Abstract
The aim of this study was to assess the factors, mechanisms and the differences between periodontal ligament (PDL) cells and denta l follicle (DF) progenitor cells towards the osteoblastic/cementoblastic differentiation and to investigate the effects of BMP-7 on developmental (DF) and mature tissue-derived (PDL) cells, respectively. Primary cell culture of PDL cells and DF progenitor cells was performed. Osteogenic differentiation was evaluated using von Kossa, Alizarin Red S and immuno-histo-chemistry staining of osteocalcin. Gene expression pattern was evaluated via real-time PCR. A series of CD surface marks were tested using flow cytometry and fluorescence-activated cell-sorting analysis was performed. Real-time RT-PCR demonstrated similar gene expression pattern of PDL cells and DF progenitor cells: the expression of OPN and OCN significantly was elevated when incubated with osteogenic components, Runx2 was unaffected, and Osteorix was hardly expressed whether in basic medium or induction medium. In addition, BMP-7 induced osteoblast/cementoblast differentiation of PDLSCs and DF progenitor cells in a dose- and time-dependent manner, as reflected by enhanced Runx2 and (OCN) mRNA transcript expression. BMP-7 triggers PDL cells and DF progenitor cells to differentiate towards an osteoblast/cementoblast phenotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The discovery of stem cells and progenitor cells opened new frontiers in tissue engineering, which aims to replace complex anatomical structures consisting of multiple cell lineages such as periodontal ligament. Dental tissues such as dental pulp (DP), human primary teeth, periodontal ligament (PDL) and dental follicle (DF) have been identified as appropriate sources of stem cells and/or progenitor cells [1–4].
PDL is a specialized connective tissue, which is responsible for the regeneration of the adjacent periodontal structures. However, PDL has a limited capacity to reconstruct the periodontal structure in response to trauma or inflammation. The true process of PDL regeneration involves the recruitment of progenitor cells or stem cells, differentiating into fibroblasts, cementoblasts or osteoblasts [5].
PDL stem cells (PDLSCs) share similar characteristics including the ability to self-renew and multi-lineage differentiation potential and express a heterogeneous assortment of markers associated with other adult mesenchymal stem cells (MSCs) including; dentin [2], bone [6], smooth muscle [7], neural tissue and endothelium [8, 9]. In addition, a recent study performed by Han et al. [10] showed that allogeneic PDLSCs have a marked ability to repair periodontal defects by forming bone, PDL and cementum-like tissue in vivo [11].
The dental follicle (DF), a loose ecto-mesenchymal connective tissue sac that surrounds the developing tooth, is believed to play a crucial role during the eruption of tooth [8]. It has been postulated that the DF cells near the forming root (innermost) develop into cementoblasts and the cells towards the alveolar bone (outermost) differentiate into osteoblasts secreting the bone matrix. In addition, DF cells, like BMSCs, can differentiate into adipocytes and neurons in vitro conditions. The differentiation of DF cells is regulated by a complex network composed of growth factors and cytokines [12, 13]. Epithelial stimuli derived from the bi-layered Hertwig’s epithelial sheath (HERS) seem to be responsible for the differentiation of follicle precursor cells into cementoblasts [14]. Enamel matrix derivatives (EMD) have been proposed to mimic the inductive roles of HERS products [15]. A previous study demonstrated that bone morphogenetic proteins (BMP) promote the DF cells differentiate into osteoblasts and/or cementoblasts phonotype [16–18].
BMPs, which belong to the transforming growth factors β (TGF-β) superfamily, are thought to be the most important growth factors in bone formation and healing [19]. BMPs and BMP receptors are known to regulate the development of calcified tissues by directing mesenchymal precursor cells differentiation. BMP-7, also known as osteogenic protein-1 (OP-1), was believed to be the crux of the complex cellular and molecular signals that regulate the topography and assembly of the extracellular matrix, delicately guiding angiogenesis [20], vascular invasion, and osteogenesis [21]. BMPs are also involved in osteo-cementoblast differentiation, periodontal development and healing. Taba et al. [22] have showed that BMP-2 and -7 stimulate cementogenesis during periodontium development. The incisive work of Ripamonti has shown that BMPs are the soluble signals that initiate regeneration of the periodontal tissues, including the induction of cementogenesis [23].
In the literature, it has been suggested that BMPs can induce PDLSCs and DF progenitors to differentiate toward the cementoblast phenotype; however, the exact mechanism remains still unknown. The aim of this study was to assess the factors, mechanisms and the differences between PDL cells and DF progenitors toward the osteoblastic/cementoblastic differentiation and to investigate the effect of BMP-7 on developmental (DF) and mature tissue-derived (PDL) cells, respectively.
Materials and methods
Cell culture
Preparation and culture of PDL cells
Human impacted third molars were surgically extracted and collected with patient’s informed consent at the department of Oral and Maxillofacial Surgery of the Christian Albrechts University in Kiel. The age of the patients varied between 15 and 36 years. The whole procedure followed an informed protocol approved by the local ethics committee.
PDL was gently scraped from the middle root surface; coronal and apical portions of the ligament were not used to avoid contamination by gingival and pulpal cells. PDL tissues were then minced and digested in a solution containing 3 mg/ml collagenase type I (Worthington Biochem Co, Lakewood, NJ, USA) and 2.5 mg/ml dispase II (Roche, Mannheim, Germany) for 1 h at 37 °C.
