Abstract
Background
Mixed cell populations from oral tissues may be superior to pure stem cells for regenerative approaches. Therefore, the aim of the present study was to explore the osteogenic potential of mixed cells derived from oral connective tissues compared to alveolar osteoblasts.
Materials and methods
Primary cells were isolated from the alveolar bone, periodontal ligament and gingiva. Following characterization by colony formation, growth capacity and flow cytometry, all cells were subjected to osteogenic differentiation induction and screened for a large panel of osteogenic markers using western blots, qPCR arrays, and matrix mineralization and alkaline phosphatase quantification.
Results
Non-induced mixed cells from gingiva showed higher colony formation efficiency but decreased proliferation compared to non-induced periodontal mixed cells, while both entities revealed similar surface markers tested in this setup. Following osteogenic induction, all cell populations individually expressed receptors with distinctively activated downstream effectors. Gene expression of induced periodontal mixed cells was similar to alveolar osteoblasts, but was differently modulated in gingival mixed cells. The latter failed to achieve osteogenic differentiation in terms of matrix mineralization and alkaline phosphatase activity, which was well observed in periodontal mixed cells and osteoblasts.
Conclusion
Mixed cells from periodontal ligament but not from gingiva feature an inherent osteogenic capacity in vitro. From these results, it can be concluded that periodontal cells do not require further stem cell enrichment in order to qualify for bone regeneration.
Clinical relevance
Our data contribute to the development of novel cell-based therapies using mixed cells from the periodontal ligament in regenerative periodontics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, stem cell-based therapies are entering the clinical trial phase [1] and for their successful implementation, it is highly important to unravel which cells may contribute to tissue regeneration. More than a decade ago, mesenchymal stem cells were described to reside in different oral tissues [2, 3], and today, for periodontal regeneration, stem cells from oral connective tissues are considered superior to mesenchymal stem cells from other sources [4,5,6]. Despite the undeniable body of evidence that oral connective tissues, i.e. periodontal ligament or gingiva, hold progenitor cells, precise isolation and characterization protocols are missing so far [7], and so-called stem cells often are isolated by mere explant culture and/or enzymatic digestion [8]. However, stem or progenitor cell isolation necessarily requires further enrichment by selecting colony-forming or specific epitope-expressing cells [9,10,11]. Without enrichment, the resulting mixed population has been shown to be predominantly comprised of fibroblasts, while containing less than 1% of stem cells [12]. As a result, a great deal of scientific papers may have used rather mixed cell populations instead of real ‘stem cells’ [13]. Nevertheless, it may be feasible to use mixed cell populations for regeneration therapies [13] because the heterogenicity of differently purified stem cells may impair their therapeutic efficacy and introduce broad variations in the clinical outcome of regenerative approaches [14]. However, mixed cell populations hold the advantage of delivering stem cells together with surrounding non-progenitor cells, which are known to govern the stem cell behaviour [15, 16]. In addition, mixed cells from the periodontal ligament and the gingiva were already found to be capable of a gradual osteogenic differentiation if bone marrow-derived mesenchymal stem cells were present [17]. Moreover, there is evidence that oral fibroblasts even display an osteogenic potential on their own [18,19,20], emphasizing their usefulness for bone generation in periodontal regeneration. In consequence, transplantation of mixed cells instead of pure stem cells could possibly enhance the therapeutic outcome of regenerative therapies. However, the osteogenic differentiation potential of mixed cell populations from distinct oral tissues is yet to be determined. Hence, the aim of the present study was to investigate the hard tissue-forming capacity of explant culture-derived mixed cells from oral connective tissues and compare them to bone cells. In this context, we hypothesized that mixed cells from the gingiva and periodontal ligament exhibit osteogenic receptors, signalling molecules, genes, matrix calcification and alkaline phosphatase activity comparable to alveolar osteoblasts in response to appropriate biochemical stimulation.
Materials and methods
Cell isolation and culture
All experiments were carried out in accordance with the guidelines of the World Medical Association Declaration of Helsinki and were approved by the Committee of Ethics of the Medical Faculty of the Albert Ludwigs University Freiburg, Germany (vote number 516/12). All cells were derived from operative biopsies (n = 12 donors) with informed and written consent. In brief, alveolar osteoblasts were derived from alveolar bone biopsies of patients undergoing corrective osteotomy surgery, and only compact bone parts were used. Periodontal mixed cells were obtained from the ligament tissues of non-carious human premolar or wisdom teeth with healthy periodontium extracted for orthodontic reasons, and the periodontal ligament was scraped off from the middle third of the tooth roots. Gingival mixed cells were derived from healthy gingival connective tissue of donors undergoing gingivoplastic surgery, and tissue fragments were carefully dissected from gingival epithelium. All tissue specimens were rinsed with 10% iodine, thoroughly washed and minced. Tissue fragments were plated as explants in MEMalpha (Lifetechnologies, Darmstadt, Germany) supplemented with 10% foetal bovine serum (Biochrom, Berlin, Germany), 1% glutamax (Life technologies), 1% amphotericin and 1% antibiotics, which was exchanged every 2–3 days until cell outgrowth. Cells were expanded by splitting (osteoblasts passaged (p)4–6 times, periodontal mixed cells p2–4 and gingival mixed cells p3–5), before osteogenic differentiation was induced for 21 days with NH OsteoDiff medium (Miltenyi, Bergisch Gladbach, Germany). All experiments were systematically carried out in replicates for the cells from all donors, except for western blot analyses, which were performed in triplicate by using the cells from two donors each for alveolar osteoblasts and periodontal and gingival mixed cells.
