Abstract
A significant volume of medical data remains unstructured. Natural language processing (NLP) and machine learning (ML) techniques have shown to successfully extract insights from radiology reports. However, the codependent effects of NLP and ML in this context have not been well-studied. Between April 1, 2015 and November 1, 2016, 9418 cross-sectional abdomen/pelvis CT and MR examinations containing our internal structured reporting element for cancer were separated into four categories: Progression, Stable Disease, Improvement, or No Cancer. We combined each of three NLP techniques with five ML algorithms to predict the assigned label using the unstructured report text and compared the performance of each combination. The three NLP algorithms included term frequency-inverse document frequency (TF-IDF), term frequency weighting (TF), and 16-bit feature hashing. The ML algorithms included logistic regression (LR), random decision forest (RDF), one-vs-all support vector machine (SVM), one-vs-all Bayes point machine (BPM), and fully connected neural network (NN). The best-performing NLP model consisted of tokenized unigrams and bigrams with TF-IDF. Increasing N-gram length yielded little to no added benefit for most ML algorithms. With all parameters optimized, SVM had the best performance on the test dataset, with 90.6 average accuracy and F score of 0.813. The interplay between ML and NLP algorithms and their effect on interpretation accuracy is complex. The best accuracy is achieved when both algorithms are optimized concurrently.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Hypothesis
Artificial intelligence software’s ability to predict radiologist intent in an oncologic diagnostic report relies on the co-dependent, combinatorial optimization of both the natural language processing (NLP) and machine learning (ML) algorithms.
Background
The advent of structured reporting may improve the availability of standardized data elements in a radiology report for text mining. However, most radiology reports remain unstructured. The lack of structure reporting can result in poor communication of abnormal radiology reports to referring physicians; this is particularly true for unstructured reports that contain complex results and convey intrinsic diagnostic uncertainty such as oncologic follow-up [1]. For named-entity recognition, regular-expression and search-based report analytics have been shown to extract specific critical diagnoses successfully [2, 3]. NLP is increasingly being used to analyze radiology reports for oncologic imaging [4, 5]. For instance, the presence of specific malignant diagnoses such as lung cancer and colon cancer has been previously examined [6].
Commonly deployed pre-processing techniques for unstructured text include stop word removal (SWR) and word stemming [7]. In SWR, the most commonly used words in a language such as “and” and “the” are removed from the dataset prior to text processing. Word stemming converts inflected forms of words into their root forms; for example, “write,” “writes,” and “writing,” share the stem “write.” For the processing of text written in English, Martin Porter’s algorithm has become the most commonly deployed stemmer [8]. Although SWR and stemming have been shown to improve NLP of English lay-language text, a paucity of literature exists describing their effect in the processing of unstructured medical text.
The “bag-of-words” model may be used to represent unstructured text as vectors. The vectors contain features based on the frequencies of tokens contained in an up to a pre-determined length N (N-gram). For instance, term frequency (TF) is a commonly deployed method of feature generation, in which terms are represented by the frequency of the N-gram. While meaningful N-grams may appear more frequently within a document, the tokens with the highest frequencies may represent an artifact of commonality. For instance, in English a word such as “the” would have very high TF. Therefore, in some use cases, each term’s TF as normalized by the inverse document frequency (TF-IDF), which has been shown to be superior to using TF alone [9]. However, as TF and TF-IDF strategies can sometimes create extraordinarily high dimensional datasets, feature hashing has been shown to be an effective strategy for dimensionality reduction [10]. Bag-of-words models have been used to successfully process radiology reports [11, 12].
ML techniques have become increasingly common in medical text processing. An abundance of research explores the use of a specific approach to address a specific problem. For example, support vector machines (SVMs) have shown success in predicting sepsis in emergency department [13]. In radiology, SVM has been used to predict significant findings in diagnostic reports [12]. Other ML techniques utilizing Bayesian probabilities, neural networks (NNs), and random decision forests (RDF) have shown some success in producing clinically useful insights from both text and imaging data [14,15,16,17].
Although pre-processing techniques, text feature representation models, and ML algorithms have all shown usefulness in the processing of medically relevant text data, there is a paucity of literature exploring the comparative effectiveness of different NLP techniques and ML algorithms performing the same task. In this study, we assess the effect of multiple NLP techniques and ML algorithms on the automatic detection of the radiologist’s intent in oncologic evaluations.
