Abstract
The epidermal growth factor receptor is a well-established cancer therapeutic target due to its stimulation of proliferation, motility, and resistance to apoptosis. Recently, additional roles for the receptor have been identified in growth of metastases. Similar to development, metastatic spread requires signaling interactions between epithelial-derived tumor cells and mesenchymal derivatives of the microenvironment. This necessitates reactivation of developmental signaling molecules, including the hypercalcemia factor parathyroid hormone-related protein. This review covers the variations of epidermal growth factor receptor signaling in cancers that produce bone metastases, regulation of parathyroid hormone-related protein, and evidence that the two molecules drive cancer-mediated diseases of bone.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The epidermal growth factor family of receptor tyrosine (Tyr) kinases is a well-established therapeutic target in a wide range of malignancies, due to its potent stimulation of proliferation, motility/invasiveness, and resistance to apoptosis in cancer cells. An expanding understanding of metastasis has revealed additional roles for the epithelial growth factor receptor (EGFR) family in later events of the process. Similar to the development of epithelial organs, metastatic spread requires signaling interactions between epithelial-derived tumor cells and various mesenchymal derivatives of the microenvironment. This necessitates the reactivation of many signaling pathways that played a prominent role in development. One of the developmental regulatory molecules activated by EGFR signaling is parathyroid hormone-related protein (PTHrP). This peptide initially came to light in the 1980s as the causative factor of cancer-associated hypercalcemia. Subsequently it was recognized that tumor-derived PTHrP was an important component of the vicious cycle that drives the growth of bone metastases from a wide variety of carcinomas. The intersection of EGFR and PTHrP signaling likely has a substantial pathological impact on epithelial-derived bone metastases resulting from breast, prostate, lung, and head and neck tumors. This review covers the emerging variations of EGFR signaling, the expression of the receptor and its ligands in cancers that produce bone metastases, its regulation of PTHrP gene expression, and evidence of how the two molecules influence primary tumor growth and bone metastases.
EGFR signaling variations
Overview of ErbB signaling
The most comprehensively studied form of EGFR signaling is the autocrine stimulation that occurs in many epithelial cancer cells. The EGF family of receptors consists of 4 members that are generically called the ErbB family: ErbB1. ErbB2, ErbB3, and ErbB4 [1]. The term ErbB was originally derived from avian erythroblastosis virus. The V-erb oncogene, present in the virus, was ultimately found to share sequence homology with the human receptor for epidermal growth factor leading it to be called ErbB1, as other members of the family were identified. ErbB2 is frequently referred to as human EGF receptor 2 (HER2) due to its sequence homology to the EGFR [2]. Signaling is generated when EGFR and ErbB4 bind to their ligands. In contrast, the ErbB2 extracellular binding domain (ECD) fails to bind any of the ErbB agonists. ErbB3 is thought to be kinase dead due to lack of key functional residues, but recently it has shown that it retains the ability to transphosphorylate its own intracellular domain [3]. When stimulated by ligands, EGFR and ErbB4 receptors can transduce their signals as homodimers or heterodimers; however, the signaling from ErbB2 or ErbB3 generally requires heterodimerization with another ErbB family member with a functional ECD or kinase [1].
The ErbB receptors are stimulated by 12 distinct ligands, providing significant complexity to ErbB signaling as several of these agonists can bind to more than one receptor. Ligands exclusive to EGFR are epidermal growth factor (EGF), transforming growth factor alpha (TGF-α), amphiregulin (AREG), and epigen (EPGN) [1, 4]. ErbB4 is specifically bound and activated by neuregulins (NRG) 3, 4, 5, [1, 4]. Heparin-binding EGF-like growth factor (HB-EGF), epiregulin (EREG), and β-cellulin (BTC) bind and activate both the EGFR and ErbB4 [1, 4]. NRG 1 and 2 bind both ErbB3 and ErbB4 [1, 4]. Thus, EGFR can potentially be bound and activated by 7 ligands, EGF, TGF-α, AREG, EPGN, HB-EGF, EREG, and BTC.