Colony-forming units and growth curves
For colony-forming unit (CFU) assays, cells were seeded in limiting dilution, i.e. 2 cells/cm2, incubated for 14 days, fixed with 4% formaldehyde and stained with 0.1% Azur II (Sigma Aldrich, Taufkirchen, Germany) for 20 min at room temperature, air-dried and photographed with a SZH10 microscope (Olympus, Münster, Germany) equipped with a CCD Colour view III camera and the resulting images were taken using the Cell* software (both Olympus, Münster, Germany). Images were analyzed using the ImageJ plugin ‘analyze particles’ following threshold adjustment. Stained colonies of ≥ 50 cell were scored as CFU-F and counted. CFU-F efficiency was calculated as follows: CFU-F efficiency = (counted CFU-F/cells originally seeded) × 100. For growth curves, cells were seeded at 5 × 103 cells/well in a 24-well plate (duplicates per time point and donor), and incubated in growth medium for 1, 3, 5, 7, 10 and 14 days. Growth medium was replaced with medium supplemented with 1:10 Alamar blue®-dye (Thermo Fisher Scientific, Schwerte, Germany). Cells were incubated for 3 h, supernatant was centrifuged, absorbance was measured in triplicate in an Ensight Multimode Plate Reader (PerkinElmer, Rodgau, Germany), and data were collected with the Kaleido 2.0 software program.
Flow cytometry
For each run, a total of 1 × 106 of non-induced cells were incubated with a final concentration of 5 μg/ml each of fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, peridinin chlorophyll protein complex (PerCP)- or allophycocyanin (APC)-conjugated mouse anti-human CD10, CD13, CD14, CD19, CD34, CD44, CD45, CD73, CD90, CD105, CD146, CD166, HLA-DR, c-kit, Stro1 and alkaline phosphatase antibodies (BD Biosciences, Heidelberg, Germany) or isotype-matched controls, respectively (20 min, 4 °C in the dark). After intense washing and resuspension in phosphate-buffered saline (PBS), flow cytometric analysis was performed using a FACSCalibur (BD Biosciences) flow cytometer. Data were collected and analyzed with CellQuest software (BD Biosciences). For each run, 20.000 cell events were gated and fluorochrome spectral overlap was checked and compensated whenever required.
Western blot
Cells were lysed using the MemPer Plus kit with Halt® protease and phosphatase inhibitor (both Thermo Scientific) according to manufacturer’s instructions and quantified using a modified Lowry assay (BCA; Thermo Scientific). Lysates were mixed with Laemmli/DTT buffer, heated and separated on Criterion TGX precast gels (Bio-Rad). After electrophoresis, proteins were transferred to a nitrocellulose membrane using the Transblot Turbo Transfer system (Bio-Rad). The membranes were blocked with 5% bovine serum albumine for 2 h at least, and proteins were detected with appropriate primary antibodies and visualized with the corresponding secondary antibodies using the ECL kit (Thermo Scientific). The antibodies used were as follows: ms SMAD1/2/3, ms TGFBR2, rb EGFR, ms IGF1R antibody (all Santa Cruz Biotechnology, Heidelberg, Germany), rb BMPR1 and rb TGFBR1 antibody (both Abcam, Cambridge, UK), rb phospho-SMAD2 (Ser465/467)/SMAD3 (Ser423/425), rb phospho-SMAD1 (Ser463/465)/SMAD5 (Ser463/465)/SMAD9 (Ser465/467), rb BMPR2, rb p38, rb phospho-p38 (Thr180/Tyr182), ms MEK1, rb phospho-MEK1/2 (Ser217/221), rb ERK1/2 and rb phospho-ERK1/2 (Thr202/Tyr204) antibody (all New England Biolabs, Frankfurt, Germany). Data were collected with the ChemiDoc XRS+ system and analzyed using the ImageLab v6.0 sofware (both Bio-Rad).
qPCR arrays
Gene expression was monitored using the RT2 Profiler PCR Array technology (QIAGEN, Hilden, Germany). Total cellular RNA was purified using a guanidium–thiocyanate method (RNeasy Mini kit; QIAGEN, Hilden, Germany) and stored at − 80 °C. The RNA integrity and quantity were verified using the Experion RNA StdSens chip microfluidic technology according to manufacturer’s instructions (Bio-Rad, Munich, Germany), and cDNA was synthesized from 200 ng of total RNA each by using the RT2 PreAmp cDNA synthesis kit (QIAGEN, Hilden, Germany) in a C1000 Thermal Cycler (Bio-Rad). After genomic DNA elimination and reverse transcription, one quarter of the resulting cDNA was pre-amplified using the RT2 PreAmp Pathway Primer Mix (QIAGEN). qPCR was performed with the resulting pre-amplicons in duplicate each in a CFX96 cycler (Bio-Rad, Munich, Germany), according to manufacturer’s instructions. Genomic DNA contamination, reverse transcription performance and purity, and qPCR efficiency were routinely checked. The amplicons’ specificity was checked by melting curve examination. Data were collected with CFX96 Manager Software version 1.0 (Bio-Rad), and the respective genes of interest were normalized to a housekeeping gene pool comprising ACTB, RPL0 and HPRT. Data were analyzed and plotted using the RT2 Profiler PCR array data analysis template (https://www.qiagen.com/de/shop/genes-and-pathways/data-analysis-center-overview-page/).
Mineralization assay
Cells were fixed with ethanol, washed and incubated with 40 mM Alizarin red solution (Sigma Aldrich, pH 4.1) for 20 min at RT. After thorough washing, the air-dried specimens were evaluated with a SZH10 microscope (Olympus, Münster, Germany) equipped with a CCD Colour view III camera and the resulting images were taken and analyzed using the Cell* software (both Olympus, Münster, Germany). For quantification, Alizarin red was extracted with 10% acetic acid (30 min, RT). The solution was covered with mineral oil (Sigma Aldrich) and incubated for 10 min at 85 °C. The absorbance of the supernatant was read at 420 nm in triplicate, and data were collected and analyzed using the Magellan v6.2 software (Tecan, Crailsheim, Austria).
