Abstract
Polychlorinated biphenyls (PCBs) represent a large group of recalcitrant environmental pollutants. Up to now, many studies have focused on bioremediation of PCBs by fungal strains; however, the mechanisms of adaptation of these strains towards PCBs remain unknown despite their importance in developing effective bioremediation processes. We studied five species, each consisting of two strains isolated either from PCB-polluted or PCB-unpolluted substrates (control strains). We investigated their responses to PCB contamination by studying their tolerance to PCBs, their ability to reduce these pollutants, and their expression level of Laccase genes. In Thermothelomyces thermophila, Thermothelomyces heterothallica, Thermoascus crustaceus, and Fusarium solani, all the studied strains showed a similar tolerance and PCB degradation regardless of their origin. In Schizophyllum commune, while both strains showed similar resistance to PCBs, i.e., PCBs and their degradation products presented no toxicity for these strains, the rate of PCB degradation of the strain from a PCB-polluted environment was significantly slightly higher. The PCB degradation did not correlate with the expression level of genes encoding Laccases. These results demonstrate that the tolerance and PCB degradation by the fungal strains, which did not involve Laccase genes, required different adaptation systems which seem to be constitutive or rapidly inducible by PCB according to the fungal species.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Polychlorinated biphenyls (PCBs) are organochlorine compounds consisting of a biphenyl ring substituted by one or more chlorine atoms with 209 possible congeners that differ in the number of chlorine atoms and their position on the biphenyl ring (Marco-Urrea et al. 2015; Sharma et al. 2017). Commercial PCB mixtures have been largely used from the 1930s to the 1970s in industries with a large number of applications mainly in electrical equipment (Pieper and Seeger 2008; Passatore et al. 2014). Although their manufacture and marketing have been banned, PCBs remain persistent in the environment because of their high stability (Passatore et al. 2014). It is estimated that about 1 million tons has been produced worldwide and that about 30% of all manufactured PCBs have been released into the environment leading to soil and sediment contamination (UNEP 2011; Antolin-Rodriguez et al. 2016). PCBs represent a threat to ecosystem health due to their low degradability and high rate of toxicity, cause a wide range of toxic effects on animals and humans, and are also considered as potentially carcinogenic (Brouwer et al. 1999; Pointing 2001; Dercova et al. 2009).
Although physicochemical technologies are available for the remediation of PCB-contaminated matrices, bioremediation using microorganisms offers a beneficial alternative with low environmental impact and reduced cost. Several model fungal species, in particular white rot basidiomycetes, have been shown to efficiently degrade PCBs under laboratory conditions (Ruiz-Aguilar et al. 2002; Kamei et al. 2006). This capacity is linked to their rich and varied enzymatic systems, in particular extracellular and intracellular enzymes which, together, ensure efficient biodegradation of various xenobiotic pollutants (Harms et al. 2011; Passatore et al. 2014). However, despite their interesting metabolic potentialities, their application in the rehabilitation of polluted sites remains unsatisfactory due to their inability to develop on PCB-polluted matrices probably resulting from the lack of competition in front of indigenous microbial community or their low tolerance to PCBs (Harms et al. 2011).
The use of indigenous fungal strains for bioremediation processes could turn out to be more efficient in comparison to model strains and is promising for environmental biotechnology (Sage et al. 2014). They are naturally selected by their environments and are able to develop in the presence of toxic xenobiotics (Marco-Urrea et al. 2015). These features are probably due to their physiological adaptation which could lead to the resistance to pollutants resulting in the change in their growth rates and spore production or their ecological adaptation affecting, for example, pigment production or their genetic adaptation sustained by change in the expression of genes involved in the pollutant degradation (Colpaert et al. 2000; Sharples et al. 2001; Zafra et al. 2015). Fungal metabolic adaptations at different biological levels in response to pollutants may vary according to the species and pollutants, and understanding the involved mechanisms is crucial for the development of an effective bioremediation strategy (Sharples et al. 2001; Garon et al. 2004; Zafra et al. 2015). While several studies focused on the screening of strains capable of effectively degrading PCBs (Tigini et al. 2009; Mouhamadou et al. 2013), to our knowledge, there are still no studies dealing with the mechanisms of indigenous fungal strain adaptation in response to PCBs.