Methods
This project was reviewed by the institutional review board (IRB) and approval was waived. At our tertiary referral academic institution, all abdominal and pelvic CT and MRI reports must include one of two standardized assessment categorization schemes. For patients with no known malignancy, radiologists employ an in-house lexicon called Code Abdomen that assigns a numeric category according to the malignant likelihood of focal masses in the abdomen and pelvis. Code Abdomen is loosely based on the Breast Imaging Reporting and Data System (BI-RADS) [18]. For patients with known malignancy, radiologists use a different in-house lexicon, called Code Oncology. Code Oncology is loosely based on the categories in the Response Evaluation Criteria in Solid Tumors (RECIST) system [19]. Using Code Oncology, the interpreting radiologist assigns values in a structured reporting template to two specified categories: (a) interval evolution of existing lesions and (b) interval development of new lesions. Within the departmental voice recognition software, options are provided for the values of the fields within the structured templates; these are shown in Table 1. The options chosen for each exam are left to the discretion of the interpreting board-certified radiologist. All abdominal and pelvic CT and MRI reports are mined daily to confirm the presence of one of these two categorization schemes within the reports. Radiology trainees and staff are sent email notifications for non-compliant reports and asked to issue addendums.
Between April 1, 2015 and November 1, 2016, a total of 9418 out of 50,891 cross-sectional abdominal and pelvic CT and MRI exams contained the Code Oncology scheme. Similar to RECIST, we created four response assessment groups: progression, stable disease, improvement, and complete resolution/no cancer for overall assessment. Definitions of these groups are provided in Table 2. During the initial preliminary data analysis, we discovered a wide practice variation regarding the use of non-RECIST labels. For instance, “mixed response” is sometimes used when some lesions may have been stable to decrease in size despite increases in other lesions; in clinical practice, such cases are generally considered disease progression. Examinations with new lesions that do not clearly represent metastases are sometimes categorized as “indeterminate” and sometimes “possible new lesion.” For these reasons, examinations containing “mixed response,” “indeterminate,” and “possible new lesion” were excluded from the dataset.
The structured “Code Oncology” elements were parsed and then removed from the report text prior to pre-processing. Relevant personal health information was also removed for compliance with the Health Insurance Portability and Accountability Act (HIPAA). Pre-processing was performed within the Azure Machine Learning Studio (Microsoft Corporation, Redmond, WA), using a combination of the Python programming language (version 3.5), the Natural Language Toolkit Python package (version 3.2), and native preprocessing modules [20]. Only deidentified report text was made available through the cloud resource for data security. Text header detection was performed using regular expressions to segment the radiology report by section. Only the impression was utilized in the final comparative analysis, as the use of the impression yielded more accurate performance relative to the full report text based on our preliminary work (Table 3). If more than one impression bullet point existed, then the impression was included both in total as well as separated by each bullet point. All report text was then converted to lower case and all punctuations removed. For each section, evaluation was performed after applying an English word tokenizer both with and without SWR and both with and without applying a Porter stemmer [8].
Three forms of text feature vectorization using the bag-of-words model were compared: term frequency-inverse document frequency weighting (TF-IDF), term frequency weighting (TF), and 16-bit feature hashing. Vectorization parameters were adjusted for the overall best predictive performance defined by the ML model’s micro-average F score [21]. Parameters adjusted include N-gram (up to five-gram). For TF and TF-IDF, K-skip size, minimum N-gram document absolute frequency, and maximum N-gram document ratio were also explored for optimal performance. Filter-based feature selection was performed to select the most relevant features using mutual information [22].
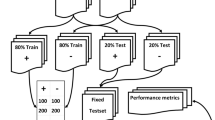
Five ML algorithms were compared in the present study, including logistic regression (LR), RDF, one-vs-all SVM, one-vs-all Bayes point machine (BPM), and fully connected NN. Input data was stratified by classification label and randomly assigned into training (70%) or testing (30%) datasets. The BPM was implemented to train for 60 iterations with bias. The training data was divided into five folds to perform an eight-run random sweep with cross-validated hyperparameter model tuning to identify the best parameter set for each of the remaining four ML algorithms. Table 4 lists all the parameters that were optimized for each ML algorithm. The performance was measured using a micro-average F score and average classification accuracy using the testing dataset [21, 23].
Results
Of the 9418 examinations performed within the study timeframe, 8614 examinations met the inclusion criteria. Of these, 2800 were manually categorized as “resolution/no cancer,” 2498 categorized as “progression,” 2132 categorized as “stable disease,” and 1184 categorized as “improvement.”