Alkaline phosphatase assay
Alkaline phosphatase (ALP) enzymatic activity was quantified using p-nitrophenol phosphate (pNPP) as substrate. Cellular proteins were purified using complete Lysis-M buffer containing protease inhibitor (Roche, Mannheim, Germany) and quantified using a modified Bradford method (Pierce 660 assay; Thermo Scientific). ALP activity was measured in triplicate for each donor and culture condition. In brief, proteins (5 μg) were mixed with ALP buffer containing 5 mM Mg acetate and 10 mM pNPP, and absorbance at OD = 405 nm was read continuously for 30 min in a microplate reader (Tecan, Crailsheim, Germany). Data were calculated by [(ODtx − ODt0) × 103 × reaction volume]/(tx × ε × λ × sample volume) with ε = 18.75 mM−1 cm−1 and λ = (ODcal − ODH2O)/ε × c.
Statistics
A repeated measures analysis was performed with a linear mixed model for each outcome of interest (growth, normalized protein expression, alkaline phosphatase activity, mineralization). The group effects and differences of least square (ls) means were calculated with their 95% confidence intervals. Several multiple comparisons of ls means in groups were performed and p values were adjusted for multiple testing by the method of Tukey-Kramer. Calculations were done using PROC MIXED from the statistical software program SAS 9.1.2 (Cary, NC 27513, USA).
Results
Characterization of non-induced mixed cells reveals low numbers of stem/progenitor cells
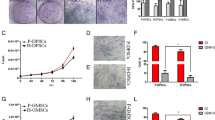
Alveolar osteoblasts and mixed cells from the periodontal ligament and from gingiva were randomly distributed in the culture plates, while no colony formation was observable (Fig. 1a). This observation was substantiated by CFU assays, which revealed a CFU-F efficiency of 0.5 ± 1.0% for alveolar osteoblasts and periodontal mixed cells, while the efficiency was increased in gingival mixed cells (7.0 ± 5.8%; Fig. 1a). All cell populations showed a significant increase in metabolic activity with increased culture duration (p < 0.001 for 3, 5, 7, 10 and 14 days compared to 1 day in all entities (Fig. 1b). However, metabolic activity gain was significantly different for all groups and all time points, with gingival mixed cells showing the highest initial gain (p < 0.001 compared to periodontal mixed cells and alveolar osteoblasts) and periodontal mixed cells showing the highest final gain (p < 0.001 compared to gingival mixed cells and alveolar osteoblasts; Fig. 1b). Further, the time-related proliferation increase, i.e., metabolic activity values normalized to metabolic activity of 1 day, was significantly higher in gingival mixed cells at 3d compared to osteoblasts (p = 0.037), while subsequently, both periodontal and gingival mixed cells proliferated significantly more intensely than osteoblasts (5 days: p < 0.001 periodontal mixed cells vs. osteoblasts, p = 0.003 gingival mixed cells vs. osteoblasts; 7 days: p = 0.02 periodontal mixed cells vs. osteoblasts, p = 0.001 gingival mixed cells vs. osteoblasts; 10 days: p < 0.001 both periodontal and gingival mixed cells vs. osteoblasts). Additionally at 10 days, periodontal mixed cells had a significantly elevated metabolic activity increase compared to gingival mixed cells (p = 0.014), while the increase at 14 days was significant only in periodontal mixed cells compared to osteoblasts (p = 0.003; Fig. 1b). With regard to surface marker expression, we found that non-induced alveolar osteoblasts strongly expressed CD13, CD44, CD73, CD90, CD105 and CD166, while the expression of CD146 and membrane-bound ALP was low (Fig. 2a, Table 1). Further, the expression of CD14, CD19, CD34, CD45, HLA-DR, c-kit and Stro1 was virtually absent, and the expression of CD10 was moderate in osteoblasts (Fig. 2a, Table 1). In comparison, non-induced periodontal and gingival mixed cells showed similar surface epitopes and highly expressed CD13, CD44, CD73, CD90, CD105 and CD166, respectively (Fig. 2b, c, Table 1). Similar to osteoblasts, periodontal and gingival mixed cells negatively expressed CD14, CD19, CD34, CD45 and HLA-DR, but both showed an intense expression of the fibroblast marker CD10 (Fig. 2b, c, Table 1). The percentage of cells with membrane-bound ALP was very low in periodontal (Fig. 2b, Table 1) and gingival mixed cells (Fig. 2c, Table 1). Regarding putative stem cell markers, periodontal mixed cells expressed intermediate levels of CD146, while the expression of Stro1 and c-kit was very low (Fig. 2b, Table 1). Of note, only 0.06 ± 0.03% of periodontal mixed cells were double-positive for Stro1 and CD146. In comparison, gingival mixed cells expressed only low levels of CD146, and the percentage of Stro1- and c-kit-positive cells was close to zero (Fig. 2c, Table 1). These data suggest that mixed cell populations from periodontal ligament and gingiva mainly comprise fibroblasts together with very low numbers of stem/progenitor cells.