The aim of this study was to investigate the responses of fungal strains to PCBs by focusing on their metabolic potential and on the expression of Laccase genes. Although studies on the involvement of Laccases in the biodegradation of PCBs are contradictory (Plackova et al. 2012; Gayosso-Canales et al. 2012), we hypothesized that the studied fungal strains could use these enzymes to degrade PCBs as they are involved in the biodegradation of a wide range of xenobiotic compounds (Harms et al. 2011; Viswanath et al. 2014). We used four Ascomycota species, Fusarium solani, Thermothelomyces thermophila, Thermothelomyces heterothallica, and Thermoascus crustaceus and one Basidiomycota species, Schizophyllum commune. Each species consisted of two strains obtained from PCB-polluted and PCB-unpolluted substrates. The effects of strain origins were evaluated on the capacity of each strain to metabolize the mixture of seven PCB congeners (Mouhamadou et al. 2013), on the tolerance of each strain and on the transcriptional level of Laccase genes.
Materials and methods
PCBs
The seven PCB congeners, PCBs 28, 52, 101, 118, 138, 153, and 180, were obtained from Sigma Aldrich Corp. (St. Louis, MI, USA). A stock solution containing 15 mg of each PCB in 105 ml of DMSO was prepared.
Fungal strains
Soils from former industrial sites contaminated with PCBs were collected and fungal strains (Table 1) were isolated from these samples using the suspension/dilution method. For instance, 1 g of contaminated soil was added to 9 ml of sterile water containing 0.05% of sodium dodecyl sulfate (w/v). After stirring, aliquots of the suspension were serially diluted and spread onto Petri dishes containing malt extract agar (1.5% w/v) supplemented with 0.05% chloramphenicol. Cultures were incubated at 25 °C and 37 °C. After isolation, fungal strains were identified according to general principles of fungal classification (Ellis 1971; Booth 1977a; Booth 1977b; Domsch et al. 1980). References of strains isolated from PCB-unpolluted substrates belonged to the laboratory’s collection (CMPG, Collection Mycology Pharmacy Grenoble).
Analyses of PCB degradation
The protocol concerning the biodegradation test was that described by Mouhamadou et al. (2013). Succinctly, the mycelium of each strain, grown in malt extract (1.5% w/v), was introduced into Erlenmeyer flasks containing 20 ml of modified GS liquid medium (Galzy and Slonimski 1957) supplemented with glucose (5 g l−1) and incubated at 25 °C for Fusarium solani and Schizophyllum commune or 37 °C for Thermothelomyces thermophila, Thermothelomyces heterothallica, and Thermoascus crustaceus on a rotary shaker (120 rpm). After 48-h incubation, the culture of each strain was spiked with 400 μl of a PCB mix giving a final concentration of 3 μg of each congener per milliliter. Each experiment containing each strain was performed in triplicate and included fungal-free flasks which are the abiotic controls. After 5 days of incubation, the mycelium was separated from the culture medium by filtering through Whatman 40 filters.
PCB extraction and HPLC analyses
Culture medium and fungal mycelium were extracted separately. Culture medium was extracted three times with 20 ml of hexane with agitation (250 rpm, room temperature) for 30 min. To minimize the phenomenon of PCB biosorption, mycelium was first homogenized in the serum bottles with a Potter homogenizer in 20 ml of hexane, vigorously agitated for 10 min, and incubated 24 h at room temperature with agitation (250 rpm). The organic phases from culture medium and mycelium were evaporated under vacuum using a rotary evaporator and adjusted to a volume of 5 ml.