After digestion, tissue samples were seeded into culture flasks with alpha-modification of Eagle’s Medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10 % fetal bovine serum (Hyclone), 2 mM Glutamine (Sigma-Aldrich, Steinheim, Germany), 100 U/ml penicillin and 100 µg/ml streptomycin (Biochrom KG Seromed, Berlin, Germany) at 37 °C in 5 % CO2 in a humidified atmosphere. After single cells were attached to the plastic bottom of the flask, non-adherent cells were removed by changing the medium. When confluent, the cells surrounding the explants were sub-cultured with 0.25 % trypsin and 0.02 % EDTA. Cells of passage 4–5 were used for the experiment.
Preparation and culture of DF cells
The DF cells were obtained from the attached dental follicle of the tooth at the early onset of the root formation (assessed by radiographic and clinical observation). DF cells were prepared and cultured the same way as the PDL cells. Under these culture conditions, epithelium cells did not survive. Cells of passage 4–5 were used for the experiment.
Cell colony test
PDL cells and DF cells were seeded in a dilution of 10 cells/cm2 on 35–mm cell culture dishes in four parallel wells. On the 12th day, cells were washed with PBS and then stained with 3 g/l crystal violet solution (Merck).
The frequency of CFU-F (colony forming fibroblast units) was assessed. Aggregates of >50 cells were scored as colonies.
Alkaline phosphatase (ALP) staining
After cultured in standard medium for 14 days, cells were fixed in 4 % paraformaldehyde (PFA), and then incubated with SIGMA FAST BCIP for 20 min.
Cell differentiation
Osteogenic differentiation
-
Osteogenic medium
To induce osteogenic differentiation, cells were seeded at a density of 2 × 103/cm2 and maintained with α-MEM supplemented with 10 % FBS, 100 nM dexamethasone (Serva), 10 mM β-Glycerolphosphate (Sigma), 52 mg/l Ascorbin-2-Phosphate (Sigma) and 230 mg/l CaCl2 (Sigma). Control cultures were maintained without osteogenic supplements. After 14 or 21 days, the cells were fixed and subjected to Alizarin Red S and von Kossa staining to assess for mineral deposition. Control cultures were maintained without osteogenic supplements.
-
Von Kossa staining
Von Kossa staining was used to detect mineralization in vitro. After 21 days cultured in medium with osteogenic supplements, the cells were fixed in 4 % paraformaldehyde (PFA) in PBS, and then treated with silver nitrate solution for 15 min in the dark, and then exposed to light for an hour or so. The calcium will be reduced by the strong light and replaced with silver deposits, visualized as metallic silver. Experiments were performed twice with comparable results.
-
Alizarin Red S staining
The principle of Alizarin Red S staining is that calcium will form an alizarin red S-calcium complex in a chelation process. Briefly, after stimulated in osteogenic medium for 14 days, the cells were fixed in 4 % paraformaldehyde (PFA) in PBS after incubation in osteogenic medium for 14 days, and then treated with 40 mM Alizarin red S (Sigma) solution. The alizarin red S solution was fresh, and adjusted the PH to 4.1–4.3 with 0.1 M NH4OH solution. Experiments were performed twice with comparable results.
-
Immunohistochemistry staining of osteocalcin (OCN)
After PDL cells and DF progenitor cells were cultured for 21 days in osteogenic inductive medium, cells were trypsinized and reseeded into 8-chamber slides (2 × 103/cm2). The cells were allowed to attach 24 h before immuno-histo-chemical staining of OCN. Briefly, the monolayers cells were exposed to methanol containing 0.6 % H2O2 to block the endogen peroxidase and incubated with Rabbit anti-human osteocalcin polyclonal primary antibody (LINARIS, Wertheim-Bettinger, Germany) in 1 % BSA for 2 h at room temperature. For negative control, only 1 % BSA was added. After rinsing 3–5 min in TBS, the slides were incubated with goat anti-rabbit secondary antibody in 1 % BSA for 30 min. After rinsing in TBS, the slides were incubated in StrepABC Complex/HRP. The samples were subsequently incubated in DAB in the dark. Finally, the nucleuses were stained Hemalaun nach Meyer. Slides were mounted in Kaiser’s Glyceringelatine. Brown cytoplasms were assessed as osteocalcin positive.
Adipogenic differentiation
-
Adipogenic medium
To induce adipogenic differentiation, cells were seeded at a density of 2 × 104/cm2 and maintained with α-MEM supplemented with 10 % FCS, 1 µM dexamethasone (Serva), 0.5 mM 1-methyl-3-isobutylxanthine (Sigma), 60 µM Indomethacin (Sigma) and 20 U/l Insulin (Sigma). Control cultures were maintained without osteogenic supplements. After 21 days, the cells were fixed and subjected to Oil Red O staining to evaluate adipogenesis. Control cultures were maintained without adipogenic supplements.
-
Oil Red O staining
After 21 days, the cells were fixed, incubated with 2 ml 60 % isopropyl alcohol per well, and stained with 2 ml 0.3 % Oil Reo O working solution. Photos were taken within half an hour. The fat lipids were stained red.
Fluorescence-activated cell-sorting analysis (FACS)
The principle of flow cytometry is utilization of a laser beam to separate cell fractions from a larger population by employing the differential light-absorbing or fluorescing properties of certain cell subpopulations.
Cultured PDL cells and DF cells of passage 4 were re-suspended at 1.0 × 106 cells/ml medium. In each tube, 100 μl cell suspension and 10 μl antibodies (with different fluoresceins were added and incubated at 4 °C for 20 min after vortexing. The addition of the antibodies is shown in Table 1.
