T. longibrachiatum. Only three isolates completely consumed the PCP from the media: T. longibrachiatum and M. plumbeus > P. glandicola, up to PCP initial concentration of 10 and 5 mg/l, respectively. The moderate (~50–75%) and the weakest (<45%) PCP consumers were P. decumbens > P. variabile > P. fennelliae and C. sitophila (PCP up to 15, 10 and 5 mg/L) and P. adametzii and P. corylophilum > C. herbarum (PCP up to 15 and 10, respectively). P. restrictum and P. diversum were unable to degrade PCP, although they can still grow in the presence of 5 mg/l of PCP.
Cluster analysis of PCP biotic degradation in co-metabolic conditions (GMM)
The relationships between the capacities of the isolates to tolerate and degrade PCP co-metabolically were established by using a hierarchical cluster analysis. In the resulting tree (depicted in Fig. 2), the branch lengths reflect the degree of dissimilarity between the fungal strains. There are six main clusters (cutoff rescaled distance of six) that separate the isolates by their PCP bioremediation potential (accounting for, equally, the species tolerance level and PCP degradation capacity): high (I and II), moderate (III, IV), low (V) and null (VI). In fact, cluster VI can be regarded as the outlier group due to the apparent inability of the isolates, P. restrictum and P. diversum, to degrade PCP. The fungal species with the strongest bioremediation potential are the most tolerant ones: P. brevicompactum, E. hirayamae, P. janczewskii, P. olsonii and P. glabrum, with the exception of T. longibrachiatum. The latter has only scarcely grown in the presence of 20 mg/l of PCP, as shown by the high Gr content.
Cluster analysis of the growth and bioremediation behaviour of the cork-colonising fungal strains during co-metabolic degradation of PCP. Data were analysed by hierarchical cluster, using the complete linkage between the groups calculated by the squared Euclidian distance method. The resulting six clusters of strains are labelled
PCP biotic degradation by fungi in metabolic conditions
PCP metabolism by some of the tested fungal isolates was observed in MM containing PCP up to initial levels of 5 mg/l. Interestingly, amongst the 17 isolates tested only M. plumbeus, P. glandicola, C. sitophila, P. janczewskii and T. longibrachiatum were able to consume PCP and have tolerated the toxic in a distinctive way (Table 2). From these, only C. sitophila and T. longibrachiatum failed to grow in the presence of 5 mg/l, yet completely metabolised PCP at an initial concentration of 2.5 and 1 mg/l, respectively.
Analysis of PCP metabolic intermediates
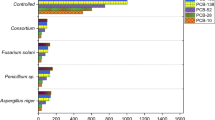
Each fungal culture produced a PCP-specific chromatographic footprint (UV 230 nm), which was highly consistent between replicates (see "Detection of PCP degradation intermediates" above). They can be regarded qualitatively as the metabolic fingerprinting of the extracellular metabolites of the fungus, which can be computed so as to select solely the peaks, species specific, which are PCP dependent. The putative identity of PCP degradation intermediates (in MM and GMM) was made through analysis of the similarity of the chromatographic peaks and retention times (T r) with that of the standard compounds. Only in co-metabolic conditions could the newly formed peaks be associated to the standards, as depicted in Fig. 3, e.g., P. variabile. The putative identification of CHQ was possible for the cultures of C. sitophila, P. corylophilum, P. glabrum, P. glandicola, P. janczewskii and P. variabile; 2,6-dichloro-1,4-benzoquinone (DCBQ) in the cultures of C. herbarum, P. decumbens and T. longibrachiatum; and tetrachloro-1,4-benzoquinone (TeCBQ) in the cultures of P. adametzii.