PCB analyses were performed in the “Institut de Chimie Moléculaire of Grenoble.” Aliquots of 1 μl from culture medium or from mycelium were analyzed using a gas chromatograph (5977-7890B), equipped with a HP-5MS 5 phenyl methyl silox column (ID, 0.25 mm; length, 30 m; film, 0.25 mm) and a quadrupole detector (Agilent Technologies, USA). The carrier gas was helium. The injector temperature and the transfer line temperature were 250 °C and 280 °C, respectively. The initial column temperature was 65 °C held for 2 min. The gradient setting was 30 °C min−1 to 240 °C, held for 2 min at 240 °C, then 10 °C min−1 to 340 °C. The final temperature was 340 °C held for 5 min.
To determine the level of PCB removal, we determined the total residual PCB level in the fungal cultures by adding the residual levels of PCBs of the mycelium and of the culture medium and compared this rate with that of the abiotic control.
Expression level of Laccase genes
As most of the studied fungal species are described as laccase producers (Hamato et al. 1999; Wu et al. 2010; Babot et al. 2011), we investigated the expression level of the genes encoding this enzyme. Each strain was cultivated in the presence of PCBs as described in the biodegradation test. In parallel, control cultures were carried out by incubating each strain only in the culture medium on the one hand and in the presence of DMSO on the other hand. After 7-day incubation, mycelium was recovered by filtering through Whatman 40 filters and 200 mg of mycelium was used for the RNA extraction using FastRNA Spin kit (MP Biomedicals, France) according to the manufacturer’s recommendations. Four micrograms of total RNA was reverse transcribed using the SuperScript III kit (Fisher Scientific) and subsequently, the RT products were amplified by specific primers designed in this study. These primers (Table 2) were defined by aligning orthologous sequences available in the GenBank database and encoding Laccases. From these alignments, we have targeted the conserved regions and defined couples of primers that could amplify DNA fragments of between 110 and 220 bp.
From these primers, the expression level on Laccase gene was quantified via qPCR on an iQ5 system as described by Mouhamadou et al. (2017). cDNA of each strain obtained from the reverse transcription was mixed with 0.3 mM of iQ SYBR Green Supermix (Biorad, USA) and 0.3 mM of each of the forward and reverse primers in a total volume of 25 μl. A melting curve analysis was performed to check for the unique presence of the targeted PCR product. For each species, a dilution series of the pooled PCR products from the two strains, precisely quantified using a qubit fluorometer (Invitrogen, USA), were run in tandem in qPCR with the cDNA of each strain. Each reaction was run in triplicate.
Statistical analysis
The effects of substrate origin, strain type, and incubation type on the expression level of Laccase gene were tested using a three-way ANOVA. As strong interactions were found between the three explaining factors, we tested the effect of substrate origin and incubation type using a two-way ANOVA followed by least square difference post hoc tests. The effects of substrate origin and incubation type on the growth of mycelium were tested using a two-way ANOVA followed by Tukey’s test. All the analyses were performed in R version 3.2.2 (R Core Team 2015), using the lsmeans (Lenth 2016) for least square difference post hoc tests and agricolae (Mendiburu 2018) for Tukey’s post hoc test. When necessary, data were transformed to conform to assumptions of normality and homoscedasticity.
Results
Tolerance of fungal strains to PCBs
The study of the tolerance of each fungal strain to PCBs was performed by comparing the biomass weight formed during the culture of the strains in the presence/absence of PCBs. For each of the four Ascomycota species, no significant biomass differences were observed in fungal cultures in the presence/absence of PCBs according to the strain origin (Table 3). In the Basidiomycota species Schizophyllum commune (Table 3), the strain isolated from PCB-polluted substrate showed no difference in biomass in the presence/absence of PCBs, whereas in that from PCB-unpolluted substrate, despite the positive effect of the solvent on the growth of this strain, there was no significant difference in its growth in the presence/absence of PCBs.