Isolation, colony formation and growth of human alveolar osteoblasts and mixed cells from periodontal ligament and gingiva. a After removal of tissue impurities like bone marrow or gingival epithelium, cells were isolated from compact alveolar bone, periodontal ligament, or gingival connective tissue, respectively. All cells appeared equally distributed throughout the petri dish (scale bar = 50 μm), and virtually no colonies were visible for osteoblasts and periodontal mixed cells, while single colonies emerged in gingival mixed cell populations (scale bar = 5000 μm). b All cell populations showed increased metabolic activity by time, while periodontal (white triangles connected by a solid line) and gingival mixed cells (grey diamonds connected by a dashed line) proliferated significantly more than osteoblasts (black circles connected by a dotted line; blue = periodontal mixed cells vs. osteoblasts, red = gingival mixed cells vs. osteoblasts, green = periodontal vs. gingival mixed cells)
Immunophenotypic characterization of osteoblasts and mixed cells from periodontal ligament and gingiva. Flow cytometry screening of mesenchymal surface epitopes associated with hard tissue cells revealed that similar to alveolar osteoblasts (a), periodontal (b) and gingival mixed cells (c) expressed CD13, CD44, CD73, CD90, CD105 and CD166, while lacking CD14, CD19, CD34, CD45 and HLA-DR. a Alveolar osteoblasts expressed intermediate levels of CD10, low levels of CD146 and ALP, while Stro1 and c-kit-expression were absent. b The percentage of CD10-positive cells was high in periodontal mixed cells that expressed intermediate levels of CD146 with very low levels of ALP, Stro1 and c-kit. c Gingival mixed cells expressed comparably high levels of CD10, but low levels of CD146, while the expression of ALP and Stro1 was very low and c-kit was absent
Mixed periodontal and gingival cells individually express biomolecules associated with responsiveness to osteogenic induction
Western blot analyses revealed that all cell types expressed similar levels of BMPR1, which were elevated following 21 days of osteogenic induction (Fig. 3a, Table 1). While appearing generally weaker in soft tissue-derived cells, BMPR2 proteins were increased in gingival mixed cells following osteogenic induction in contrast to alveolar osteoblasts and periodontal mixed cells (Fig. 3a). The expression intensity of TGFBR1 was comparable to non-induced controls, while appearing slightly decreased in induced mixed cells from the periodontal ligament (Fig. 3a). Similarly, TGFBR2 was slightly decreased in all cell types under study following 21 days of osteogenic induction (Fig. 3a). While the expression of IGF1R was clearly stronger in non-induced osteoblasts, the opposite trend became obvious both for mixed cells from the periodontal ligament and gingiva (Fig. 3a). The expression of EGFR was strong and quite uniform for osteoblasts and gingival mixed cells irrespective of osteogenic induction, while appearing decreased in induced mixed cells from the periodontal ligament (Fig. 3a). SMAD molecules were strongly expressed in alveolar osteoblasts and gingival mixed cells, but were low in periodontal mixed cells, with no significant expression changes following osteogenic induction in all cell types (Fig. 3b). MEK1/2 was intensely expressed in alveolar osteoblasts and gingival mixed cells, while only faint bands appeared for periodontal mixed cells which remained unchanged by osteogenic induction (Fig. 3b). The expression of ERK1/2 was not affected by osteogenic induction in osteoblasts, but was obviously decreased in mixed cells from the periodontal ligament (p = 0.020) and gingiva (p = 0.003, Fig. 3b). Changes in p38 expression following osteogenic induction were found non-significant for all cell types under study (Fig. 3b). Regarding the activation status of downstream signalling effectors, we found only weak signals for phosphorylated MEK1/2, which appeared stable in induced osteoblasts as well as in induced periodontal and gingival mixed cells (Fig. 3b, Table 1). Phosphorylated ERK1/2 was robustly detectable in non-induced controls, but significantly downregulated for all cell types following osteogenic induction (Fig. 3b, p = 0.007 for alveolar osteoblasts, p = 0.005 for periodontal mixed cells and p < 0.001 for gingival mixed cells, respectively). Phosphorylated p38 was most strongly expressed in periodontal mixed cells, while it remained unchanged in induced osteoblasts. Although only low amounts of active p38 were found in non-induced gingival mixed cells, it was significantly decreased following osteogenic induction (p = 0.027). The changes in SMAD protein phosphorylation were found non-significant for all cell types; however, both active SMAD1/5/9 and active SMAD2/3 appeared decreased or constant, respectively, in alveolar osteoblasts and gingival mixed cells undergoing osteogenic differentiation, and showed an upward trend in periodontal mixed cells. To estimate the involvement of the pathways under study, ratios of active/non-active downstream signalling effectors were calculated. For all cell types, changes in phosphorylated SMAD2/3 normalized to SMAD1/2/3 and phosphorylated SMAD1/5/9 normalized to SMAD1/2/3 were non-significant (Fig. 3b). Similarly, phosphorylated MEK1/2 normalized to MEK1/2 showed an upward trend in all cell types (Fig. 3b). However, phosphorylated ERK1/2 normalized to total ERK1/2 was significantly decreased in induced osteoblasts (p = 0.003), and induced mixed cells from the periodontal ligament (p = 0.005) and gingiva (p < 0.001, Fig. 3b) following osteogenic induction. Again, phosphorylated p38 normalized to total p38 was significantly decreased only in induced gingival mixed cells (p < 0.001, Fig. 3b).
Osteogenesis-related receptor (a) and downstream signaling effector expression and activation (b) in oral cells. a Western Blot analyses showed that all cells under study expressed similar levels of BMPR1 and TGFBR2 under control conditions (−), which were simultaneously increased or decreased, respectively, if cells were cultured in osteogenic differentiation medium (+). However, differences were obvious for BMPR2, TGFBR1, EGFR and IGF1R expression. The housekeeping protein ACTB was expressed uniformly for all cell types irrespective of osteogenic induction. b Analysis of phosphorylated mitogen-activated protein kinases (bar graphs, y-axis: ratio of normalized volume intensities, x-axis: cell types and culture conditions) revealed that phosphorylated MEK1/2 was increased in induced osteoblasts while rather decreased in periodontal and gingival mixed cells. Phosphorylated ERK1/2 was decreased in all cell types following osteogenic induction, while phosphorylated p38 was differentially regulated. While phosphorylated SMAD1/5/9 and phosphorylated SMAD2/3 appeared decreased in induced alveolar osteoblasts and gingival mixed cells, both were rather increased in periodontal mixed cells following osteogenic induction (norm. = normalized to)
Gene expression profile reflects the differential osteogenicity of periodontal and gingival mixed cells
We were next interested in the expression of genes related to osteogenesis. In alveolar osteoblasts, the vast majority of genes investigated was regulated significantly both in biological and statistical terms following osteogenic induction, i.e. showing a ≥ 1.5-fold up- or downregulation, respectively, with p < 0.05 (Fig. 4a, Suppl. Table 1). Regarding osteogenic soluble factors, the gene expression of the following markers was found to be significantly upregulated in alveolar osteoblasts following osteogenic induction: BGN, BMP6 and TGFB1; while BMP4, FGF2 and TGFB3 appeared significantly decreased (Fig. 4a, Suppl. Table 1). Further, mRNA expression of some osteoblast-featured receptors was regulated in induced osteoblasts, i.e. EGFR, FLT1, IGF1R and TGFBR1 were decreased and FGFR1 together with TGFBR2 were increased (Fig. 4a, Suppl. Table 1). Transcripts of the respective downstream effectors involved in osteogenesis, i.e. SMAD1 and SMAD3, were downregulated in alveolar osteoblasts after 21 days of osteogenic induction (Fig. 4a, Suppl. Table 1). Further, the gene expression of the following transcription factors and differentiation markers associated with osteogenesis appeared significantly increased: BGLAP, RUNX2, and OSX (Fig. 4a, Suppl. Table 1).