All antibodies used in the current experiment were purchased from Beckman Coulter (USA). Due to the limited knowledge regarding the surface marks of PDLSCs and DF progenitors, a series of CD surface marks were tested using flow cytometry (FC-500 Flow Cytometer) IgGs in the first line were isotypes of FITC, PE, PC5, PC7 and ECD, respectively.
Real- time RT- PCR
RNA extraction
PDL cells and DF cells at the fourth passage were harvested, seeded at a density of 2 × 103 cells/cm2 into 6-well plastic culture plate, and maintained in 2 ml medium. Total RNA was extracted on 14th or 21st days, using the Qiagen Rneasy® Plus Mini kit (Qiagen GmbH, Hilden, Germany), and quantified by spectrometry at 260 and 280 nm. Genomic DNA contamination was eliminated by gDNA Eliminator Spin column.
Reverse transcription
Total RNA was reverse-transcribed to cDNA using Qiagen QuantiTech™ Reverse Transcription Kit (Qiagen GmbH, Hilden, Germany). Briefly, the genomic DNA Elimination reaction was prepared, incubated for 2 min at 42 °C, and then placed immediately on ice. At the same time, the master mix was prepared on ice. A template DNA was added to each tube containing reverse-transcription master mix and incubated for 15 min at 42 °C and followed by incubation for 3 min at 95 °C to inactivate the Quantiscript Reverse Transcriptase.
Real-time polymerase chain reaction (Real-time PCR)
Real-time PCR was performed on a light-cycler (Roche, Mannheim, Germany) machine using the software 3.5.3. The commercial available PCR primers were purchased from Qiagen QuantiTect primer assay (Qiagen GmbH, Hilden, Germany). PCR amplification was carried out in a total volume of 20 µl in PCR master mix containing 10 µl of QuantiTect SYBR® Green (Qiagen GmbH, Hilden, Germany), 2 µl of 10 × QuantiTect primer assay and 2 µl of the reverse transcription product filled up to 20 µl with RNase-free H2O. The optimal annealing temperature was 55 °C for 40 cycles. The PCR solutions without template were used as negative control.
Calculation of real time PCR results
To normalize the content of cDNA samples, the comparative threshold (Ct) cycle method was used. The number of target genes copies vs. the number of house-keeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was compared. For comparative analysis of gene expression, data were obtained using ΔCt method derived from a mathematical elaboration. During PCR analysis, reactions were run in triplicate and performed three times on separate occasions.
Bone morphogenetic protein-7 effect on differentiation parameters
To determine the effects of BMP-7 on gene expression of PDL cells and DF progenitor cells, cells were plated in 6 well plates, at a density of 103 cells/cm2 in α-MEM with 10 % FBS. Upon confluence, media were replaced with α-MEM with 1 % FBS. After incubation with α-MEM with 1 % FBS for 24 h, cells were further stimulated with α-MEM supplemented osteo-inductive medium containing 1 % FBS and BMP-7 and incubated for 1, 3, 5 day. Media were changed on the 2nd and 4th days. For initial experiments, BMP-7 was used at a concentration of 50 ng/ml. To determine the effects of different concentration of BMP-7, media were supplemented with BMP-7 (0, 10, 25, 50, or 100 ng/ml), and cells were harvested on the 5th day.
Statistical analysis
All the cell and biological analysis results were expressed as mean and standard deviation (SD) and tested for statistical significance with Student’s two-tailed t test. P ≤ 0.05 was considered to indicate statistical significance. All statistical tests were carried out using SPSS (version 12.0; SPSS, Chicago, IL, USA).
Results
Primary cell culture
Primary cell culture of PDL cells
The single PDL cell suspensions were plated in 75 cm2 cell flasks. Three days later, the cells attached to the plastic bottom of the flasks displayed typical fibroblastic morphology in standard medium and shared common features with bone marrow stromal stem cells. Sub-confluence was observed after 10–14 days, indicating the intense cell proliferation. Cells were passaged at a density of 30 cells/cm2. The same cell characteristics were observed in the secondary cultures (Fig. 1a). The low seeding density allowed the PDL-derived cells to form single clonogenic cell clusters of fibroblast-like cells (Fig. 1b). PDL cells were obtained through single colony selection and limited dilution method. It took 12 days to get 70–80 % confluence at the first passage.
a Morphological analysis of primary human PDL cells on passage 1 by phase-contrast microscopy. PDL cells were spindle and fibroblast like. Cell culture was performed in standard medium. Scale bar 50 μm. b Cell clusters derived from PDL formed a single colony stained with crystal violet solution at day 8. Scale bar 100 μm
Primary cell culture of DF progenitor cells
DF progenitor cells were isolated by plastic adherence from freshly extracted dental follicle tissues. Only a small number of single dental follicle cells attached to the plastic surface and grew as fibroblastic cells. Non-adherent cells were removed by changing the medium (Fig. 2a). The DF progenitor cells were characterized by typical fibroblast-like morphology. The DF cells were sub-cultured at the same density with PDL cell of 30 cells/cm2 and had the ability to form colonies after 10 days (Fig. 2b).
a Morphological analysis of primary human DF cells on passage 1 by phase-contrast microscopy. DF cells were spindle and fibroblast like. Cell culture was performed in standard medium. Scale bar 50 μm. b Cell clusters derived from DF formed a single colony stained with crystal violet solution at day 8. Scale bar 100 μm
Cell colony test
Cell colony test was performed 12 days after seeding with a density of 10 cells/cm2. The values are expressed as CFUs/per 105 cells. Data were obtained from 3 independent assays. PDL cells had a higher colony forming ability than DF progenitor cells. * = P < 0.05 (Fig. 3).