PCB removal by fungal strains
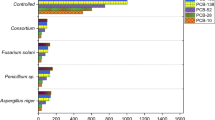
We determined the percentage of depletion of PCBs by comparing the quantity of residual PCBs in the culture medium and in the mycelium (biosorption) of each strain with the quantity of residual PCBs in abiotic control. All strains were able to efficiently reduce PCBs (Fig. 1). In Ascomycota, strains of the same species showed no significant differences in the rate of PCB removal. It ranged from 84.79 to 88.11% in Fusarium solani, from 79.86 to 86.61% in Thermothelomyces heterothallica, from 95.22 to 95.91% in Thermoascus crustaceus, and from 93.44 to 94.64% in Thermothelomyces thermophila. In the Basidiomycota, Schizophyllum commune, the two studied strains isolated from PCB-unpolluted and PCB-polluted substrates reduced PCBs at 80.91% and 86.20%, respectively, but these rates are significantly different (F = 16.93; p = 0.015).
Percentage of PCB biodegradation in the five studied fungal strains (n = 3, mean ± SE). These values were determined by taking into account the PCB recovery yield (80%) and the abiotic losses (9%). The results of a one-way ANOVA are represented by an asterisk symbol which indicates significant substrate effect within each fungal strain
Expression level of Laccase genes
Primers were defined to investigate the expression level of Laccase transcripts. With these primers, using the genomic DNA of each strain as template, only the results of Thermothelomyces heterothallica could not be obtained. For the other strains, one specific PCR product was obtained at the expected size indicating the amplification of a single Laccase gene. Results of RT-qPCR (Fig. 2) showed that the expression level of Laccase gene was significantly different between strains (F = 13,185; p < 0.001) with the following patterns: Thermoascus crustaceus < Thermothelomyces thermophila < Schizophyllum commune < Fusarium solani. In the three-way ANOVA showing strong interactions (F = 6.45; p < 0.001) between substrate origin (PCB-unpolluted vs PCB-polluted), strain type, and incubation type (control vs solvent vs PCB), we realized two-way ANOVA for each strain to disentangle substrate origin and incubation type effects (Table 4). In Fusarium solani, only a substrate origin effect was observed (Table 4) with a higher expression level of Laccase gene in PCB-polluted substrates (Fig. 2). In the three other strains, strong interactive effects were observed (Table 4) but with different patterns (Fig. 2). For Thermothelomyces thermophila, the expression level of Laccase gene was lower in the presence of PCBs in the strains isolated from PCB-polluted substrates than in that from PCB-unpolluted substrates (Fig. 2). For Thermoascus crustaceus, the expression level of Laccase gene was higher in the presence of PCBs than that in the absence of PCBs in the strain isolated from PCB-unpolluted substrate but was similar to all other treatments (Fig. 2). For Schizophyllum commune, the expression level of Laccase gene was lower in the strains isolated from PCB-polluted substrate than that in those from PCB-unpolluted substrates. In these PCB-polluted substrates, the expression level of Laccase gene was higher in the presence than that in the absence of PCBs (Fig. 2).
Transcript level of the Laccase genes in the four studied fungal strains (n = 3, mean ± SE). The results of least square difference post hoc tests are represented by different letters when significant differences (p < 0.05) between means were obtained. Lowercase letters illustrate the interactive effects between substrate and incubation types; and uppercase letters only illustrate a substrate effect
Finally, the interactions between the rate of PCB removal and the expression level of Laccase gene were studied using a linear regression analysis and no correlation was found between whatever the fungal species studied (Fig. 3).
Discussion
Up to now, the mechanisms of adaptation of fungal strains to PCBs remain unknown and yet important to develop effective bioremediation processes. To bridge this gap, we investigated the responses of four Ascomycota and one Basidiomycota species towards PCBs by examining their tolerance, their ability to reduce these pollutants, and the expression level of gene encoding Laccases, non-specific enzymes involved in the biodegradation of environmental pollutants. Each species consisted in two strains provided from PCB-polluted and PCB-unpolluted substrates.
