Abstract
Magnetic resonance imaging (MRI) protocoling can be time- and resource-intensive, and protocols can often be suboptimal dependent upon the expertise or preferences of the protocoling radiologist. Providing a best-practice recommendation for an MRI protocol has the potential to improve efficiency and decrease the likelihood of a suboptimal or erroneous study. The goal of this study was to develop and validate a machine learning-based natural language classifier that can automatically assign the use of intravenous contrast for musculoskeletal MRI protocols based upon the free-text clinical indication of the study, thereby improving efficiency of the protocoling radiologist and potentially decreasing errors. We utilized a deep learning-based natural language classification system from IBM Watson, a question-answering supercomputer that gained fame after challenging the best human players on Jeopardy! in 2011. We compared this solution to a series of traditional machine learning-based natural language processing techniques that utilize a term-document frequency matrix. Each classifier was trained with 1240 MRI protocols plus their respective clinical indications and validated with a test set of 280. Ground truth of contrast assignment was obtained from the clinical record. For evaluation of inter-reader agreement, a blinded second reader radiologist analyzed all cases and determined contrast assignment based on only the free-text clinical indication. In the test set, Watson demonstrated overall accuracy of 83.2% when compared to the original protocol. This was similar to the overall accuracy of 80.2% achieved by an ensemble of eight traditional machine learning algorithms based on a term-document matrix. When compared to the second reader’s contrast assignment, Watson achieved 88.6% agreement. When evaluating only the subset of cases where the original protocol and second reader were concordant (n = 251), agreement climbed further to 90.0%. The classifier was relatively robust to spelling and grammatical errors, which were frequent. Implementation of this automated MR contrast determination system as a clinical decision support tool may save considerable time and effort of the radiologist while potentially decreasing error rates, and require no change in order entry or workflow.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
The appropriate use of magnetic resonance imaging (MRI) represents a significant challenge in the current healthcare landscape. MRIs are costly, are time-consuming, and require considerable effort in protocoling and interpretation [1, 2]. Protocols can often be erroneous or suboptimal given the wide variety of possible options in many cases [3,4,5]. Providing a best-practices recommendation for an MRI protocol has the potential to improve efficiency and decrease the likelihood of a suboptimal or erroneous study. Therefore, there is need for an algorithm capable of interpreting the clinical indication for the study and automatically providing an appropriate protocol. Ideally, such an algorithm would err on the side of caution in providing contrast and also be capable of flagging a study for further evaluation by a radiologist when unsure.
We set out to develop such an algorithm based on novel natural language processing (NLP) techniques and compare our results to more traditional methods. Briefly, NLP is an established field of computer science that deals with the interaction between computers and human language [6, 7]. In recent years, the field has undergone considerable change attributable to improved technology, processing power, and increased accessibility of machine learning. Multiple applications have been developed within radiology alone, including text mining of clinical narratives, coding, classification, detection of critical observations, and quality assessment [8,9,10,11,12,13,14,15]. A powerful tool—IBM’s Watson supercomputer—gained fame as the Jeopardy! champion in 2011 and has since branched out into various machine learning tasks, including natural language classification [16]. However, to our knowledge, no such application for MRI protocoling has yet been developed.
The goal of this study was to use IBM Watson to create a natural language classifier that could automatically assign the use of intravenous contrast for musculoskeletal MRI protocols based upon the free-text clinical indication of the study.
Methods
This IRB-approved study included a retrospective analysis of 1544 musculoskeletal MRI exams from a tertiary referral hospital, including their free-text protocols and free-text clinical indications. Study types included all musculoskeletal MRIs, including MRIs of the spine. Original protocols were assigned by radiology residents and fellows under the supervision of attending radiologists.
A robustly labeled dataset was created by classifying each MRI protocol as “with contrast” (WC) or “non-contrast” (NC) using semi-automated techniques with manual verification. Twenty-four examinations were excluded due to unresolvable ambiguity in the final protocol regarding the use of contrast, so as to not include training examples for which ground truth could not be determined. The most common example of this was a protocol that instructed the MRI technologist to call the radiologist after initial non-contrast sequences to assess the need for administering contrast (i.e., “MRI lumbar spine non-con, call radiologist after non-consequences to determine need for contrast”). For analysis of inter-reader agreement, each MRI was also classified by a blinded second radiologist with 4 years of experience. Classifications were assigned based solely on the provided clinical indication without access to additional patient data.