ALP staining
When PDL cells and DF cells were cultured in stem cell growth medium, they formed cell clusters after 2 weeks, and these clusters stained for ALP (Fig. 4).
Cell differentiation potential
Osteogenic differentiation
-
Von Kossa staining
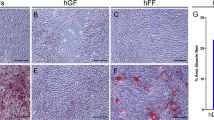
Following the exposure to the osteogenic induction medium for 3 weeks, PDL cells formed calcified nodules, which were stained black by von Kossa staining against red cytoplasm (Fig. 5a). Similarly, DF progenitor cells differentiated into osteoblasts/cementoblasts and developed mineralization nodules (clusters) (Fig. 6a). The cells in the control medium were not stained (Figs. 5b, 6b).
Alizarin Red S staining
To assess the calcification activity of PDL cells and DF progenitor cells, two cells were cultured in osteogenic induction medium and control medium for 2 weeks. All cultures were exposed to Alizarin Red S staining. Both PDL cells and DF progenitor cells formed calcified deposits, which were stained red (Figs. 7a, 8a). None of the cells cultured in control medium showed Alizarin red S reaction (Figs. 7b, 8b).
-
OCN staining
After 21 days in osteogenic inductive medium, PDL cells and DF progenitor cells were re-seeded into 8-chamber slides. Immuno-histochemical staining showed that PDL cells and DF progenitor expressed cementoblastic/osteoblastic marker OCN (Fig. 9a, b). It has been also observed that PDL cells and DF progenitor cells grew bigger and polygonal in shape, which is a typical morphology of osteoblasts, after osteogenic induction. Figure 9c and d was negative control of PDL cells and DF progenitor cells, respectively.
Adipogenic differentiation
Following the exposure to the adipogenic induction medium for 3 weeks, cultured PDL cells and DF progenitor cells displayed enlarged cell body and formed Oil Red O-positive lipid cluster (Fig. 10a, b, d, e). Standard culture medium did not induce any Oil Red O-positive fat drops (Fig. 10c, f).
Adipogenic differentiation of a, b PDL cells and d, e DF progenitor cells. Oil O red-positive lipid clusters were spotted in PDL cells and DF progenitor cells after 3 week of induction. b and e are higher magnification of PDL cells and DF progenitor cells, respectively. No oil O red-positive lipids were found in normal growth medium (c, f)
FACS results
As demonstrated in Figs. 11 and 12, both PDL cells and DF progenitor cells expressed a series of mesenchymal stem cells markers, such as CD105, CD29, CD90 and CD73 and did not express any hematopoietic cell lineage markers including CD45, CD19, HLA-DR, CD14 and CD34. 7-AAD was reference staining for dead and/or apoptotic cells.
Gene expressions during osteogenic and adipogenic differentiation by means of real-time PCR
Gene expressions during osteogenic differentiation by means of real-time RT-PCR
PDL cells were harvested after 2 week of culture time for real-time RT-PCR analysis. As shown in Fig. 13, the expression of Runx2 was unaffected during osteogenic differentiation, whereas expressions of OPN, Osteorix and OCN were significantly elevated in induction group (P < 0.05).
Real-time RT-PCR for relative gene expression of PDL cells derived from 2-week culture. Three replicates were made for each experiment and gene expression relative to GAPDH reference gene expression was calculated for normalization. Each bar represents the average of three experiments. Error bar denotes the range among three experiments. a OPN, b osteorix, c Runx2, d OCN
In DF cells, the expressions of Runx2 and Osteorix were unaffected during osteogenic differentiation, whereas expressions of OPN and OCN were significantly elevated in the induction group (P < 0.05) (Fig. 14).
Real-time RT-PCR for relative gene expression of DF cells derived from 2-week culture. Three replicates were made for each experiment and gene expression relative to GAPDH reference gene expression was calculated for normalization. Each bar represents the average of three experiments. Error bar denotes the range among three experiments. a OPN, b osteorix, c Runx2, d OCN
Gene expressions during adipogenic differentiation by means of Real-time-PCR
For gene expression analysis during adipogenic induction, PDL and DF cells were collected after 3 weeks of culture. As shown in Figs. 15 and 16, the expression of LPL and PPARG was significantly enhanced in the induction group (P < 0.05).
Real-time RT-PCR for relative gene expression of PDL cells derived from 3-week culture. Three replicates were made for each experiment and gene expression relative to GAPDH reference gene expression was calculated for normalization. Each bar represents the average of three experiments. Error bar denotes the range among three tests. a LPL, b PPARG
Real-time RT-PCR for relative gene expression of DF cells derived from 3-week culture. Three replicates were made for each experiment and gene expression relative to GAPDH reference gene expression was calculated for normalization. Each bar represents the average of three experiments. Error bar denotes the range among three tests. a LPL, b PPARG
Effects of BMP-7 on PDL and DF progenitor cells gene expression
Effects of BMP-7: dose response
-
Effects of BMP-7 on PDL cells gene expression: dose response
To determine the dose of BMP-7 required to increase for specific markers, PDL cells at a confluence were treated with BMP-7 at concentrations from 0 to 100 ng/ml for 5 days and RNA was extracted for analysis of mineral-associated genes. As shown in Fig. 17, exposure of PDL cells to BMP-7 resulted in up-regulation of OCN and Runx2mRNA in a dose-dependent fashion. As shown in Fig. 18, highest expression of OCN transcripts was noted at 50 ng/ml. A similar pattern was found for Runx2 mRNA levels, peaking at 50 ng/ml. Based on these results, 50 ng/ml BMP-7 was selected for use in further experiments.