All the studied strains showed high efficiency in PCB removal in liquid medium comparable to that obtained with basidiomycete ligninolytic fungi (Kubatova et al. 2001; Kamei et al. 2006; Cvancarova et al. 2012; Federici et al. 2012) or autochthonous strains isolated from highly polluted PCB substrates (Tigini et al. 2009; Mouhamadou et al. 2013). First, we considered that the substantial losses of PCBs were essentially due to the fungal biodegradation as reported by Mouhamadou et al. (2013) since the extraction of PCBs from the mycelium corresponding to the fungal biosorption was drastic by crushing the mycelium in hexane and by incubating it 24 h in hexane with vigorous agitation. Second, in the four species belonging to Ascomycota, we did not detect any effects of the origin of strains on their resistance to PCBs and their ability to degrade PCBs. These results seem to rule out or to minimize the need of prior adaptation of the studied strains towards PCBs for its degradation. Comparable trends, characterizing white rot fungi, had also been reported in some species belonging to Zygomycota where strains isolated from polycyclic aromatic hydrocarbon (PAH)-contaminated soils had an equivalent efficiency of PAH biodegradation in comparison to that of the same species isolated from PAH-uncontaminated soils (Garon et al. 2004). It is possible to speculate that the systems of resistance/degradation of PCBs seem to be constitutive or rapidly inducible by PCBs as described in bacterial communities which adapt rapidly to new xenobiotics through, for example, the acquisition of mobile genetic elements (Top and Springae 2003).
In the Basidiomycota Schizophyllum commune, the fact that both strains reduce PCBs at a rate higher than 80%, regardless of their origin, would suggest that they would probably bear a constitutive mechanism of PCB biodegradation. However, a slightly greater efficiency in the PCB biodegradation of the strain isolated from PCB-contaminated habitat vs control strain suggests either individual variation in the biodegradation that may involve two catabolic pathways with different efficiencies or an adaptation to the PCBs enhancing the efficiency of the strain from PCB-polluted habitat. The latter hypothesis could be excluded as both strains exhibited the same resistance to PCBs.
We investigated the expression level of one of the Laccase genes to examine the possible link between resistance/PCB degradation observed in the studied strains and this enzyme. We found that 8 out of the 10 analyzed strains expressed the genes. In Thermothelomyces heterothallica, we were unable to obtain amplification products corresponding to this gene probably because of the non-specificity of our primers. The transcript level of the Laccase gene did not correlate with PCB removal. We hypothesized that this enzyme was not involved in PCB biodegradation, consistent with studies which challenge the implication of Laccase in the fungal PCB biodegradation (Tigini et al. 2009; Plackova et al. 2012). In addition, the variability in the expression of this gene in the studied strains confirms this hypothesis and is in line with studies which demonstrated that some Laccase genes were down or upregulated by xenobiotics while the others were constitutively expressed (Canero and Roncero 2008). This suggests that the transcripts detected in our study were probably expressed from different genes.
Our study showed that all the studied fungal strains were tolerant to PCBs and able to efficiently degrade them in liquid medium whatever their origin. This degradation, which did not involve Laccases, seems to require different adaptation systems according to the strains. Further studies are needed to demonstrate such adaptation systems across a large number of fungal species. These could be of interest for optimizing the use of fungal strains in environmental biotechnology.