Chromatographic profiles (between 6 and 32 min) of P. variabile culture extracts after growth in GMM (negative control) and in GMM containing PCP (initial concentrations of 5 and 10 mg/l). Some of the newly formed peaks are labelled with an arrow, and the peak eluting at 12.6 min was putatively identified as CHQ. PCP decay in media, under co-metabolic conditions, was 74 and 22% for PCP initial concentrations of 5 and 10 mg/l, respectively. The peaks eluting at 30.3 and 17.5 min correspond to PCP and chloramphenicol, respectively
Discussion
The species diversity amongst the cork-colonising fungal communities is largely influenced by the origin of the cork, explaining some of the discrepancy found in the strains reported by different authors [1, 2, 8, 11, 17, 26–29]. In this study, the fungal strains used were isolated from cork bark samples, which were collected during plank preparation for the manufacture of stoppers. Within this community Penicillium spp. is the dominant genus, agreeing with the previous reports on cork microbiology [26]. From the 17 fungal strains tested, only 1 belongs to the Zygomycota phylum (Mucor sp.), while the remaining 16 belong to the Ascomycota phylum (13 Penicillium spp., one Trichoderma sp., one Cladosporium sp. and one Chrysonilia sp.).
The capacity of ascomycetes and zygomycetes fungi to tolerate and degrade PCP in a submerged culture has seldom been studied. Our aim was to evaluate individually the significance of these strains for PCP degradation. The maximum concentration of PCP used in the cultures was 20 mg/l providing the PCP in a bio-available fraction (PCP solubility in water is ~18 and 20 mg/l at 20 and 25°C, respectively). The degree of abiotic degradation in the non-inoculated controls (after incubation in the dark) was higher than that previously reported by other authors, 8% after 12 days [24] or null after 8 days [34], probably due to the high incubation periods used here (40–60 days). Also, the abiotic decay was found to be higher in the GMM (~30%) than in the MM media (7.5%). PCP photolytical degradation in water has been reported to produce lower chlorinated phenols and chlorinated dihydroxybenzenes, which can be ultimately mineralised to CO2 [37]. The 230-nm chromatographic peak lists of the abiotic decay controls were not altered after incubation, suggesting PCP volatilisation or transformation to compounds not absorbing at 230 nm. This was apparently favoured in the GMM, relative to the MM.
From the 17 strains tested, all were observed to tolerate PCP co-metabolically (Table 1). PCP is known to strongly affect spore germination, even in cultures of the highly efficient PCP degrading white rot fungus P. chrysosporium, which has been reported to stop germination in the presence of 4 mg/L of PCP, with this effect only being circumvented when the toxic was added to the growing mycelium [16]. Surprisingly, in this work, the spores of all the strains tested were able to germinate in the presence of 5 mg/L of PCP (GMM) (Table 1). Below the species toxic MICs, increasing PCP concentration in media resulted in unchanged or apparently reduced biomass (data not shown). The latter is demonstrated in Fig. 1 for C. sitophila (5 mg/l) and M. plumbeus (5 and 10 mg/l) and has been previously observed in cultures of the basidomycetes P. sordida and P. chrysosporium, where high concentrations of PCP have greatly decreased mycelial extension rates [12].
The phylum Ascomycota is taxonomically a heterogeneous group, consequently reflecting a rather diverse behaviour in the degradation capacity of PCP [23, 24]. In fact, while analysing their PCP bioremediation potential under co-metabolic conditions, the 16 tested ascomycetes have clustered in 6 different groups (Fig. 2) with the strains P. diversum and P. restrictum as the outlier group. The latter were able to germinate in 5 mg/l PCP, but completely failed to degrade it. Other environmental strains of the phylum Ascomycota have previously been reported to efficiently degrade PCP co-metabolically (e.g., Trichoderma sp. [5, 6], some Penicillium spp. [24] and Cladosporium sp. [3]).
Mucor plumbeus, the only zygomycete strain in this cork-colonising community, completely removed PCP in the GMM (up to 10 mg/l). The phylum Zygomycota is, relative to the Ascomycota, taxonomically a more homogenous group and, although devoid of ligninolytic activities, has been previously reported to extensively deplete PCP [22]. For example in the case of Rhizopus nigricans, while failing to sporulate in PCP liquid medium, 12.5 mg/l PCP could be efficiently co-metabolised [34]. Furthermore, the zygomycete M. ramosissimus was also observed to efficiently degrade PCP [32].