-
Effects of BMP-7 on DF cells gene expression: dose response
To determine the dose of BMP-7 required to increase for specific markers, DF cells at confluence were treated with BMP-7 at concentrations from 0 to 100 ng/ml for 5 days. Results from this experiment revealed that DF cells respond to BMP-7 in a dose-dependent fashion (Fig. 18). As shown in Fig. 18, exposure of DF cells to BMP-7 resulted in an increase of OCN and Runx2 in mRNA. Moreover, strong expression of OCN transcripts was noted at 100 ng/ml. Highest Runx2 mRNA levels were peaking at 50 ng/ml.
Effects of BMP-7: time response
-
Effects of BMP-7 on PDL cells gene expression: time response
Figure 19 illustrated the time response of PDL cells to BMP-7. PDL cells were exposed to BMP-7 (50 ng/ml) for 1, 3 and 5 days. OCN mRNA levels were elevated within 1 day in BMP-7 group, and increased with time. Runx2 mRNA levels showed minimal changes within 3 days; however, a marked increase was noted on the 5th day.
-
Effects of BMP-7 on DF cells gene expression: time response
The time response of DF cells to BMP-7 was investigated. DF cells were exposed to BMP-7 (50 ng/ml) for 1, 3 and 5 days, followed by RNA extraction and real-time RT-PCR. As shown in Fig. 20, OCN mRNA levels were enhanced in cells exposed to BMP-7 at all the time points investigated. Regarding Runx2 transcripts, BMP-7 showed no obvious effect within 3 days; however, a significant increase was noted within time.
Discussion
The characterization of PDL cells and DF progenitor cells
The results reported herein were consistent with previous studies, which have shown that PDL and DF stem cells could differentiate into osteoblasts when placed in an osteogenic induction medium and stained with Alizarin Red and von Kossa [2, 24].
Flow cytometry is practical and reliable way for cell sorting. However, no single antigenic marker has been considered to be specific for MSCs and, therefore, combinations of antibodies must be utilized. In this study, surface marker expression of PDL cells and DF progenitor cells was extensively investigated via FACS method. According to the previous reports, MSCs display similar phenotypes based on their surface marker expression, but variations exist among populations due to different origins of MSCs. For example, whereas CD49d was present in adipose-derived stem cells (ASCs), the same marker was absent in bone marrow stromal stem cells (BMSSCs) [25]. Similarly, Zuk et al. [26] reported that CD106 was absent in ASCs, but present in BMSSCs. Our report demonstrated a spectrum of surface marker expression data, and facilitated a comparison of surface marker expression profiles between different stem cell sources. To the best of our knowledge, this is the first report concerning CD73 and CD14 as progenitor cells surface antigen of PDL and DF cells.
In the current study, 4 or 5 passages were used for all the tests, thus subculture promotes the shift from PDLSCs to a mature osteoblast phenotype which is displayed by decrease of proliferative activity paralleled by an increase in ALP-specific activity and OCN secretion [27].
Gene expressions during differentiation induction
A number of previous studies documented that PDL cells expressed a series of osteoblastic or cementoblastic-related genes such as Runt-related transcription factor-2 (Runx2), Col I [28], osteopontin (OPN) [29], ALP [30], OCN [31], periostin [32] and genes related to bone metabolism regulation such as receptor activator of NF-kappa B ligand (RANKL) and osteoprotegerin (OPG) [33]. The variety of the gene expression profile is most likely due to the heterogeneity of PDL tissue cells. In the current study, in agreement with previous reports, it has been demonstrated that PDL cells express ALP (data not shown), OCN and Runx2. The expression of OPN and OCN was significantly elevated. Osteorix was weakly expressed, while Runx2 was unaffected. It is well known that OCN is a downstream target gene of Runx2 and we have found that OCN mRNA level was elevated independent of Runx2. This finding suggested that other transcription factors are involved in the differentiation of PDL cells toward osteoblasts lineage. Some investigators have proclaimed that phosphorylation of Runx2 might play a role in OCN level. This assumption was supported by the studies of Xiao et al. [33] in which they have demonstrated that the phosphorylation of Runx2 enhanced OCN activity in preosteoblast MC3T3-E1 cells, mediated by MAPK pathway. In the current study, DF progenitor cells displayed similar gene expression pattern: expression of ALP (data not shown), OCN and Runx2, trace expression of OPN and Osteorix. OPN and OCN transcript levels were significantly elevated, and Runx2 was unaffected following exposure to the osteogenic medium. Osteorix was hardly expressed in both control and the osteogenic induction group. These data were in agreement with the results of the study performed by Morsczeck et al. [34].
It is well known that BMP-7 is an important inducer of adipocyte differentiation [35]. In the present study, both PDL cells and DF progenitor cells showed no signs of adipocyte markers, such as LPL and PPARG. When incubated with adipogenic elements, PDL cells and DF progenitor cells developed into oil red O-positive fat drops, which correlated with a significant up-regulation in the expression of two adipocyte-specific transcripts, LPL and PPARG. As indicated by increased expression of adipogenic and lipogenic genes (PPARG and LPL), BMP-7 seemed to induce PDL and DF cells to differentiate into the adipo-lineage compared to the control group.
Horiuchi et al. [32] have stated that cementum-derived protein (CP23) is expressed in PDL cells and cementoblasts, but not in osteoblasts. In addition, it has been shown that PDLSCs express periostin and scleraxis mRNA, in contrast to BMSSCs [36]. In the present study, PDL cells and DF progenitor cells expressed unique genes other than BMSSCs or osteoblasts, for example, CP23.