References
Antolin-Rodriguez JM, Sánchez-Báscones M, Martin-Ramos P, Bravo-Sanchez C (2016) Estimation of PCB content in agricultural soils associated with long-term fertilization with organic waste. Environ Sci Pollut Res 23:12372–12383
Babot ED, Rico A, Rencoret J, Kalum L, Lund H, Romero J, del Río JC, Martínez AT, Gutiérrez A (2011) Towards industrially-feasible delignification and pitch removal by treating paper pulp with Myceliophthora thermophila laccase and a phenolic mediator. Bioresour Technol 102:6717–6722
Booth C (1977a) Fusarium laboratory guide to the identification of the major species. Commonwealth Mycological Institute, Kew
Booth C (1977b) The genus Fusarium. Commonwealth Mycological Institute, Kew
Brouwer A, Longnecker MP, Birnbaum LS, Cogliano J, Kostyniak P, Moore J, Schantz S, Winneke G (1999) Characterization of potential endocrine-related health effects at low-dose levels of exposure to PCBs. Environ Health Perspect 107:639–649
Canero DC, Roncero MIG (2008) Functional analyses of laccase genes from Fusarium oxysporum. Phytopathology 98:509–518
Colpaert JV, Vandenkoornhuyse P, Adriaensen K, Vangronsveld J (2000) Genetic variation and heavy metal tolerance in the ectomycorrhizal basidiomycete Suillus luteus. New Phytol 147:367–379
Core Team R (2015) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Cvancarova MC, Kresinova Z, Alena Filipova A, Covino AS, Cajthaml T (2012) Biodegradation of PCBs by ligninolytic fungi and characterization of the degradation products. Chemosphere 88:1317–1323
Dercova K, Seligova J, Dudasova H, Mikulasova M, Silharova K, Tothova L, Hucko P (2009) Characterization of the bottom sediments contaminated with polychlorinated biphenyls: evaluation of ecotoxicity and biodegradability. Int Biodeterior Biodegrad 63:440–449
Domsch KH, Gams W, Anderson TH (1980) Compendium of Soil Fungi. Academic Press, London.
Ellis MB (1971) Dematiaceous Hyphomycetes, principal mycologist. Commonwealth Mycological Institute, Kew
Federici E, Giubilei M, Santi G, Zanaroli G, Negroni A, Fava F, Petruccioli M, D’Annibale A (2012) Bioaugmentation of a historically contaminated soil by polychlorinated biphenyls with Lentinus tigrinus. Microb Cell Factories 23:11–35
Galzy P, Slonimski P (1957) Variations physiologiques de la levure au cours de la croissance sur l’acide lactique comme seule source de carbone. Comptes Rendus de l’Academie des Sciences 245D:2423–2426
Garon D, Sage L, Seigle-Murandi F (2004) Effects of fungal bioaugmentation and cyclodextrin amendment on fluorine degradation in soil slurry. Biodegradation 15:1–8
Gayosso-Canales M, Rodríguez-Vazquez R, Esparza-García FJ, Bermúdez-Cruz RM (2012) PCBs stimulate laccase production and activity in Pleurotus ostreatus thus promoting their removal. Folia Microbiol 57:149–158
Hamato O, Sekine H, Nakano E, Abe K (1999) Cloning and expression of a cDNA encoding the Laccase from Schizophyllum commune. Biosci Biotechnol Biochem 63:58–64
Harms H, Schlosser D, Wick LY (2011) Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat Rev Microbiol 9:177–192
Kamei I, Kogura R, Kondo R (2006) Metabolism of 4,40-dichlorobiphenyl by white-rot fungi Phanerochaete chrysosporium and Phanerochaete sp. MZ142. Appl Microbiol Biotechnol 72:566–575
Kubatova A, Erbanova P, Eichlerova I, Homolka L, Nerud F, Sasek V (2001) PCB congener selective biodegradation by the white rot fungi Pleurotus ostreatus in contaminated soil. Chemosphere 43:207–215
Lenth RV (2016) Least-squares means: the R package lsmeans. J Stat Softw 69:1–33
Marco-Urrea E, Inmaculada Garcıa-Romera I, Aranda E (2015) Potential of non-ligninolytic fungi in bioremediation of chlorinated and polycyclic aromatic hydrocarbons. New Biotechnol 32:620–628