mRNA profile of osteogenic marker genes in alveolar osteoblasts (a) and periodontal (b) and gingival mixed cells (c). Volcano plots of mRNA expression following osteogenic induction display statistical significance (y-axis) versus fold regulation change (x-axis) compared to matching controls (middle black line) with 1.5 as fold-change cutoff (vertical black lines on the left and right) and p < 0.05 as the p value cutoff (horizontal green line). a In osteoblasts, a broad panel of genes associated with osteogenesis was significantly upregulated following osteogenic induction, i.e. TGFBR2, TGFB1, BGN, BMP6, OSX, SPP1 and BGLAP, while TGFBR1, EGFR, IGF1R, FGFR2, FLT1, FGF2, IGF1, BMP4, SMAD1 and SMAD3 were significantly downregulated. b In periodontal mixed cells, similar genes associated with osteogenesis were in part significantly upregulated. c mRNA expression in gingival mixed cells resembled the osteoblast pattern in particular with respect to downregulated genes. Common for all cell types, TGFBR2 was significantly upregulated, while EGFR, IGF1R, SMAD1 and SMAD3 were significantly downregulated following osteogenic induction
Induced periodontal mixed cells showed a fairly equal gene expression pattern particularly for positively regulated genes (Fig. 4b, Suppl. Table 1). Here, even more genes encoding for osteogenic soluble factors were modulated, with BMP6 and TGFB1 being significantly upregulated, and BMP4 and FGF2 being downregulated (Fig. 4b, Suppl. Table 1). Of note, the gene expression of molecules that govern osteoclast regulation, i.e. RANKL and OPG, was meaningfully increased compared to non-induced periodontal mixed cells (+ 6.1 and + 3.9-fold, respectively) without reaching statistical significance (Suppl. Table 1). Comparable to induced alveolar osteoblasts, the gene expression of TGFBR2 was significantly upregulated, while the transcription of EGFR and IGF1R was downregulated in induced periodontal mixed cells (Fig. 4b, Table 1). Further, the gene expression of BMPR1B was significantly increased, but the gene expression of ACVR and BMPR1A was decreased (Fig. 4b, Suppl. Table 1). While no significant expression changes were found for SMAD genes, the transcription of ALP and IHH was tremendously increased, and OSX and COL1A1 genes were significantly stronger expressed upon osteogenic induction (Fig. 4b, Suppl. Table 1).
With respect to transcripts showing similarity to alveolar osteoblasts, mixed cells from gingiva displayed preference to the spectrum of downregulated genes following osteogenic induction. Except for BMP2 and CSF1 gene expression, the regulation pattern in gingival mixed cells of osteogenic soluble factors nearly matched with that observed for alveolar osteoblasts (Fig. 4c, Suppl. Table 1). Comparable to alveolar osteoblasts and periodontal mixed cells, the gene expression of TGBR2 was significantly increased while the gene expression EGFR and IGF1R was decreased in induced gingival mixed cells (Fig. 4c, Suppl. Table 1). Further, the gene expression of FLT1, ACVR and BMPR1A was decreased in induced gingival mixed cells comparable to the other cell types, whereas FGFR1 transcription was decreased only in gingival mixed cells (Fig. 4c, Suppl. Table 1). Apart from SMAD4, the expression of all other SMAD genes under study was significantly decreased in gingival mixed cells following osteogenic induction (Fig. 4c, Suppl. Table 1). Gingival mixed cells showed some distinctive features with regard to the gene expression of osteogenic transcription factors and differentiation markers, which is substantiated by a significant downregulation of NFKB1, ALP and BGLAP (Fig. 4c, Suppl. Table 1). Of note, the expression of osteogenic hallmark genes such as RUNX2, OSX and SPP1 appeared either clearly weaker or inverse in gingival mixed cells (Suppl. Table 1).
Oral cells show an individual matrix mineralization capacity
After 21 days of osteogenic induction, mineralization events were found in all cell types in a gradual manner, i.e. osteoblasts showed the most intense and uniform alizarin red stainings, followed by periodontal mixed cells with a patchy alizarin red staining and finally, by gingival mixed cells which showed the lowest staining intensity (Fig. 5)a. This observation was confirmed by alizarin red quantification, which revealed that induced alveolar osteoblasts (p < 0.001) and periodontal mixed cells (p < 0.0001) showed significant matrix mineralization compared to controls (Fig. 5b). Although a clear trend in matrix mineralization was observable for gingival mixed cells, it was not significant compared to matched controls (Fig. 5b). Consequently, intercellular comparison revealed significantly more calcium deposits in induced osteoblasts (p = 0.0002) and periodontal mixed cells (p = 0.0406) compared to gingival mixed cells, while osteoblasts and periodontal mixed cells showed fairly equal levels of matrix mineralization (Fig. 5b). With respect to intracellular alkaline phosphatase (ALP) activity, we found that control osteoblasts showed some baseline ALP activity, which was significantly increased upon osteogenic induction (p = 0.023, Fig. 5c, compare black with white dots). This significance also applied for periodontal mixed cells (p = 0.004, Fig. 5c), while gingival mixed cells showed no elevation of their ALP activity (Fig. 5c). These data suggest a differential osteogenic capacity for the cells from the alveolar bone or from periodontal ligament and gingiva, respectively, in response to osteogenic induction.