Considering the previous studies reporting that the prolonged passaging during the course of clone selection (over 35 days) may result in loss of differentiation commitment [37]. PDL cells and DF cells were utilized at the 4th passage and cells were harvested on the 14th day for osteogenic induction and on the 21st day for adipogenic induction for Real-time RT-PCR analysis. A study performed by Vacanti et al. [38] also showed that the prolonged passaging would enhance the adipogenic potential of long-term MSC cultures at the expense of the osteochondrogenic potential.
Analysis of surface epitopes by FACS
Five different fluorescein-labeled antibodies were used to detect the surface antigens of the cells, which enabled the cross reference possible.
FACS analysis of PDL cells demonstrated that the percentage of positivity for CD14, CD19, CD34, CD45 and HLA-DR (hematopoietic stem cell markers) was lower than 2 %, which suggests that the PDL cells were not contaminated by hematopoietic and angiogenic lineages.
The percentage of positive PDL cells isolated in the current study was higher than 98 %. This result was consistent with previous studies [39–41]. In the present study, CD105 (adhesion molecule) was detected positive in 98 % of PDL cells, which was significantly higher than the previous study performed by Nagatomo [42]. This might be attributed to the declined CD 105 expression during osteogenic differentiation process [43].
According to previous reports, PDLSCs exhibited lower cell surface expression of both STRO-3 [44] and CD106 [45] than BMSSCs. Such observations might be attributed to the differences in the potential and origin of cell type.
CD73 is a cell adhesion molecule, expressed on BMSSCs [46]. To the best of our knowledge, the expression of CD73 on the PDL cells is reported for the first time herein.
In the present study, single colony selection and limited dilution method to purify PDL cells and DF progenitor cell was utilized, which has proved to be a practical and reliable method, at least according to the FACS results. We ran a FACS test of PDL cells at the 3rd passage and observed that the proportion of CD105 positive was only 25.6–30.9 % (data not shown).
Effect of BMP-7 on PDL cells and DF progenitor cells
Recent studies have showed that dental follicle compartments exhibit similar properties to mesenchymal progenitor cells which are present in adult PDL, suggesting that PDLSCs may be derived from dental follicles [47, 48]. Existing data also indicate that BMPs increase Runx2 (Cbfa1) transcript level and subsequently BSP and OCN expression [49]. Owing to that, the current study also examined Runx2 and OCN mRNA expression and showed that BMP-7 significantly promoted both PDL cells and DF progenitor cells to differentiate towards an osteoblast/cementoblast lineage as measured by increased expression of Runx2 and OCN mRNA transcripts in a time- and dose-dependent manner. More importantly, elevation of Runx2 mRNA paralleled BMP-7-induced/enhanced expression of OCN mRNA and indicated BMP-7-mediated increase in Runx2 expression, which might be partially responsible for the differentiation of PDL cells and DF progenitor cells. As mentioned before, Runx2 mRNA transcription level remained unchanged during osteogenic induction, but its expression level significantly increased following administration of BMP-7.
Regarding the signaling mechanisms responsible for BMPs to promote differentiation of PDLSCs and/or DF progenitor cells, MAPK pathway was considered to play a pivotal role [50]. Moreover, Zhao et al. [50] also postulated that collagen–integrin interactions are important for BMP-mediated cell differentiation, which was consistent with some previous studies.
It is well defined that BMPs active their targets by binding and bringing together a class of transmembrane serine/threonine kinase receptors, types IA, IB, and type II [51]. In addition, Zhao et al. [50] spotted that BMPR (BMP receptors) IA and II were present on DF cells. BMPRs-Ib and II were also detected in mouse DF and PDL cells [52].
According to the literature, PDL cells exposed to BMP-2 did not express an osteoblast/cementoblast phonotype [50, 53]. However, current study indicated that compared to DF progenitor cells, PDLSCs exhibited a slightly higher response to BMP-7. This apparent discrepancy might be attributed to the different composition of PDL cells selected for the experiments. It was very likely that they selected a population of PDL cells that had more fibroblastic versus osteoblastic characteristics and the cells used in the current study contained a large proportion of stem cell or progenitor cells, as evaluated by FACS results.
Conclusion
An increasing attention was given to stem cell-based tissue engineering in the management of periodontal diseases. PDLSCs and DF progenitor cells are promising seeded cells, with the ability to differentiate into all three types cells of periodontium. According to the results provided herein, it can be concluded that BMP-7 triggers PDL cells and DF progenitor cells to differentiate towards an osteoblast/cementoblast phenotype.
References
Alhadlaq A, Mao JJ. Mesenchymal stem cells: isolation and therapeutics. Stem Cells Dev. 2004;13:436–48.
Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–55.
Zhang W, Walboomers XF, Shi S, Fan M, Jansen JA. Multilineage differentiation potential of stem cells derived from human dental pulp after cryopreservation. Tissue Eng. 2006;12:2813–23.
Yao S, Pan F, Prpic V, Wise GE. Differentiation of stem cells in the dental follicle. J Dent Res. 2008;87:767–71.
Shi S, Bartold PM, Miura M, Seo BM, Robey PG, Gronthos S. The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod Craniofac Res. 2005;8:191–9.
Lekic P, Rojas J, Birek C, Tenenbaum H, McCulloch CA. Phenotypic comparison of periodontal ligament cells in vivo and in vitro. J Periodontal Res. 2001;36:71–9.
Arora PD, McCulloch CA. Dependence of collagen remodelling on alpha-smooth muscle actin expression by fibroblasts. J Cell Physiol. 1994;159:161–75.