The set of text preprocessing techniques which yielded the best predictive accuracy and F score is referred hereafter as “reference preprocessing settings” consisting of tokenized unigrams and bigrams with TF-IDF, SWR, Porter stemming, and filter-based feature selection limited to the top 1000 features. Using the reference preprocessing techniques on the testing dataset, the BPM algorithm achieved an 89.5% average classification accuracy. After hyperparameter model tuning, the best performing multi-class LR algorithm, RDF algorithm, fully connected NN, and SVM achieved an average predictive accuracy of 90.2, 90.0, 88.3, and 90.6%, respectively. Table 5 displays the results from training and testing accuracy as well as F scores.
With other elements of the reference preprocessing techniques held constant, SWR slightly improved the micro-average F score for all ML algorithms relative to no SWR. Word stemming slightly improved the performance of BPM, NN, and SVM but did not impact or minimally degraded the F score of LR and RDF. TF-IDF was superior to TF alone for BPM, NN, and SVM but slightly decreased accuracy in RDF and had no effect in LR. Using feature hashing rather than TF-IDF improved the runtime of model training but decreased micro-average F score for BPM, LR, and SVM, with minimal performance effect on RDF and NN. Table 6 demonstrates the relative contribution of each of the NLP parameters.
A combination of unigrams and bigrams outperforms other lengths of contiguous word series for all ML algorithms except for RDF, which performed best with a combination of unigrams, bigrams, as well as trigrams (Fig. 1). Table 7 lists the top 15 most discriminating word features ranked by mutual information. While LR and NN performed best with all the N-gram features, the other ML algorithms performed best when only the top 1000 features are used based on the filter-based selection. The effect of filter-based feature selection on F score of all five ML algorithms is shown in Fig. 2.
Increasing the number of included relevant N-gram tokenized features improved the performance of neural network and random decision forest but has detrimental effect on support vector machine, logistic regression, and Bayes point machines. Report text was processed using tokenized unigrams and bigrams with TF-IDF, SWR, and Porter stemming
Discussion
The present study uses standardized reporting structures embedded within formal diagnostic reports as the ground truth for ML. Our results show that the performance of radiology report classification is likely dependent on both the ML algorithm and on the NLP parameters. Modern NLP includes increasingly complex manipulations and vectorization approaches such as Word2Vec and Stanford University’s GloVe [24, 25]. However, the present study focuses on traditional text preprocessing techniques and adds to current literature by assessing multiple ML algorithms simultaneously regarding their performance on the same diagnostic radiology reports. Our findings agree with existing literature in electronic report text mining that SVM performs well in classification tasks [6]. Specifically, the best predictive performance was achieved using SVM with the reference preprocessing techniques.
The present study further assessed the effect of optimizing NLP parameters by assessing the impact of each modification on five different ML algorithms. SWR generally improves the F scores of all ML algorithms except for BPM, although the precise underlying reason is unclear. However, as with many real-word use cases of NLP and ML, the optimal parameter settings are often difficult to determine due to the complexity of the task and rely on empiric experimentation, for which hyperparameter tuning has been well-established in the literature. The use of TF-IDF rather than TF alone had a modest to equivocal effect on F scores across the board. Our findings are compatible with the published literature on the use of inverse document frequency to normalize the TF [9, 26]. The use of 16-bit feature hashing significantly improved the runtime of all five algorithms but decreased the F score of BPM, LR, and SVM. It had little to no impact on the RDF and fully connected NN algorithms.
The relative performance of SVM decreases when more features are included. Specifically, with greater than 2500 text features, RDF outperforms SVM when other parameters are held constant. When the full set of 4122 text features are used, both RDF and LR perform better than SVM. The interval decreases in performance in SVM, LR, and BPM—but not RDF or NN—as the size of the feature set increases is likely related to overfitting. The problem of overfitting in SVM has been well studied in text categorization and other non-linear classification tasks, particularly when the number of features is large [27, 28]. Additionally, our findings agree with existing literature that RDF and NNs can be relatively resistant to performance penalties from overfitting [16, 29].
The natural language used in radiologic reporting is inherently ambiguous due to the complexity of human illness and the variable language used to describe it. The top 20 “high-confidence misses” by the best-performing SVM is listed in Supplemental Table 1. Preliminary manual analysis of the “high-confidence misses” by the best-performing SVM with reference preprocessing techniques showed a variety of possible causes for predictive error. In some cases, the algorithm inappropriately overweighed portions of the impression describing improvement and underweighed a separate section of impression documenting disease progression. In other cases, the interpreting radiologists made a human error in manual categorization of existing and new disease, while the algorithm correctly identified the disease states. In yet other cases, both designations could have been considered correct. Although the present study focuses on the differential performance of NLP and ML algorithms, a thorough, detailed analysis of these discrepancies arising from the best performing algorithm is a direction of future pursuit.