From the final 1520 MRI exams, the dataset was randomly divided into training/validation and test sets containing 1240 and 280 studies, respectively (Fig. 1). Data pre-processing was conducted using the natural language processing package in the statistical programming language R [17]. The free-text fields were stripped of punctuations, whitespace, and commonly used words that do not add to the clinical meaning (e.g. “reason,” “with,” “and,” “eval,” “for,” “MRI,” “has,” “please,” etc.). Numbers and punctuation were removed, and if applicable, each word was converted to its radical form.
Flowchart demonstrating data processing of 1544 free-text MRI protocols with their respective clinical indications. Initial labels used in the training and test set were assigned using regular expression searches and manually verified by the authors. MRI protocols with ambiguous contrast assignment were excluded from the dataset
Traditional machine learning was performed with eight different models using a personal laptop. Using the natural language processing libraries in R, including “RTextTools,” we pre-processed the texts, created a document term matrix with term-frequency weighting, and then trained the classification models [18]. Machine learning algorithms used for the models were support vector machine (SVM), scaled linear discriminant analysis (SLDA), boosting, bootstrap aggregating (Bagging), classification and regression tree (CART), random forest, Lasso and elastic-net regularized generalized linear model (GLMNET), and maximum entropy [18,19,20,21,22,23,24,25,26]. A majority-vote ensemble of all eight models was created to further enhance labeling accuracy.
Deep learning-based natural language classification was conducted using a proprietary natural language classifier from IBM Watson [16, 27]. The Watson algorithm uses hypothesis generation, string analysis, and deep learning-based word-scoring to generate a prediction for class NC and WC [27]. Performance of the classifier was evaluated with the test set. Inter-reader agreement was calculated using pairwise Cohen’s kappa between the original protocol and Watson, the second reader and Watson, and the original protocol and second reader. In addition, Watson’s performance was evaluated for the subset of cases in which the second reader and original protocol agreed.
Every disagreement between the original protocol and Watson was analyzed to attempt to ascertain the source of error. All data handling was done in “R: A language and environment for statistical computing,” including generation of descriptive statistics and other text mining tasks based on traditional machine learning algorithms.
Results
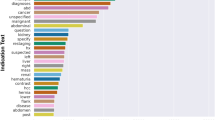
Of the 1520 final included MRI examinations, 650 (42.8%) protocols were class WC and 870 (57.2%) were class NC. A total of 86.2% studies involved the spine, 3.0% involved the upper extremity, and 10.8% involved the lower extremity (Supplemental Table 1). The three most common words in the clinical indication were “pain, weakness, and injury,” likely relating to origination from a level 1 trauma center with a high proportion of uninsured care, drug abuse, motor vehicle collisions, and gunshot wounds (Fig. 2).
Training time with IBM Watson was 46 min compared to 10 s for the eight traditional machine learning algorithms, in total. Performance on the test set was 1 min and 46 s for Watson and nearly instantaneous for traditional machine learning algorithms. These training and testing times are provided for qualitative understanding of the time required to implement these algorithms and not intended for direct comparison since hardware configurations were not the same.
Inter-reader agreement between Watson and the original protocol, between Watson and the second reader, and between the second reader and original protocol was 0.66 [0.58–0.75], 0.77 [0.69–0.84], and 0.79 [0.76–0.82], respectively.
Performance of Watson compared to the original protocol, second reader, and subset of cases for which the second reader and original protocol agreed is presented in Table 1 and corresponding confusion matrices in Table 2. When compared to the original protocol, Watson correctly assigned 129/140 cases in class NC and 104/140 cases in class WC, resulting in a sensitivity of 0.743, specificity of 0.921, positive predictive value (PPV) of 0.904, negative predictive value (NPV) of 0.782, and overall accuracy of 0.832. Accuracy for the subset of non-spine cases in the test set (n = 15) was comparable at 0.800.