Osteogenic differentiation visualized by Alizarin red stains (a) and quantification (b) as well as alkaline phosphatase activity (c). a Osteogenic induction yielded intensely coloured Alizarin red stains (ARS) in alveolar osteoblasts, followed by periodontal mixed cells, and, to a lower extent, by gingival mixed cells. b Quantification of extracted ARS revealed that in osteoblasts and periodontal mixed cells, the increase in ARS was significant following osteogenic induction, while gingival mixed cells showed less calcium deposits (box plot graphs, y-axis: ARS level [mM], x-axis: osteogenic induction vs. controls). c Alkaline phosphatase line graphs show that following osteogenic induction (white dots representing means ± s.e.m.), the enzymatic activity was significantly increased in alveolar osteoblasts and periodontal mixed cells compared to non-induced controls (black dots), while being stable in gingival mixed cells irrespective of osteogenic induction (i, y-axis: ALP activity [U/L × μg], x-axis:time [min])
Discussion
Since mixed cells from the periodontal ligament and the gingiva prove promising for the regeneration of periodontal hard tissues, i.e. bone and cementum, they were characterized and tested for their osteogenic differentiation capacity in this study. We found that periodontal and gingival mixed cell populations comprise only low numbers of stem/progenitor cells, as substantiated by minute percentages of colony-forming and/or Stro1- and c-kit-positive cells. While mixed cells from the periodontal ligament displayed an osteogenic differentiation capacity comparable to osteoblasts, gingival mixed cells did not achieve in vitro hard tissue formation. Both mixed cell population showed some unique features: a great deal of non-induced periodontal cells positively expressed CD146, while induced gingival cells showed enhanced expression of BMP receptors, decreased levels of active p38 kinase, inverse regulation of some osteogenic marker genes and no remarkable matrix mineralization or alkaline phosphatase activity.
If dealing with mixed cells, the precise nature and composition of the cell population is unknown. The phenotypic characterization in the present study revealed an epitope spectrum that is in line with current reports [12, 21, 22], emphasizing the very low percentage of stem/progenitor cells within the mixed cell populations. Stro1 is considered the ‘gold standard’ marker for stem cells from oral tissues including dental pulp, periodontal ligament, gingiva and bone marrow [2]. Originally known as marker for stem cells with cardiomyogenic potential [23], CD117/c-kit has been used to isolate progenitors from dental pulp [24] and gingiva [25]. CD90, also known as Thy-1, is highly expressed by several cell types including fibroblasts, as well as immune and neuronal cell lines [26, 27], and in our hands by all kinds of oral tissue-derived cells, including oral fibroblasts [17] and both Stro1-positive and Stro1-negative dental pulp cells (unpublished data). CD146 is a cell adhesion molecule that labels endothelial cells and pericytes which co-express CD73, CD90 and CD105 [28, 29]. The latter maintain the hematopoietic stem cell niche and act in a pro-angiogenic manner, while exhibiting a limited multipotency [14]. In periodontal ligament cells, CD146 is accepted as a marker for stem/progenitor cell enrichment only if co-expressed with Stro1 [30, 31], and in gingiva-derived cells, CD146 expression is associated with the absence of Stro1 [32]. In this regard, the finding that high percentages are positive for CD146 suggests that mixed cells could contribute to tissue regeneration, but also implies that CD146 together with CD73, CD90 and CD105 are unspecific stem cell markers in the oral tissue context. The immunophenotyping results, together with the low colony efficiency, suggest that periodontal and gingival mixed cells are comprised of fibroblasts rather than progenitor cells.
Following osteogenic induction, the expression pattern of the receptor and downstream effector configuration of periodontal and gingival mixed cells points to a major role of non-canonical signaling pathways in oral cells [33]. Since SMAD signaling is predominant during osteogenic differentiation in long bone-derived osteoblasts [34, 35], the present findings disclose a unique feature of oral cells which may be attributed to their developmental origin as neural crest descendants [36, 37]. Given the individual downstream effector activation patterns in each cell type under study, it is not surprising that the expression of osteogenic genes and the final differentiation performance are distinctive. In the present study, gene expression was measured after 21 days, and time-dependent variations in mRNA transcription may account for some counter-regulation of the investigated biomolecules at the mRNA level compared to the corresponding protein status. Nevertheless, both protein and gene expression analysis showed disparities between gingival and periodontal mixed cells, and a main finding is that gingival mixed cells fail to accomplish osteogenic differentiation in response to biochemical induction. Of note, gingival cells were previously found to be superior to periodontal cells in terms of hard tissue formation in the presence of mesenchymal stem cells [17]. This means that gingival cells are able to adopt an osteogenic phenotype, following interaction with mesenchymal stem cells, despite their failure of hard tissue formation in response to biochemical induction. The discrepancy between the previously published observations and the present findings suggests that (i) gingival mixed cells alone may not be useful for bone and/or cementum regeneration, because (ii) bona fide cell-to-cell interactions with mesenchymal stem cells are obviously required for triggering their osteogenesis [17]. More importantly, our results show that mixed periodontal cells are in fact capable of hard tissue formation to comparable degrees as osteoblasts, which renders them an attractive cell source for bone and cementum regeneration. Periodontal mixed cells are easily available from extracted teeth, and their use holds great advantages compared to stem cells from the periodontal ligament for the following reasons: (i) no depletion of the initially small cell numbers derived from the very small periodontal ligament biopsies is required for stem cell enrichment, with the consequence that (ii) the time span of in vitro cell manipulation that precedes therapeutic administration is considerably reduced, (iii) thereby decreasing the incidence of undesired genetic and epigenetic changes [38] together with cell senescence [39].