Mendiburu FD (2018) Agricolae: statistical procedures for agricultural research. R package version 1.2–9. https://cran.r-project.org/web/packages/agricolae/index.html
Mouhamadou B, Faure M, Sage L, Marçais J, Souard F, Geremia R (2013) Potential of autochthonous fungal strains isolated from contaminated soils for degradation of polychlorinated biphenyls. Fungal Biology 117:268–274
Mouhamadou B, Sage L, Perigon S, Seguin V, Bouchart V, Legendre P, Caillat M, Yamounia H, Garon D (2017) Molecular screening of xerophilic Aspergillus strains producing mycophenolic acid. Fungal Biology 121:103–111
Passatore L, Rossettic S, Juwarkard AA, Massacci A (2014) Phytoremediation and bioremediation of polychlorinated biphenyls (PCBs): state of knowledge and research perspectives. J Hazard Mater 278:189–202
Pieper DH, Seeger MJ (2008) Bacterial metabolism of polychlorinated biphenyls. J Mol Microbiol Biotechnol 15:121–138
Plackova M, Svobodova K, Cajthaml T (2012) Laccase activity profiling and gene expression in PCB-degrading cultures of Trametes versicolor. Int Biodeterior Biodegrad 71:22–28
Pointing SB (2001) Feasibility of bioremediation by white rot fungi. Appl Microbiol Biotechnol 57:20–33
Ruiz-Aguilar GML, Fernandez-Sanchez JM, Rodriguez-Vazquez R, Poggio-Varaldo H (2002) Degradation by white-rot fungi of high concentrations of PCB extracted from a contaminated soil. Adv Environ Res 6:559–568
Sage L, Périgon S, Faure M, Gaignaire C, Abdelghafour M, Mehu J, Geremia RA, Mouhamadou B (2014) Autochthonous ascomycetes in depollution of polychlorinated biphenyls contaminated soil and sediment. Chemosphere 110:62–69
Sharma JK, Gautam RK, Nanekar SV, Weber R, Singh BK, Singh SK, Juwarkar AA (2017) Advances and perspective in bioremediation of polychlorinated biphenyl-contaminated soils. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-017-8995-4
Sharples JM, Meharg AA, Chambers SM, Cairney JWG (2001) Arsenate resistance in the ericoid mycorrhizal fungus Hymenoscyphus ericae. New Phytol 151:265–270
Tigini V, Prigione V, Di Toro S, Fava F, Varese GC (2009) Isolation and characterization of polychlorinated biphenyl (PCB) degrading fungi from a historically contaminated soil. Microb Cell Factories 8:5
Top EM, Springae D (2003) The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr Opin Biotechnol 14:262–269
UNEP (United Nations Environment Program) (2011) Report of the conference of the parties to the Stockholm convention on persistent organic pollutants on the work of its fifth meeting, 5th: Geneva
Viswanath B, Rajesh B, Janardhan A, Kumar AP, Narasimha G (2014) Fungal laccases and their applications in bioremediation. Enzyme Research 2014:163242
Wu YR, Luo ZH, Kwok-Ke Chow R, Vrijmoed LL (2010) Purification and characterization of an extracellular laccase from the anthracene-degrading fungus Fusarium solani MAS2. Bioresour Technol 101:9772–9777
Zafra G, Absalón AE, Cortés-Espinosa DV (2015) Morphological changes and growth of filamentous fungi in the presence of high concentrations of PAHs. Braz J Microbiol 46:937–941
Acknowledgments
We are deeply grateful to Viviane Barbreau for her critical reading of the manuscript and interesting remarks. We address our special thanks to Nael Mouhamadou, Ilian Mouhamadou, and Lila Mouhamadou for their help and unconditional support.
Funding
This research was financed by UMR 5553 CNRS/UGA “Projet interne”.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editor: Philippe Garrigues
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Périgon, S., Massier, M., Germain, J. et al. Metabolic adaptation of fungal strains in response to contamination by polychlorinated biphenyls. Environ Sci Pollut Res 26, 14943–14950 (2019). https://doi.org/10.1007/s11356-019-04701-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-019-04701-5