The “Code Oncology” reporting structure at our institution was developed to help clarify otherwise potentially confusing reports due to the complexity of medical language used in radiologic reports. Nevertheless, the structured report coding is for each study relies on the radiologist’s manual curation. Therefore, the development of an algorithm’s capable of identifying the radiologist’s diagnostic intent has several clinical implications. First, our preliminary analysis of the “high-confidence misses” reveals that in some minority of cases, human radiologists may accidentally assign an erroneous code to a report, and the algorithm serves as an error-correction mechanism. Additionally, we plan to implement the superior algorithm clinically so that accurate, automatic assignment “Code Oncology” can be in for approximately 90% of the cases, allowing radiologists to reduce human coding errors and to improve efficiency.
Additional next steps include the application of additional NLP algorithms. For instance, convolutional neural networks (CNN) have shown remarkable success in image recognition and classification and have been applied to natural language feature extraction using medical literature such as semantic models [30, 31]. Additionally, the use of skip-gram models in the future may yield improved performance over TF, TF-IDF, and hashing mechanics. The present study is limited by the size of its annotated dataset and the computational power of the hardware. Improvements to the present technique may be achieved by using gradient descent or grid-based methods for hyperparameter tuning. Due to the use of k-fold cross-validation and hyperparameter optimization for model training, we were unable to include CNNs as a comparative ML algorithm. A future analysis in this field would include applying convolutional models on significantly larger training sets by using graphical processing units.
Conclusion
Although NLP and ML algorithms have the potential to accurately classify the radiologist’s diagnostic intent in the oncologic interpretation, the overall performance depends on the combinatorial optimization of both the NLP and ML algorithms. We demonstrated that (1) the best predictive performance was achieved using SVM with the reference preprocessing techniques, (2) SWR generally improves the F scores of all ML algorithms except for BPM, and (3) the relative performance of SVM decreases with more features included.
References
Schwartz LH, Panicek DM, Berk AR, Li Y, Hricak H: Improving communication of diagnostic radiology findings through structured reporting. Radiology. 260(1):174–181, 2011
Lakhani P, Kim W, Langlotz CP: Automated extraction of critical test values and communications from unstructured radiology reports: an analysis of 9.3 million reports from 1990 to 2011. Radiology. 265(3):809–818, 2012
Lakhani P, Kim W, Langlotz CP: Automated detection of critical results in radiology reports. J Digit Imaging. 25(1):30–36, 2012
Cai T, Giannopoulos AA, Yu S, Kelil T, Ripley B, Kumamaru KK et al.: Natural language processing technologies in radiology research and clinical applications. Radiogr Rev Publ Radiol Soc N Am Inc. 36(1):176–191, 2016
Yim W-W, Yetisgen M, Harris WP, Kwan SW: Natural language processing in oncology: a review. JAMA Oncol. 2(6):797–804, 2016
Kocbek S, Cavedon L, Martinez D, Bain C, Mac Manus C, Haffari G et al.: Text mining electronic hospital records to automatically classify admissions against disease: measuring the impact of linking data sources. J Biomed Inform., 2016
Rajaraman A, Ullman JD: Mining of massive datasets [Internet]. Cambridge: Cambridge University Press, 2011, [cited 2017 May 24]. Available from: http://ebooks.cambridge.org/ref/id/CBO9781139058452
Porter MF: An algorithm for suffix stripping. Program. 14(3):130–137, 1980
Wu HC, Luk RWP, Wong KF, Kwok KL: Interpreting TF-IDF term weights as making relevance decisions. ACM Trans Inf Syst. 26(3):1–37, 2008
Weinberger K, Dasgupta A, Langford J, Smola A, Attenberg J: Feature hashing for large scale multitask learning. In ACM Press; 2009 [cited 2017 May 24]. p. 1–8. Available from: http://portal.acm.org/citation.cfm?doid=1553374.1553516
Hassanpour S, Langlotz CP. Unsupervised Topic Modeling in a Large Free Text Radiology Report Repository. J Digit Imaging. 29(1):59–62, 2016.
Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artif Intell Med. 66:29–39, 2016.