All of the ErbB agonists are synthesized as integral plasma membrane-bound proteins [5]. In some cases, these transmembrane ligands can stimulate ErbB signaling on adjacent cells, through a mechanism called juxtacrine signaling [6, 7]. Typically, autocrine ErbB signaling is thought to require proteolytic cleavage of the EGFR ligand ECD at the cell surface, termed “protein ectodomain shedding”. Shedding of membrane-associated ligands and the subsequent release of soluble factors is required for receptor binding on the same cell or on a neighboring cancer cell. The proteases that mediate the process are from the ADAM family (a disintegrin and metalloproteinase) [8, 9]. The cleavage of AREG, EREG, HB-EGF, EPGN, TGFα, and NRG 1&2 is mediated by ADAM 17, while BTC and EGF are cleaved by ADAM 10 [8]. It is also likely that there are other metalloproteinases that are capable of cleaving ligands [10]. Thus, for autocrine signaling to occur, the receptor, the ligand, and the activated protease must be present in the tumor cell.
EGF-induced EGFR homodimer signaling
Due to its abundance in mouse salivary glands and the relative ease of purification from this source, EGF was the first ligand identified [11]. Although ligands typically range from 160 and 250 amino acids, EGF is by far the largest protein among them, containing over 1200 amino acids [12]. The ECD of EGF is composed of more than 1000 amino acids and nine EGF repeats. It is the terminal EGF repeat near the transmembrane domain that is a functional ligand when it is proteolytically cleaved and released [12]. This 52-amino acid-EGF domain has been extensively studied and is a high-affinity ligand for EGFR.
EGF has historically been used for receptor binding, signaling, trafficking, and cell fate studies and is in many ways considered the prototype for receptor tyrosine kinase signaling [13–15]. Ligation of EGF to EGFR exposes the dimerization arm of the ECD that permits interaction with either another EGFR to create a homodimer or with another ErbB member to create a heterodimer. Ligand binding also induces a conformational change in the receptor that activates the intracellular cytoplasmic kinase domain, which in turn can catalyze autophosphorylation of tyrosine residues on the adjacent C-terminal tail of the dimerized ErbB receptor. The 10 phosphorylated tyrosine residues serve as docking sites for adapter proteins or other signal transduction components, resulting in activation of Ras, MAPK, src, STAT 3/5 and PLCγ/PKC, and the PI3-kinase AKT pro-survival pathway. Activation of these signaling pathways by EGFR dimers has a profound impact on cellular proliferation, resistance to apoptosis, differentiation, as well as motility-/migration-associated behaviors. Phosphorylation also plays a central role in trafficking of the receptor after ligand binding. Phosphorylation of residue tyrosine-974 triggers endocytosis of the receptor in clathrin-coated pits, and phosphotyrosine-1045 binds to c-Cbl that facilitates subsequent degradation [4, 16].
The binding of EGF to EGFR shows a degree of sensitivity to pH, and disengagement of the ligand leads to recycling of internalized receptors to the plasma membrane [17]. Trafficking studies suggest that ~50 % of EGF-stimulated EGFR is degraded in the lysosome, while the remainder is recycled back to the plasma membrane [17]. Thus, activation of EGFR by EGF directly stimulates a myriad of intricate cellular signaling pathways, many of which converge on elements of the ERK/MAPK pathway (see Fig. 1a). The rate of EGFR signaling can be regulated by receptor turnover. The rapid turnover of EGF-activated EGFR is believed to decrease the stimulation of cellular proliferation pathways, permitting a balance with various differentiation-inducing stimuli present in normal tissue [1, 17, 18]. In cancers, autocrine EGFR homodimer signaling is substantially attenuated in many ways, but overall shifts the cell fate balance toward cellular proliferation and survival rather than differentiation, apoptosis, and senescence.