Very efficient PCP degradation was observed in the cultures of four ascomycetes, P. glandicola, C. sitophila, P. janczewskii and T. longibrachiatum and the zygomycete M. plumbeus, under metabolic conditions (MM) (Table 2). These strains tolerated the toxic in a distinctive way, but all could completely remove PCP from the media at an initial concentration of 1 mg/l. This has been reported in cultures of the brown rot basidomycete Gloephyllum striatum, which is devoid of ligninolytic activities, but in a submerged culture degraded 1.3 mg/l PCP more efficiently in metabolic than co-metabolic conditions [9].
The chromatographic profiles of the fungal cultures, where the recovered PCP was significantly reduced, revealed the appearance of a variable number of new peaks indicating their active biotransformation (ca. 10 and 3 novel peaks per degrading culture in co- and metabolic conditions, respectively). However, from our results, it cannot be excluded that another process (such as bioaccumulation) also may be involved. At this stage, efforts to identify the intermediate products of PCP degradation were made solely by comparing the chromatographic features of the peaks with that of pure compounds. Under co-metabolic conditions some PCP sub-products were putatively indentified: TeCBQ (P. adametzii), DCBQ (C. herbarum, P. decumbens and T. longibrachiatum) and CHQ (C. sitophila, P. corylophilum P. glabrum, P. glandicola, P. janczewskii and P. variabile) (Fig. 3), thus suggesting a PCP degradation pathway similar to that previously described for P. chrysosporium [20]. On the other hand, under metabolic conditions, none of the newly formed peaks could be associated to the standard compounds.
The potential of cork-colonising fungi to degrade chlorophenols has previously been analysed (e.g., [1, 14, 18, 28]). In one study, the majority of the fungal strains tested have transformed 2,4,6-trichlorophenol (TCP) (1 mg/l) in the corresponding anisole [1]. On the contrary, in an earlier study, some ascomycetes strains, including C. sitophila, degraded the chlorophenol in a rather heterogeneous fashion, but its methylation was not the preferential pathway [28]. This discrepancy is due to the different experimental conditions used, namely the nitrogen source used. A similar effect was reported in different studies on P. chrysosporium that degraded efficiently both chlorophenols: TCP [21] and PCP [12, 20], forming the corresponding anisoles as vestigial or major intermediates under high-carbon-low-nitrogen conditions and high-carbon-high-nitrogen conditions, respectively. Interestingly, in the study reported here, pentachloroanisole was not detected in any of the culture extracts, neither in GMM or MM, with both media containing nitrate as the sole nitrogen source.
Conclusion
The purpose of this study was to assess the behaviour of each of the 17 fungal species tested from the cork-colonising community during co- and direct metabolism of PCP. Our data further emphasise the significance of fungi belonging to a phylum other than Basidiomycota to have an interesting potential to be used in PCP biodegradation processes. In fact, 15 of the 17 strains tested could degrade PCP to a certain extent. Additionally, PCP in media was completely degraded co-metabolically by T. longibrachiatum, P. glandicola and M. plumbeus and metabolically by those and P. janczewskii and C. sitophila. Additional studies will be carried out to investigate in the most efficient fungal strains the identity of PCP degradation intermediates, aiming to better understand its degradation pathways.