The performance of Watson compared to the second reader was higher, with a sensitivity of 0.812, specificity of 0.952, PPV of 0.939, NPV of 0.849, and overall accuracy 0.886. If only considering the subset of cases for which the second reader agreed with the original protocol (n = 251), Watson demonstrated a sensitivity of 0.836, specificity of 0.961, PPV of 0.953, NPV of 0.861, and accuracy of 0.900.
Of the 47 total errors, Watson disagreed with both the original protocol and second reader in 25 cases (Table 3). In the remaining 22 errors, Watson disagreed with the original protocol but agreed with the second reader (Table 4). False-positives in class NC included a spinous process fracture and epidural abscess evaluation in a dialysis patient. False-negatives in class WC included patients with malignancy as well as cases for which contrast was explicitly requested in the clinical indication but without a stated clinical reason. The classifier was otherwise robust to numerous spelling and grammatical errors, including concatenation of two words which may be an artifact of our data storage and retrieval system (Supplemental Table 2).
The eight traditional machine learning algorithms achieved overall accuracy rate ranging from 70 to 75% as singleton, described in Supplemental Table 3. Boosting methodology demonstrated the worst performance and maximum entropy demonstrated the best. Majority-vote ensemble was performed on the eight models, which yielded an overall accuracy of 0.800.
Discussion
IBM Watson’s Natural Language Classifier enabled relatively accurate assignment of intravenous contrast for MRI examinations using only the free-text clinical indication and required little to no technical knowledge. Overall performance was similar to an ensembling of eight traditional NLP models using a term-document matrix. Analysis of errors for Watson can be subdivided into two categories. Twenty-two of 47 errors for which Watson disagreed with the original protocol but agreed with the second reader can be attributed to additional clinical data that influenced the original protocol but was unavailable to Watson and the second reader. It is promising that, in the absence of this extra information, Watson protocoled these studies according to the standard practices of the blinded second reader. This is further demonstrated by a significant increase in accuracy to 0.900 in the subset of test cases for which the original protocol and second reader agreed.
In contrast, analysis of the 25/47 errors for which Watson disagreed with both the original protocol and second reader is more difficult. Some errors were relatively straightforward such as spelling, grammar, and ambiguity of language in the clinical indication. Of these, spelling and grammar errors could be mitigated by running intelligent preprocessing or “spell-check” although this may prove difficult with medical terminology. However, other errors were more difficult to troubleshoot, highlighting the downside of a “black-box” algorithm. For example, the study provided in Table 3, “POST OP FOR REMOVAL OF THORACIC TUMOR Reason: POST-OP FOR THORACIC TUMOR,” was appropriately assigned contrast in the original protocol and by the second reader. However, for reasons that are unclear, Watson did not assign contrast in this case and gave an overall low confidence score of 0.53. We could postulate that a low prevalence of thoracic spine tumor follow-ups biased the classifier to assign class NC; however, this is only speculative.
When evaluating the types of errors made by Watson, the false-negative rate (erroneously not assigning contrast) was three to four times higher than the false-positive rate (erroneously assigning contrast), regardless of which ground truth was used for comparison. Contrast assignment errors have varying degrees of clinical consequences, though we believe not providing contrast to be the safer of the two error types. For example, non-administration of contrast for a tumor follow-up would require a patient to return for additional sequences and may delay diagnosis; however, this is typically not considered acutely dangerous. Conversely, inappropriate administration of gadolinium-based contrast to an end-stage renal disease patient can result in debilitating or fatal nephrogenic systemic fibrosis and Watson did make one such critical error in the test set. Finally, we note that these results were achieved without incorporating the requested study type (e.g., “MRI lumbar spine without contrast”). We suspect that the inclusion of this data would improve overall accuracy; however, this may be at the expense of biasing the algorithm into misclassifying the relatively infrequent cases in which the clinician ordered an incorrect study (i.e., request for a non-contrast study for osteomyelitis).