EGF and ligand-independent EGFR signaling. a Schematic of EGFR dimer induced by EGF biding. Sites of cytoplasmic tyrosine (Y) phosphorylation are indicated, as are cytosolic effector proteins that bind to these phosphorylated tyrosine residues and some of the effector signaling pathways. b Schematic of potential ligand-independent EGFR signaling induced by mutations and oxidation. The major effector pathways stimulated by these forms of EGFR signaling are indicated
Heterodimerization and EGFR signaling
The most well understood of potential receptor interactions is the EGFR signaling that occurs when the receptor heterodimerizes with ErbB2 [19, 20]. Despite being unable to bind ligands, the ErbB2 dimerization arm is constitutively exposed, allowing this receptor to more efficiently dimerize with other liganded ErbB family members [4]. The ErbB2 heterodimers attenuate EGFR signal transduction in several ways [21–25]. First, the affinity of this ErbB2 complex for ligands is enhanced. Second, more efficient signal transduction occurs because the ErbB2 phosphotyrosine domains of heterodimers bind most adapter proteins with a higher affinity than those of ErbB homodimers. Third, ErbB2/EGFR heterodimers are slowly endocytosed and are more frequently recycled to the plasma membrane than the EGF-stimulated homodimers. By virtue of its strong interactions with adapter proteins and recycling to the plasma membrane, an EGFR/ErbB2 heterodimer can amplify and extend the duration of EGFR ligand signaling, leading to proliferation and survival at the expense of other cell fates [21–24].
In contrast to the fairly well-established understanding of ErbB2 heterodimers, there have been few studies on the EGFR heterodimerization with ErbB4 or ErbB3. Co-immunoprecipitation experiments have confirmed the presence of the ErbB4/EGFR in lung epithelial cells and type II pneumocytes; however, the specific function of this complex was not determined [26, 27]. Co-expression of ErbB4 and EGFR plasmids in model NIH 3T3 fibroblasts or CHO lines provided evidence of dimerization of these receptors and suggested that this complex could induce cellular transformation in the presence of either EGF or NRG1. Further analysis of the CHO system found that the ErbB4/EGFR heterodimer specifically induced B-Raf kinase activity, which may induce transformation by increasing the activity of the ERK/MAPK pathway [28]. Recently, ErbB3/EGFR heterodimers have been identified in pancreatic cancer cell lines [29, 30]. It appears that the ErbB3/EGFR complex may be a more effective stimulus for proliferation in pancreatic cancer cell lines than EGFR homodimers [29]. Additionally, these studies suggest the ligand amphiregulin (AREG) is able to stimulate activity of the ErbB3/EGFR heterodimer [29, 30]. Unfortunately, the comprehensive binding, signal transduction, and trafficking studies completed for ErbB2-containing receptor complexes have not been completed for EGFR/ErbB4 or ErbB3 heterodimers. Expanded knowledge of the roles of ErbB heterodimers is essential for the effective use of ErbB-targeted therapeutics in cell types that express multiple ErbB receptors.
Ligand-independent signaling
The identification and characterization of the v-erbB oncogene provided the first insight into ligand-independent ErbB signaling [31]. Amino acid sequence comparison of v-erbB and the human EGFR revealed that the viral oncogene lacked a large portion of the ligand binding ECD [32]. Soon afterward, experiments indicated that the ability of the V-erb oncogene to transform cells or display tyrosine kinase activity did not require EGF stimulation, suggesting that the ErbB receptor signaled independently of ligand [31, 33].
A truncated EGFR lacking much of the ECD, termed variant III or EGFRvIII, was identified in gliomas and glioblastomas [34, 35]. Similar to v-erbB, EGFRvIII failed to bind to ligand, but still displayed kinase activity [34, 35]. Activated EGFRvIII induced anchorage-independent growth in numerous cell types without the addition of ligands such as EGF, and EGFRvIII-related mutants can induce tumorigenesis in various tissues in transgenic mice [36, 37]. Unlike the wild-type EGFR, ras-MAPK signaling is not typically elevated by the forced expression of EGFRvIII [38]. However, signaling via STAT and PI3-kinase AKT and mTOR pathways may be enhanced by the truncated receptor [31, 39]. Taken together, EGFR mutants lacking large segments of the ECD and that fail to bind ligands appear to be constitutively active.