Abbreviations
- PCP:
-
Pentachlorophenol
- HQ:
-
Hydroquinone
- CHQ:
-
Chlorohydroquinone
- TeCHQ:
-
Tetrachlorohydroquinone
- DCBQ:
-
2,6-Dichloro-1,4-benzoquinone
- TeCBQ:
-
Tetrachloro-1,4-benzoquinone
- PCA:
-
Pentachloroanisole
References
Alvarez-Rodriguez ML, Lopez-Ocana L, Lopez-Coronado JM, Rodriguez E, Martinez MJ, Larriba G, Coque JJR (2002) Cork taint of wines: role of the filamentous fungi isolated from cork in the formation of 2,4,6-trichloroanisole by O-methylation of 2,4,6-trichlorophenol. Appl Environ Microbiol 68(12):5860–5869
Basílio MC, Gaspar R, Silva Pereira C, San Romão MV (2006) Penicillium glabrum cork colonising isolates—preliminary analysis of their genomic similarity. Rev Iberoam Micol 23(3):151–154
Borazjani H, Ferguson B, Hendrix F, McFarland L, McGinnis G, Pope D, Strobel D, Wagner J (1989) Cladosporium sp., a potential fungus for bioremediation of wood-treating wastes. Abs Papers Am Chem Soc 197:80-ENVR
Carlile M, Watkinson S, Gooday G (2001) The fungi. Elsevier Academic Press, Amsterdam, p 588
Cserjesi AJ (1967) Adaptation of fungi to pentachlorophenol and its biodegradation. Can J Microbiol 13(9):1243–1245
Cserjesi AJ, Johnson EL (1972) Methylation of pentachlorophenol by Trichoderma virgatum. Can J Microbiol 18(1):45–47
Czaplicka M (2004) Sources and transformations of chlorophenols in the natural environment. Sci Total Environ 322(1–3):21–39
Danesh P, Velez Caldas F, Figueiredo Marques J, San Romao M (1997) Mycobiota in Portuguese ‘normal’ and ‘green’ cork throughout the manufacturing process of stoppers. J Appl Microbiol 82(6):689–694
Fahr K, Wetzstein HG, Grey R, Schlosser D (1999) Degradation of 2,4-dichlorophenol and pentachlorophenol by two brown rot fungi. FEMS Microbiol Lett 175(1):127–132
Johnson ME, Szekely A, Warnock DW (1998) In vitro activity of voriconazole, itraconazole and amphotericin B against filamentous fungi. J Antimicrob Chemother 42:741–745
Lacey J (1973) The air spora of a Portuguese cork factory. Ann Occup Hyg 16(3):223–230
Lamar RT, Larsen MJ, Kirk TK (1990) Sensitivity to and degradation of pentachlorophenol by Phanerochaete spp. Appl Environ Microbiol 56(11):3519–3526
Lyytikäinen M, Sormunen A, Peräniemi S, Kukkonen J (2001) Environmental fate and bioavailabilty of wood preservatives in freshwater sediments near an old sawmill site. Chemosphere 44(3):341–350
Maggi L, Mazzoleni V, Fumi MD, Salinas MR (2008) Transformation ability of fungi isolated from cork and grape to produce 2,4,6-trichloroanisole from 2,4,6-trichlorophenol. Food Addit Contam 25(3):265–269
McLellan L, Carvalho M, Silva Pereira C, Hursthouse A, Morrison C, Tatner P, Martins I, Romão MVS, Leitão M (2007) The environmental behaviour of polychlorinated phenols and its relevance to cork forest ecosystems: a review. J Environ Monitor 9:1055–1063
Mileski GJ, Bumpus JA, Jurek MA, Aust SD (1988) Biodegradation of pentachlorophenol by the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 54(12):2885–2889
Oliveira AC, Peres CM, Correia Pires JM, Silva Pereira C, Vitorino S, Figueiredo Marques JJ, Barreto Crespo MT, San Romao MV (2003) Cork stoppers industry: defining appropriate mould colonization. Microbiol Res 158(2):117–124
Prak S, Gunata Z, Guiraud JP, Schorr-Galindo S (2007) Fungal strains isolated from cork stoppers and the formation of 2,4,6-trichloroanisole involved in the cork taint of wine. Food Microbiol 24:271–280
Rabinovich ML, Bolobova AV, Vasil’chenko LG (2004) Fungal decomposition of natural aromatic structures and xenobiotics: a review. Appl Biochem Microbiol 40(1):1–17
Reddy GVB, Gold MH (2000) Degradation of pentachlorophenol by Phanerochaete chrysosporium: intermediates and reactions involved. Microbiology 146:405–413