Horng S, Sontag DA, Halpern Y, Jernite Y, Shapiro NI, Nathanson LA: Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PloS One. 12(4):e0174708, 2017
Morid MA, Fiszman M, Raja K, Jonnalagadda SR, Del Fiol G: Classification of clinically useful sentences in clinical evidence resources. J Biomed Inform. 60:14–22, 2016
Polak S, Mendyk A: Artificial neural networks as an engine of Internet based hypertension prediction tool. Stud Health Technol Inform. 103:61–69, 2004
Tong W, Xie Q, Hong H, Shi L, Fang H, Perkins R et al.: Using decision forest to classify prostate cancer samples on the basis of SELDI-TOF MS data: assessing chance correlation and prediction confidence. Environ Health Perspect. 112(16):1622–1627, 2004
Wang Z, He Y, Jiang M: A Comparison among Three Neural Networks for Text Classification. In IEEE; 2006 [cited 2017 May 29]. Available from: http://ieeexplore.ieee.org/document/4129218/
Zafar HM, Chadalavada SC, Kahn CE, Cook TS, Sloan CE, Lalevic D et al.: Code abdomen: an assessment coding scheme for abdominal imaging findings possibly representing cancer. J Am Coll Radiol JACR. 12(9):947–950, 2015
Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L et al.: New guidelines to evaluate the response to treatment in solid tumors. J Natl Cancer Inst. 92(3):205–216, 2000
Bird S, Klein E, Loper E: Natural language processing with Python, 1st edition. Beijing: O’Reilly, 2009, 479 p
Lipton ZC, Elkan C, Naryanaswamy B: Optimal thresholding of classifiers to maximize F1 measure. Mach Learn Knowl Discov Databases Eur Conf ECML PKDD Proc ECML PKDD Conf. 8725:225–239, 2014
Bennasar M, Hicks Y, Setchi R: Feature selection using joint mutual information maximisation. Expert Syst Appl. 42(22):8520–8532, 2015
Hripcsak G, Rothschild AS: Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc JAMIA. 12(3):296–298, 2005
Pennington J, Socher R, Manning C. Glove: global vectors for word representation. In association for computational linguistics; 2014 [cited 2017 Sep 2]. p. 1532–43. Available from: http://aclweb.org/anthology/D14-1162
Mikolov T, Chen K, Corrado G, Dean J. Efficient Estimation of Word Representations in Vector Space. ICLR Workshop. 2013 Jan 16;
Zhang W, Yoshida T, Tang X. TFIDF, LSI and multi-word in information retrieval and text categorization. In IEEE; 2008 [cited 2017 May 29]. p. 108–13. Available from: http://ieeexplore.ieee.org/document/4811259/
Dietrich R, Opper M, Sompolinsky H: Statistical mechanics of support vector networks. Phys Rev Lett. 82(14):2975–2978, 1999
Joachims T: Text categorization with support vector machines: learning with many relevant features. In: Nédellec C, Rouveirol C Eds. Machine learning: ECML-98 [Internet]. Berlin: Springer Berlin Heidelberg, 1998, pp. 137–142 [cited 2017 May 29] Available from: http://springerlink.bibliotecabuap.elogim.com/10.1007/BFb0026683
Liu X, Song M, Tao D, Liu Z, Zhang L, Chen C et al.: Random forest construction with robust semisupervised node splitting. IEEE Trans Image Process Publ IEEE Signal Process Soc. 24(1):471–483, 2015
Wang J, Zhang J, An Y, Lin H, Yang Z, Zhang Y et al.: Biomedical event trigger detection by dependency-based word embedding. BMC Med Genomics 9 Suppl 2:45, 2016
Wei W, Marmor R, Singh S, Wang S, Demner-Fushman D, Kuo T-T et al.: Finding related publications: extending the set of terms used to assess article similarity. AMIA Jt Summits Transl Sci Proc AMIA Jt Summits Transl Sci. 2016:225–234, 2016
Funding
This study received no funding support from a grant agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Po-Hao Chen is a co-founder of Alphametric Health LLC. Maya Galperin-Aizenberg, Hanna Zafar, and Tessa S. Cook declare that they have no conflicts of interest.
Informed Consent
For this type of study formal consent is not required.
Electronic Supplementary Material
ESM 1
(DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Chen, PH., Zafar, H., Galperin-Aizenberg, M. et al. Integrating Natural Language Processing and Machine Learning Algorithms to Categorize Oncologic Response in Radiology Reports. J Digit Imaging 31, 178–184 (2018). https://doi.org/10.1007/s10278-017-0027-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-017-0027-x