A second class of EGFR mutations were discovered in large clinical trials testing the efficacy of the inhibitor gefitinib on non-small cell lung cancer (NSCLC) patients. These mutations conferred sensitivity to gefitinib and the treatment resulted in unprecedented reduction of tumor burden and in some cases durable remissions [40–42]. These were somatic in-frame point mutations and deletions (either exon 19 deletions or the exon 21 arginine-for-leucine substitution at amino acid 858, or L858R) clustered around the ATP-binding pocket within the tyrosine kinase domain of the EGFR [42]. The mutations induce changes in the conformation of the ATP-binding pocket that may permit greater accessibility of EGFR inhibitors. However, the efficacy of the tyrosine kinase inhibitors is mainly attributed to the ability of these compounds to exploit “oncogene addiction” in the cells bearing EGFR mutations [42, 43]. Cancer cell dependency appears to result from high levels of stimulation of the pro-survival PI3K-AKT signaling pathway downstream of the mutant EGFR [42, 43]. It is thought that this pathway is perhaps activated by EGFR–ErbB3 heterodimers, which would provide increased docking sites for PI3K [42] (see Fig. 1b). It has been proposed that the efficient stimulation of downstream signaling pathways such as AKT result from the disruption of the auto-inhibitory interactions, which restrain basal kinase activity of the mutated EGFR [44]. This implies that the EGFR kinase domain mutations found in NSCLC patients may be constitutively active.
Recent data from research on the connection between cigarette smoking and EGFR activation in the initiation of lung cancer has alluded to a scenario where the receptor may signal in a ligand-independent manner. It has been known for some time that agents such as cigarette smoke that induce oxidative stress can also induce ligand-independent phosphorylation of EGFR in a respiratory epithelial cell line [45, 46]. These agents induce an EGFR conformation that is distinct from that formed by the binding of EGF, and it is speculated that the oxidized receptor does not dimerize [45, 47]. Cigarette smoke also induces receptor association with unique proteins such as c-src and caveolin-1 [46, 48]. Although the phosphorylation of EGFR stimulated by oxidative stress is relatively extensive, the Cbl binding 1045 residue is not phosphorylated and the receptor is not rapidly degraded [45]. Consistent with the relatively extensive phosphorylation of the EGFR induced by oxidative stress, MAPK signaling is also activated downstream of the receptor [48]. In addition, the receptor activated by oxidizing agents is trafficked through rab 11-containing endosomes [46, 48]. Finally, EGFR tyrosine kinases fail to block signaling by the cigarette smoke-activated EGFR [45]. Thus, oxidation of the EGFR stimulates a form of ligand-independent signaling that may play a substantial role in promoting the formation of cancers within the lung and other tissues subjected to agents such as cigarette smoke.
Ligands and EGFR signaling
There are 7 ligands that bind and activate EGFR. It is likely that specific differences in signaling induced by each of the ligands will emerge, but at this point, the ligands can be organized into two main groups based on the receptor trafficking they induce after internalization in clathrin-coated pits (see Fig. 2). First, there are the ligands that have high affinity for the receptor, both on the plasma membrane and in the low pH of the late endosome. These tend to induce high levels of tyrosine phosphorylation of the c-terminal tail of the receptor, and after internalization from the plasma membrane induce rapid degradation [49–52]. These ligands induce the following trafficking: the early endosome, late endosome/multivesicular body, intraluminal vesicle of the multivesicular body, and finally to the lysosome where the receptor is degraded [49–52]. The second group consists of ligands that have either reduced affinity for the receptor, or their binding is disrupted by low pH in the endosome. These induce low levels of receptor tyrosine phosphorylation. These ligands are taken into early endosome where the ligand dissociates from the receptor. As a result, the majority of the EGFR gets rapidly recycled back to the plasma membrane after internalization [17, 49]. It is thought that due to recycling of the receptor, the low-affinity ligands tend to stimulate longer duration signaling and have a greater impact on mitogenesis than the high-affinity ligands.