Although performance between traditional NLP techniques and Watson was similar, one immediate advantage of the traditional machine learning models was an extremely short training time and minimal hardware requirements. Typical deep learning algorithms require powerful graphical processing unit to speed up the process of assigning weights to the neural network, but most traditional machine learning algorithms can be easily run on a basic laptop. Even with minimal hardware, training time was faster in the range of multiple orders of magnitude when compared to Watson. Conversely, a clear advantage of Watson is that it required no pre-processing or programming experience, and only minimal understanding of machine learning fundamentals such creation of valid training and test sets. This convenience, however, comes at a cost of the black-box problem. IBM Watson is a closed cloud service with proprietary algorithms that cannot be released in detail, and as such, most troubleshooting would need to be done by IBM staff. Algorithmic errors may remain obscure indefinitely depending on IBM’s willingness to modify the architecture for individual use cases. Furthermore, any updates to the service may inadvertently result in changes to the model, potentially resulting in detrimental errors that can lead to patient harm. On the other hand, in-house and locally run machine learning algorithms can be more easily accessed and modified by an on-site expert.
In regards to clinical implementation, a major strength of our approach is that referring clinicians need not alter their normal workflow of ordering MRIs. Many previous paradigms have employed a structured approached, wherein the requesting clinician must answer a series of questions to arrive at a pre-determined clinical indication that has a known protocol. However, this imposes additional work on ordering clinicians who may already be suffering from “click-fatigue.” Additionally, these systems are error-prone because they rely on the requesting clinician following instructions and clicking the correct boxes. Our approach does not change referring clinician workflow and allows them to order MRIs with free-text clinical indications as they normally would.
An intrinsic limitation in the scalability of our methods at our institution was the assignment of MRI protocols (which serves as ground truth) as free-text. Many systems, including the current system at our institution, now allow the radiologist to select the protocol from a pre-defined list based on the ordered study type. This dataset would be much cleaner and circumvent the issue of manually classifying each protocol for training. With a large enough sample across multiple subspecialties, it may be possible to assign full MRI protocols rather than just contrast. It is also conceivable that such a model could smooth over the variability of individual radiologists’ protocoling patterns. Additional clinical data such as allergies, renal function, and pregnancy status could be incorporated as a fail-safe against dangerous false-positives.
Despite stated limitations, IBM Watson’s Natural Language Classifier allows automated contrast assignment using solely free-text clinical indications. The performance of the algorithm was somewhat limited by heterogeneity in the training data; however, this can be addressed in future iterations. If successfully integrated into clinical workflow, it may improve efficiency and one day serve as a decision support tool for contrast assignment. Such a tool could also be modified for use by the ordering clinician as a form of clinical decision support in determining the correct study to order.
Conclusion
We demonstrate that a natural language classification algorithm can be trained with IBM Watson to automatically determine the need for intravenous contrast in musculoskeletal MRIs. We propose that this work be further extended to assign full protocols across a range of subspecialties, helping to improve efficiency and potentially decrease error rate.
Abbreviations
- IRB:
-
Institutional Review Board
- MRI:
-
Magnetic resonance imaging
- NC:
-
Non-contrast
- NLP:
-
Natural language processing
- WC:
-
With contrast
References
Boland GW, Duszak, Jr R, Kalra M: Protocol design and optimization. Journal of the American College of Radiology. 11(5):440–441, 2014. https://doi.org/10.1016/j.jacr.2014.01.021
Ginat DT, Uppuluri P, Christoforidis G, Katzman G, Lee S-K: Identification of neuroradiology MRI protocol errors via a quality-driven categorization approach. J Am Coll Radiol. 13(5):545–548, 2016. https://doi.org/10.1016/j.jacr.2015.08.027