Reddy GVB, Gelpke MDS, Gold MH (1998) Degradation of 2,4,6-trichlorophenol by Phanerochaete chrysosporium: involvement of reductive dechlorination. J Bacteriol 180(19):5159–5164
Seigle-Murandi F, Guiraud P, Steiman R, Benoitguyod JL (1992) Phenoloxidase production and vanillic acid metabolism by Zygomycetes. Microbiologica 15(2):157–165
Seigle-Murandi F, Steiman R, Benoitguyod JL, Guiraud P (1993) Fungal degradation of pentachlorophenol by micromycetes. J Biotechnol 30(1):27–35
Seigle-Murandi F, Toe A, Benoitguyod JL, Steiman R, Kadri M (1995) Depletion of pentachlorophenol by Deuteromycetes isolated from soil. Chemosphere 31(2):2677–2686
Semple KT, Reid BJ, Fermor TR (2001) Impact of composting strategies on the treatment of soils contaminated with organic pollutants. Environ Pollut 26:9–283
Serra R, Peterson S, Venancio A (2008) Multilocus sequence identification of Penicillium species in cork bark during plank preparation for the manufacture of stoppers. Res Microbiol 159(3):178–186
Silva Pereira C, Marques JJF, San Romão MV (2000) Cork taint in wine: scientific knowledge and public perception—a critical review. Crit Rev Microbiol 26(3):147–162
Silva Pereira C, Pires A, Valle MJ, Vilas-Boas L, Figueiredo Marques JJ, San Romão MV (2000) Role of Chrysonilia sitophila on the quality for cork stoppers for sealing wine bottle. J Ind Microbiol Biotech 24:256–261
Silva Pereira C, Soares GAM, Oliveira AC, Rosa ME, Pereira H, Moreno N, Romão MVS (2006) Effect of fungal colonization on mechanical performance of cork. Int Biodeter Biodegr 57(4):244–250
Soares GAM, Basílio MC, Tenreiro R, San Romão MV (2003) Diversity of Penicillium spp. colonising cork slabs: a classical and molecular approach. In: Lima N, Smith D (eds) Biological resource centres and the use of microbes. Micoteca da Universidade do Minho, Braga, pp 161–170
Solyanikova IP, Golovleva LA (2004) Bacterial degradation of chlorophenols: pathways, biochemical and genetic aspects. J Environ Sci Health Part B 39(3):333–351
Szewczyk R, Bernat P, Milczarek K, Dlugonski J (2003) Application of microscopic fungi isolated from polluted industrial areas for polycyclic aromatic hydrocarbons and pentachlorophenol reduction. Biodegradation 14(1):1–8
Taylor TR, Tucker T, Whalen MM (2005) Persistent inhibition of human natural killer cell function by ziram and pentachlorophenol. Environ Toxicol 20(4):418–424
Tomasini A, Flores V, Cortes D, Barrios-Gonzalez J (2001) An isolate of Rhizopus nigricans capable of tolerating and removing pentachlorophenol. World J Microbiol Biotechnol 17(2):201–205
Tortella GR, Diez MC, Duran N (2005) Fungal diversity and use in decomposition of environmental pollutants. Crit Rev Microbiol 31(4):197–212
van Leeuwen JA, Nicholson BC, Hayes KP, Mulcahy DE (1997) Degradation of chlorophenolic compounds by Trichoderma harzianum isolated from Lake Bonney, South-Eastern South Australia. Environ Toxicol Water Qual 12(4):335–342
Wong AS, Crosby DG (1981) Photo-decomposition of pentachlorophenol in water. J Agric Food Chem 29(1):125–130
Acknowledgments
MBC is grateful to FC&T for the fellowship SFRH/BD/18205/2004. The work was partially supported by FC&T (POCTI/AMB/57374/2004 and POCTI/QUI/56229/2004) and by NATO (ESP.MD.SFPP 981674). The authors wish to acknowledge M. Carmo Basílio and Dr. M. Vitória San Romão for providing the fungal strains used in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. B. Carvalho and I. Martins have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Carvalho, M.B., Martins, I., Leitão, M.C. et al. Screening pentachlorophenol degradation ability by environmental fungal strains belonging to the phyla Ascomycota and Zygomycota. J Ind Microbiol Biotechnol 36, 1249–1256 (2009). https://doi.org/10.1007/s10295-009-0603-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-009-0603-2