Taken together, our work emphasizes the importance of a precise characterization of oral tissue-derived cell populations [40], and provides a methodological platform in doing so. The present study further reveals that periodontal and gingival mixed cells react individually to osteogenic induction with regard to receptors and signaling effectors equipment as well as mRNA transcription, and the degree of matrix mineralization. By delivering insights in the markers that mixed cell populations express in response to osteogenic triggers, our results provide the basis for the decision-making in future research strategies and clinical therapies, because they provide first evidence that mixed cells from the periodontal ligament are suitable for periodontal regeneration purposes. This knowledge will contribute to anticipate the clinical outcome of hard tissue-inducing strategies, and opens the road for the development of novel cell-based therapies using mixed cell populations.
References
Hu L, Liu Y and Wang S (2017) Stem cell-based tooth and periodontal regeneration. Oral Dis . doi: 10.1111/odi.12703
Egusa H, Sonoyama W, Nishimura M, Atsuta I, Akiyama K (2012) Stem cells in dentistry—part I: stem cell sources. J Prosthodont Res 56:151–165. https://doi.org/10.1016/j.jpor.2012.06.001
Volponi AA, Sharpe PT (2013) The tooth—a treasure chest of stem cells. Br Dent J 215:353–358. https://doi.org/10.1038/sj.bdj.2013.959
Hynes K, Menicanin D, Gronthos S, Bartold PM (2012) Clinical utility of stem cells for periodontal regeneration. Periodontol 59:203–227. https://doi.org/10.1111/j.1600-0757.2012.00443.x
Han J, Menicanin D, Gronthos S, Bartold PM (2014) Stem cells, tissue engineering and periodontal regeneration. Aust Dent J 59(Suppl 1):117–130. https://doi.org/10.1111/adj.12100
Tassi SA, Sergio NZ, Misawa MYO, Villar CC (2017) Efficacy of stem cells on periodontal regeneration: systematic review of pre-clinical studies. J Periodontal Res 52:793–812. https://doi.org/10.1111/jre.12455
Avinash K, Malaippan S, Dooraiswamy JN (2017) Methods of isolation and characterization of stem cells from different regions of oral cavity using markers: a systematic review. Int J Stem Cells 10:12–20. https://doi.org/10.15283/ijsc17010
Fawzy El-Sayed KM, Dorfer CE Gingival mesenchymal stem/progenitor cells: a unique tissue engineering gem. Stem Cells Int 2016, 2016:7154327–7154316. https://doi.org/10.1155/2016/7154327
Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi S (2004) Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 364:149–155. https://doi.org/10.1016/s0140-6736(04)16627-0
Shi S, Gronthos S (2003) Perivascular niche of postnatal mesenchymal stem cells in human bone marrow and dental pulp. J Bone Miner Res 18:696–704. https://doi.org/10.1359/jbmr.2003.18.4.696
Lekic P, Rojas J, Birek C, Tenenbaum H, McCulloch CA (2001) Phenotypic comparison of periodontal ligament cells in vivo and in vitro. J Periodontal Res 36:71–79
Fournier BP, Larjava H, Hakkinen L (2013) Gingiva as a source of stem cells with therapeutic potential. Stem Cells Dev 22:3157–3177. https://doi.org/10.1089/scd.2013.0015
About I (2018) "The stem cell fashion": do we need only stem cells for tissue regeneration? Clin Oral Invest 22:553–554. https://doi.org/10.1007/s00784-017-2316-7
Mo M, Wang S, Zhou Y, Li H, Wu Y (2016) Mesenchymal stem cell subpopulations: phenotype, property and therapeutic potential. Cell Mol Life Sci 73:3311–3321. https://doi.org/10.1007/s00018-016-2229-7
Jones DL, Wagers AJ (2008) No place like home: anatomy and function of the stem cell niche. Nat Rev Mol Cell Biol 9:11–21. https://doi.org/10.1038/nrm2319
Xiao L, Nasu M (2014) From regenerative dentistry to regenerative medicine: progress, challenges, and potential applications of oral stem cells. Stem Cells Cloning 7:89–99. https://doi.org/10.2147/sccaa.s51009
Proksch S, Steinberg T, Vach K, Hellwig E, Tomakidi P (2014) Shaping oral cell plasticity to osteogenic differentiation by human mesenchymal stem cell coculture. Cell Tissue Res 356:159–170. https://doi.org/10.1007/s00441-013-1777-5
Mostafa NZUH, Varkey M, Dederich DN, Doschak MR, El-Bialy TH (2011) In vitro osteogenic induction of human gingival fibroblasts for bone regeneration. Open Dent J 5:139–145
Zhou Y, Hutmacher DW, Sae-Lim V, Zhou Z, Woodruff M, Lim TM (2008) Osteogenic and adipogenic induction potential of human periodontal cells. J Periodontol 79:525–534. https://doi.org/10.1902/jop.2008.070373
Ivanovski S, Li H, Haase HR, Bartold PM (2001) Expression of bone associated macromolecules by gingival and periodontal ligament fibroblasts. J Periodontal Res 36:131–141
Otabe K, Muneta T, Kawashima N, Suda H, Tsuji K, Sekiya I (2012) Comparison of gingiva, dental pulp, and periodontal ligament cells from the standpoint of mesenchymal stem cell properties. Cell Med 4:13–21. https://doi.org/10.3727/215517912X653319
Kundrotas G (2012) Surface markers distinguishing mesenchymal stem cells from fibroblasts. Acta Med Lituan 19:75–79