Coura GS, Garcez RC, de Aguiar CB, Alvarez-Silva M, Magini RS, Trentin AG. Human periodontal ligament: a niche of neural crest stem cells. J Periodontal Res. 2008;43:531–6.
Grimm WD, Dannan A, Becher S, Gassmann G, Arnold W, Varga G, Dittmar T. The ability of human periodontium-derived stem cells to regenerate periodontal tissues: a preliminary in vivo investigation. Int J Periodontics Restorative Dent. 2011;31:e94–101.
Han J, Menicanin D, Marino V, Ge S, Mrozik K, Gronthos S, Bartold PM. Assessment of the regenerative potential of allogeneic periodontal ligament stem cells in a rodent periodontal defect model. J Periodontal Res. 2014;49:333–45.
Tour G, Wendel M, Moll G, Tcacencu I. Bone repair using periodontal ligament progenitor cell-seeded constructs. J Dent Res. 2012;91:789–94.
Bosshardt DD, Schroeder HE. Cementogenesis reviewed: a comparison between human premolars and rodent molars. Anat Rec. 1996;245:267–92.
Thesleff I, Mikkola M. The role of growth factors in tooth development. Int Rev Cytol. 2002;217:93–135.
Diekwisch TG. The developmental biology of cementum. Int J Dev Biol. 2001;45:695–706.
Hakki SS, Berry JE, Somerman MJ. The effect of enamel matrix protein derivative on follicle cells in vitro. J Periodontol. 2001;72:679–87.
Handa K, Saito M, Tsunoda A, Yamauchi M, Hattori S, Sato S, Toyoda M, Teranaka T, Narayanan AS. Progenitor cells from dental follicle are able to form cementum matrix in vivo. Connect Tissue Res. 2002;43:406–8.
Morsczeck C, Gotz W, Schierholz J, Zeilhofer F, Kuhn U, Mohl C, Sippel C, Hoffmann KH. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24:155–65.
Luan X, Ito Y, Dangaria S, Diekwisch TG. Dental follicle progenitor cell heterogeneity in the developing mouse periodontium. Stem Cells Dev. 2006;15:595–608.
Reddi AH. BMPs: from bone morphogenetic proteins to body morphogenetic proteins. Cytokine Growth Factor Rev. 2005;16:249–50.
Ramoshebi LN, Ripamonti U. Osteogenic protein-1, a bone morphogenetic protein, induces angiogenesis in the chick chorioallantoic membrane and synergizes with basic fibroblast growth factor and transforming growth factor-beta1. Anat Rec. 2000;259:97–107.
Ripamonti U. Soluble osteogenic molecular signals and the induction of bone formation. Biomaterials. 2006;27:807–22.
Taba M Jr, Jin Q, Sugai JV, Giannobile WV. Current concepts in periodontal bioengineering. Orthod Craniofac Res. 2005;8:292–302.
Ripamonti U. Recapitulating development: a template for periodontal tissue engineering. Tissue Eng. 2007;13:51–71.
Saito M, Handa K, Kiyono T, Hattori S, Yokoi T, Tsubakimoto T, Harada H, Noguchi T, Toyoda M, Sato S, Teranaka T. Immortalization of cementoblast progenitor cells with Bmi-1 and TERT. J Bone Miner Res. 2005;20:50–7.
Strem BM, Hicok KC, Zhu M, Wulur I, Alfonso Z, Schreiber RE, Fraser JK, Hedrick MH. Multipotential differentiation of adipose tissue-derived stem cells. Keio J Med. 2005;54:132–41.
Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, Alfonso ZC, Fraser JK, Benhaim P, Hedrick MH. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13(12):4279–95.
Lossdorfer S, Fiekens D, Salik MI, Gotz W, Jager A. Subculture affects the phenotypic expression of human periodontal ligament cells and their response to fibroblast growth factor-2 and bone morphogenetic protein-7 in vitro. J Periodontal Res. 2008;43:563–9.
Lukinmaa PL, Waltimo J. Immunohistochemical localization of types I, V, and VI collagen in human permanent teeth and periodontal ligament. J Dent Res. 1992;71:391–7.
Lekic P, McCulloch CA. Periodontal ligament cell population: the central role of fibroblasts in creating a unique tissue. Anat Rec. 1996;245:327–41.
Yamashita Y, Sato M, Noguchi T. Alkaline phosphatase in the periodontal ligament of the rabbit and macaque monkey. Arch Oral Biol. 1987;32:677–8.
Li H, Bartold PM, Young WG, Xiao Y, Waters MJ. Growth hormone induces bone morphogenetic proteins and bone-related proteins in the developing rat periodontium. J Bone Miner Res. 2001;16:1068–76.
Horiuchi K, Amizuka N, Takeshita S, Takamatsu H, Katsuura M, Ozawa H, Toyama Y, Bonewald LF, Kudo A. Identification and characterization of a novel protein, periostin, with restricted expression to periosteum and periodontal ligament and increased expression by transforming growth factor beta. J Bone Miner Res. 1999;14:1239–49.
Xiao G, Jiang D, Thomas P, Benson MD, Guan K, Karsenty G, Franceschi RT. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J Biol Chem. 2000;275:4453–9.
Morsczeck C. Gene expression of runx2, Osterix, c-fos, DLX-3, DLX-5, and MSX-2 in dental follicle cells during osteogenic differentiation in vitro. Calcif Tissue Int. 2006;78:98–102.