Ligand-dependent EGFR trafficking and degradation. a Schematic of EGFR internalization and trafficking after binding ligands that have high affinity for the receptor within endosomes. HB-EGF, BTC, and EGF lead to degradation of the receptor in the lysosome. However, the receptor remains phosphorylated and appears to be associated with signaling effector proteins, both in the early endosome and when it is on the external membrane of the multivesicular body. b Schematic of EGFR internalization and trafficking after binding ligands that have low affinity for the receptor within endosomes. TGFα EREG and EPGN induce internalization, but within the endosome, the ligand dissociates and the dimer comes apart, permitting the receptor to be rapidly recycled to the plasma membrane. However, the receptor does not remain phosphorylated and does not appear to extensively signal from internal compartments. Once back on the plasma membrane, the ligand can be rapidly reengaged and signal again
Ligands that induce EGFR degradation: EGF, HB-EGF, and β-cellulin
EGF, HB-EGF, and BTC ectodomains are classified as high-affinity ligands for EGFR on the basis of a binding assay using ErbB receptor ECDs fused to immunoglobulins. These ligands induce extensive EGFR tyrosine phosphorylation in most cell types studied [17, 53, 54]. Upon binding to and activation by exogenous HB-EGF and β-cellulin, EGFR is internalized into early endosomes and then trafficked to multivesicular bodies and ultimately lysosomes where they are rapidly degraded. EGF, BTC, and HB-EGF stimulate phosphorylation of tyrosine residue 1045 that facilitates persistent recruitment of c-Cbl a ubiquitin ligase. Ubiquitination of EGFR is essential for trafficking from the multivesicular body to the lysosome and subsequent degradation of the receptor [49–52]. The binding of HB-EGF and BTC to EGFR was more resistant to low pH than EGF [17]. BTC and HB-EGF show similar effects to those exhibited by EGF on cell proliferation and migration [55, 56].
Ligands that permit rapid receptor recycling to the plasma membrane: TGFα, EREG, and EPGN
These ligands fail to induce substantial receptor degradation; therefore, they facilitate prolonged signaling. Two distinct mechanisms have been identified which account for the lack of degradation of EGFR by these ligands. TGFα induces different trafficking of the receptor than EGF [57]. At an extracellular pH of 7.4, TGFα and EGF have similar binding affinities for EGFR [17, 57]. However, at around pH 5 such as found in the late endosome, TGFα has decreased affinity for EGFR [17, 57]. TGFα ligand–receptor disengagement permits nearly 100 % of receptors to be internalized and recycled to the plasma membrane [17]. It is thought that three additional histidines found in the receptor-binding domain of TGFα create an increased sensitivity to pH, which ultimately influences the strength of agonist–receptor interactions [57]. Site-directed mutagenesis was used to add these histidines to the EGF ectodomain, decreasing ligand–receptor binding at low pH [58, 59].
In contrast to a low pH-induced sensitivity of ligand–EGFR binding, EREG simply appears to have reduced affinity for the EGFR at physiological pH [25]. Upon binding to and activation by EREG, EGFR is rapidly endocytosed but then is efficiently recycled back to the plasma membrane [17]. It is speculated that the ligand disengages from the receptor in the endosome due to low-affinity binding [17].
EPGN was discovered in 2000 and as the last ErbB family member ligand identified, it has not been as extensively studied as other ligands. The ligand activates EGFR and does not activate ErbB3 or ErbB4 when these receptors are expressed in isolation [60, 61]. EPGN has an approximate 100-fold less affinity for EGFR relative to recombinant human EGF, and similar to TGFα, the ligation of EPGN to EGFR is sensitive to pH [60, 61]. Therefore, EPGN uses a combination of decreased affinity and pH sensitivity to facilitate EGFR recycling to the plasma membrane.
Each of these ligands induces modest levels of phosphorylation of EGFR when compared with EGF [17]. The 1045 residue is not readily phosphorylated after ligand engagement and c-Cbl does not efficiently bind the receptor activated by these ligands [17]. However, each of the ligands has been reported to induce prolonged MAPK signaling. Along with greater duration of signaling, there has been the finding that each of the ligands serves as more potent mitogens than EGF [60–63]. As detailed functional studies on the individual ligands are completed, it will be interesting to determine if the relationship between exogenous ligand-induced receptor turnover and magnitude of downstream cellular responses such as motility, invasion, and induction of gene expression is retained.
AREG
AREG should perhaps be considered independently because in many experimental circumstances, this ligand induces novel trafficking and cellular behavior as compared to the other ligands. Among the ErbB receptors, AREG appears to exclusively bind and activate EGFR. The relative strength of AREG binding to EGFR has been controversial. When Shoyab and colleagues initially identified human AREG, they reported that the fully processed ligand isolated from breast cancer cells had a reduced affinity for human EGFR when compared with EGF derived from the mouse salivary gland [64]. In contrast, studies with human recombinant ligands reported that AREG had an affinity for EGFR on the plasma membrane of cells similar to that of EGF and TGFα [65, 66]. Introducing further complexity, analyses of ligand–receptor interactions have suggested that recombinant AREG does not induce efficient dimerization of EGFR [67]. Proteolytic processing of AREG in mammalian cells may eliminate the C-terminal portion of the EGF domain that is required for high affinity for the receptor [68]. Also, that portion of the receptor-binding domain in AREG contains a methionine, whereas all other EGFR ligands have a leucine [68]. Collectively some data suggest that AREG produced by eukaryotic cells could bind EGFR with less affinity than EGF; however, this concept has not been sufficiently tested to either validate or reject it.