Bairstow PJ, Persaud J, Mendelson R, Nguyen L: Reducing inappropriate diagnostic practice through education and decision support. International Journal for Quality in Health Care. 22(3):194–200, 2010. https://doi.org/10.1093/intqhc/mzq016
Garg AX, Adhikari NKJ, McDonald H et al.: Effects of computerized clinical decision support systems on practitioner performance and patient outcomes. JAMA. 293(10):1223, 2005. https://doi.org/10.1001/jama.293.10.1223
Blackmore CC, Castro A: Improving the quality of imaging in the emergency department. Acad Emerg Med 22(12):1385–1392, 2015 https://doi.org/10.1111/acem.12816
Kim, Yoon: Convolutional Neural Networks for Sentence Classification. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2014. https://doi.org/10.3115/v1/d14-1181
Pons E, Braun LMM, Hunink MGM, Kors JA: Natural language processing in radiology: a systematic review. Radiology. 279(2):329–343, 2016. https://doi.org/10.1148/radiol.16142770
Hassanpour S, Bay G, Langlotz CP: Characterization of change and significance for clinical findings in radiology reports through natural language processing. J Digit Imaging 30(3):314-322, 2017. https://doi.org/10.1007/s10278-016-9931-8
Huang M-W, Chen C-W, Lin W-C, Ke S-W, Tsai C-F: SVM and SVM ensembles in breast cancer prediction. PLOS ONE. 12(1):e0161501, 2017. https://doi.org/10.1371/journal.pone.0161501
Lakhani P, Langlotz CP: Automated detection of radiology reports that document non-routine communication of critical or significant results. J Digit Imaging. 23(6):647–657, 2010. https://doi.org/10.1007/s10278-009-9237-1
Hassanpour S, Langlotz CP: Information extraction from multi-institutional radiology reports. Artif Intell Med. 66:29–39, 2016. https://doi.org/10.1016/j.artmed.2015.09.007
Cheng LTE, Zheng J, Savova GK, Erickson BJ: Discerning tumor status from unstructured MRI reports-completeness of information in existing reports and utility of automated natural language processing. J Digit Imaging. 23(2):119–132, 2010. https://doi.org/10.1007/s10278-009-9215-7
Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF: Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 35(8):128–144, 2008.
Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG: A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 34(5):301–310, 2001. https://doi.org/10.1006/jbin.2001.1029
LeCun Y, Bengio Y, Hinton G: Deep learning. Nature. 521(7553):436–444, 2015. https://doi.org/10.1038/nature14539
Ferrucci D, Levas A, Bagchi S, Gondek D, Mueller ET: Watson: beyond jeopardy! Artificial Intelligence. 199:93–105, 2013. https://doi.org/10.1016/j.artint.2012.06.009
R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL: https://www.R-project.org/
Jurka TP, Collingwood L, Boydstun AE, Grossman E, van Atteveldt W: RTextTools: a supervised learning package for text classification. R Journal. 5(1):6–12, 2013
Jurka T: MAXENT: an R package for low-memory multinomial logistic regression with support for semi-automated text classification. R J 4(1):56, 2012
Friedman J, Hastie T, Tibshirani R: Regularization paths for generalized linear models via coordinate descent. J Stat Soft 33(1):1–968, 2010. https://doi.org/10.1109/TPAMI.2005.127
Liaw A, Wiener M: Classification and regression by randomForest. R News 2(1):18, 2002
Feinerer I, Hornik K, Meyer D: Text Mining Infrastructure in R. J Stat Softw 25(5):1–54, 2008.
Peters A, Hothorn T, Lausen B: Ipred: improved predictors. R News, 2002. Available at https://cran.r-project.org/web/packages/ipred/vignettes/ipred-examples.pdf. Accessed 12 Sept 2017
Tuszynski J.: caTools: tools: moving window statistics, GIF, Base64, ROC AUC, Etc. R package version, 2008. Available at https://cran.r-project.org/web/packages/caTools/caTools.pdf. Accessed 12 Sept 2017
Ripley B.: Classification and regression trees. Available at https://cran.r-project.org/web/packages/tree/tree.pdf. Accessed 13 Sept 2017
Feinerer I, Hornik K, Meyer D: Text mining infrastructure in R. Journal of Statistical Software 25(5):1–54, 2008. 10.18637/jss.v025.i05
Ferrucci DA: Introduction to “this is Watson”. IBM Journal of Research and Development 56(3.4):1:1–1:15, 2012. https://doi.org/10.1147/JRD.2012.2184356
Funding
HT was supported by an NIH T32 Fellowship, 5T32EB001631-10. JHS was supported by the NVIDIA academic grant program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Trivedi, H., Mesterhazy, J., Laguna, B. et al. Automatic Determination of the Need for Intravenous Contrast in Musculoskeletal MRI Examinations Using IBM Watson’s Natural Language Processing Algorithm. J Digit Imaging 31, 245–251 (2018). https://doi.org/10.1007/s10278-017-0021-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-017-0021-3