Pouly J, Bruneval P, Mandet C, Proksch S, Peyrard S, Amrein C, Bousseaux V, Guillemain R, Deloche A, Fabiani JN, Menasche P (2008) Cardiac stem cells in the real world. J Thorac Cardiovasc Surg 135:673–678. https://doi.org/10.1016/j.jtcvs.2007.10.024
Pisciotta A, Carnevale G, Meloni S, Riccio M, De Biasi S, Gibellini L, Ferrari A, Bruzzesi G, De Pol A (2015) Human dental pulp stem cells (hDPSCs): isolation, enrichment and comparative differentiation of two sub-populations. BMC Dev Biol 15:14. https://doi.org/10.1186/s12861-015-0065-x
Gagari E, Rand MK, Tayari L, Vastardis H, Sharma P, Hauschka PV, Damoulis PD (2006) Expression of stem cell factor and its receptor, c-kit, in human oral mesenchymal cells. Eur J Oral Sci 114:409–415. https://doi.org/10.1111/j.1600-0722.2006.00388.x
Sorrell JM, Caplan AI (2009) Fibroblasts-a diverse population at the center of it all. Int Rev Cell Mol Biol 276:161–214. https://doi.org/10.1016/s1937-6448(09)76004-6
Bradley JE, Ramirez G, Hagood JS (2009) Roles and regulation of Thy-1, a context-dependent modulator of cell phenotype. Biofactors 35:258–265. https://doi.org/10.1002/biof.41
Wang Z, Yan X (2013) CD146, a multi-functional molecule beyond adhesion. Cancer Lett 330:150–162. https://doi.org/10.1016/j.canlet.2012.11.049
Mangialardi G, Cordaro A, Madeddu P (2016) The bone marrow pericyte: an orchestrator of vascular niche. Regen Med 11:883–895. https://doi.org/10.2217/rme-2016-0121
Xu J, Wang W, Kapila Y, Lotz J, Kapila S (2009) Multiple differentiation capacity of STRO-1+/CD146+ PDL mesenchymal progenitor cells. Stem Cells Dev 18:487–496. https://doi.org/10.1089/scd.2008.0113
Mrozik K, Gronthos S, Shi S, Bartold PM (2017) A method to isolate, purify, and characterize human periodontal ligament stem cells. Methods Mol Biol 1537:413–427. https://doi.org/10.1007/978-1-4939-6685-1_24
El-Sayed KM, Paris S, Graetz C, Kassem N, Mekhemar M, Ungefroren H, Fandrich F, Dorfer C (2015) Isolation and characterisation of human gingival margin-derived STRO-1/MACS(+) and MACS(−) cell populations. Int J Oral Sci 7:80–88. https://doi.org/10.1038/ijos.2014.41
Beederman MLJ, Nan G, Wang J, Liu X, Yin L, Li R, Shui W, Zhang H, Kim SH, Zhang W, Zhang J, Kong Y, Denduluri S, Rogers MR, Pratt A, Haydon RC, Luu HH, Angeles J, Shi LL, He TC (2013) BMP signaling in mesenchymal stem cell differentiation and bone formation. J Biomed Sci Eng 6:32–52
Ishibashi O, Ikegame M, Takizawa F, Yoshizawa T, Moksed MA, Iizawa F, Mera H, Matsuda A, Kawashima H (2010) Endoglin is involved in BMP-2-induced osteogenic differentiation of periodontal ligament cells through a pathway independent of Smad-1/5/8 phosphorylation. J Cell Physiol 222:465–473. https://doi.org/10.1002/jcp.21968
Torii D, Tsutsui TW, Watanabe N, Konishi K (2016) Bone morphogenetic protein 7 induces cementogenic differentiation of human periodontal ligament-derived mesenchymal stem cells. Odontology 104:1–9. https://doi.org/10.1007/s10266-014-0182-1
Luan X, Dangaria S, Ito Y, Walker CG, Jin T, Schmidt MK, Galang MT, Druzinsky R (2009) Neural crest lineage segregation: a blueprint for periodontal regeneration. J Dent Res 88:781–791. https://doi.org/10.1177/0022034509340641
Matsubara T, Suardita K, Ishii M, Sugiyama M, Igarashi A, Oda R, Nishimura M, Saito M, Nakagawa K, Yamanaka K, Miyazaki K, Shimizu M, Bhawal UK, Tsuji K, Nakamura K, Kato Y (2005) Alveolar bone marrow as a cell source for regenerative medicine: differences between alveolar and iliac bone marrow stromal cells. J Bone Miner Res 20:399–409. https://doi.org/10.1359/jbmr.041117
Paldi A (2013) Effects of the in vitro manipulation of stem cells: epigenetic mechanisms as mediators of induced metabolic fluctuations. Epigenomics 5:429–437. doi: 10.2217/epi.13.35
Rodier F, Campisi J (2011) Four faces of cellular senescence. J Cell Biol 192:547–556. https://doi.org/10.1083/jcb.201009094
Choi JK, Hwang HI, Jang YJ (2015) The efficiency of the in vitro osteo/dentinogenic differentiation of human dental pulp cells, periodontal ligament cells and gingival fibroblasts. Int J Mol Med 35:161–168. https://doi.org/10.3892/ijmm.2014.1986
Acknowledgments
The authors are grateful to Anja Kaufmann for her excellent technical assistance.
Funding
The corresponding author is indebted to the Baden Württemberg Stiftung for the financial support of this research project by the ‘Eliteprogramme for Postdocs’.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Committee of Ethics of the Medical Faculty of the Albert Ludwigs University Freiburg, Germany, and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
ESM 1
(PDF 46.5 kb)
Rights and permissions
About this article
Cite this article
Proksch, S., Kirsch, K., Vach, K. et al. Comparative differentiation analysis of distinct oral tissue-derived cells in response to osteogenic stimulation. Clin Oral Invest 23, 1077–1089 (2019). https://doi.org/10.1007/s00784-018-2529-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-018-2529-4