Wei X, Li G, Yang X, Ba K, Fu Y, Fu N, Cai X, Li G, Chen Q, Wang M, Lin Y. Effects of bone morphogenetic protein-4 (BMP-4) on adipocyte differentiation from mouse adipose-derived stem cells. Cell Prolif. 2013;46:416–24.
Tomokiyo A, Maeda H, Fujii S, Wada N, Shima K, Akamine A. Development of a multipotent clonal human periodontal ligament cell line. Differentiation. 2008;76:337–47.
Bonab MM, Alimoghaddam K, Talebian F, Ghaffari SH, Ghavamzadeh A, Nikbin B. Aging of mesenchymal stem cell in vitro. BMC Cell Biol. 2006;7:14.
Vacanti V, Kong E, Suzuki G, Sato K, Canty JM, Lee T. Phenotypic changes of adult porcine mesenchymal stem cells induced by prolonged passaging in culture. J Cell Physiol. 2005;205:194–201.
Sakaguchi Y, Sekiya I, Yagishita K, Ichinose S, Shinomiya K, Muneta T. Suspended cells from trabecular bone by collagenase digestion become virtually identical to mesenchymal stem cells obtained from marrow aspirates. Blood. 2004;104:2728–35.
Ikeda E, Hirose M, Kotobuki N, Shimaoka H, Tadokoro M, Maeda M, Hayashi Y, Kirita T, Ohgushi H. Osteogenic differentiation of human dental papilla mesenchymal cells. Biochem Biophys Res Commun. 2006;342:1257–62.
Lindroos B, Maenpaa K, Ylikomi T, Oja H, Suuronen R, Miettinen S. Characterisation of human dental stem cells and buccal mucosa fibroblasts. Biochem Biophys Res Commun. 2008;368:329–35.
Nagatomo K, Komaki M, Sekiya I, Sakaguchi Y, Noguchi K, Oda S, Muneta T, Ishikawa I. Stem cell properties of human periodontal ligament cells. J Periodontal Res. 2006;41:303–10.
Bruder SP, Horowitz MC, Mosca JD, Haynesworth SE. Monoclonal antibodies reactive with human osteogenic cell surface antigens. Bone. 1997;21:225–35.
Gronthos S, Fitter S, Diamond P, Robey PG, Shi S. A novel monoclonal antibody (STRO-3) identifies an isoform of tissue nonspecific alkaline phosphatase expressed by multipotent bone marrow stromal stem cells. Stem Cells Dev. 2007;16:953–63.
Simmons PJ, Masinovsky B, Longenecker BM, Berenson R, Torok-Storb B, Gallatin WM. Vascular cell adhesion molecule-1 expressed by bone marrow stromal cells mediates the binding of hematopoietic progenitor cells. Blood. 1992;80:388–95.
Barry F, Boynton R, Murphy M, Haynesworth S, Zaia J. The SH-3 and SH-4 antibodies recognize distinct epitopes on CD73 from human mesenchymal stem cells. Biochem Biophys Res Commun. 2001;289:519–24.
Kemoun P, Laurencin-Dalicieux S, Rue J, Vaysse F, Romeas A, Arzate H, Conte-Auriol F, Farges JC, Salles JP, Brunel G. Localization of STRO-1, BMP-2/-3/-7, BMP receptors and phosphorylated Smad-1 during the formation of mouse periodontium. Tissue Cell. 2007;39:257–66.
Kemoun P, Laurencin-Dalicieux S, Rue J, Farges JC, Gennero I, Conte-Auriol F, Briand-Mesange F, Gadelorge M, Arzate H, Narayanan AS, Brunel G, Salles JP. Human dental follicle cells acquire cementoblast features under stimulation by BMP-2/-7 and enamel matrix derivatives (EMD) in vitro. Cell Tissue Res. 2007;329:283–94.
Lee MH, Javed A, Kim HJ, Shin HI, Gutierrez S, Choi JY, Rosen V, Stein JL, van Wijnen AJ, Stein GS, Lian JB, Ryoo HM. Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor beta1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J Cell Biochem. 1999;73:114–25.
Zhao M, Xiao G, Berry JE, Franceschi RT, Reddi A, Somerman MJ. Bone morphogenetic protein 2 induces dental follicle cells to differentiate toward a cementoblast/osteoblast phenotype. J Bone Miner Res. 2002;17:1441–51.
Chen D, Zhao M, Mundy GR. Bone morphogeneticproteins. Growth Factors. 2004;22:233–41.
Kemoun P, Laurencin-Dalicieux S, Rue J, Vaysse F, Romeas A, Arzate H. Localization of STRO-1, BMP-2/-3/-7, BMP receptors and phosphorylated Smad-1 during the formation of mouse periodontium. Tissue Cell. 2007;39:257–66.
Song DS, Park JC, Jung IH, Choi SH, Cho KS, Kim CK, Kim CS. Enhanced adipogenic differentiation and reduced collagen synthesis induced by human periodontal ligament stem cells might underlie the negative effect of recombinant human bone morphogenetic protein-2 on periodontal regeneration. J Periodontal Res. 2011;46:193–203.
Acknowledgments
The authors would like to thank Prof. Dr. Cengiz Han Acikel from Department of Biostatistics at Gülhane Military Medical Academy for his special effort in conducting the statistical analysis of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Açil, Y., Yang, F., Gulses, A. et al. Isolation, characterization and investigation of differentiation potential of human periodontal ligament cells and dental follicle progenitor cells and their response to BMP-7 in vitro. Odontology 104, 123–135 (2016). https://doi.org/10.1007/s10266-015-0198-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10266-015-0198-1