More recent studies have focused on the distinct downstream signaling and cellular behavior caused by AREG. Unlike exogenous EGF treatment, AREG stimulation of model cell lines and breast cancer cell lines does not induce efficient phosphorylation of many of the tyrosine residues in the C-terminal tail of EGFR [17, 69–71]. Notably, the Cbl binding 1045 tyrosine residue is not efficiently phosphorylated by AREG and this ligand fails to induce rapid degradation of EGFR. Trafficking studies indicate that EGFR with the AREG ligand is rapidly internalized, but slowly recycled back to the plasma membrane. It appears that AREG may be unique among the ligands in that it induces EGFR trafficking through endosomes containing Rab 4 and Rab 11, markers for membrane recycling vesicles [17, 71]. In addition, AREG binding to EGFR is very resistant to acid pH, suggesting that the ligand does not disengage in the endosome like TGFα [17]. AREG induces prolonged phosphorylation of ERK relative to EGF [70, 72]. This altered signaling appears to be the basis of the loss of cell–cell adhesion and increased motility-/migration-associated behaviors in breast and other epithelial cells with AREG [72, 73].
Some of the unique biochemical properties of AREG, in combination with receptor-trafficking data, suggest that the ligand may induce a novel mechanism of EGFR signaling. Much of the EGF–EGFR signaling occurs in the endosome [52]. In contrast, AREG ligation of EGFR causes increased receptor localization to cell–cell junctions, and does not appear to elicit signaling from endosomes [71, 74]. In addition, the slow recycling of the receptor through the Rab 4 and Rab 11-containing vesicles is very different from the rapid recycling of the EGFR out of EEA1 endosomes observed for TGFα- or EREG-treated cells [17]. Trafficking through Rab 11-containing vesicles is similar to that observed for EGFR activated by cigarette smoke, which is proposed to signal as a monomer rather than a dimer [46, 48]. The lack of efficient EGFR dimerization, in addition to the differential trafficking, phosphorylation and localization of the receptor, has led us to speculate that AREG may induce a unique signaling complex that is distinct from those activated by the other ErbB ligands.
Juxtacrine signaling by ligands
HB-EGF has a heparin-binding region on the N-terminal to the EGF domain. This region has been shown to interact with heparin-sulfated plasma membrane proteins such as the tetraspanin, CD9, and the extracellular matrix binding/cell differentiation marker protein, CD44 [6, 55]. In particular, the heparin-mediated interaction between HB-EGF and CD9 appears to be crucial to juxtacrine signaling by the pro-ligand [75]. The association between the heparin-binding domain and the cell membrane-associated heparin-sulfated proteoglycans appears to be crucial to localizing HB-EGF to regions of cell–cell contact. Also, the interaction with these heparin-sulfated proteoglycans appeared to prevent proteolytic cleavage of the pro-ligand, whereas exogenous heparin increased shedding of HB-EGF [7, 75]. In contrast to the impact of the shed ligand, juxtacrine signaling by the pro HB-EGF stifles cellular proliferation [75]. Similar to HB-EGF, AREG contains a heparin-binding domain N-terminal to the receptor-binding region [64, 76]. There have been reports that AREG also induces juxtacrine signaling, but the effect on cell behavior is unclear [72].
Activation of the EGFR initiated by the seven ligands likely provides a mechanism for significant heterogeneity of downstream signaling. The mechanistic basis of differences in individual ligand signaling, as well as the downstream cellular and molecular consequences, may serve as predictive factors for response to various types of EGFR-targeted therapeutics.
EGFR receptor and ligand expression in cancers that induce bone pathologies
Non-small cell lung cancer
Non-small cell lung cancer comprises two major and one minor histological subtypes of lung cancers. The predominant major subtype is adenocarcinoma (AC), which are thought to arise from any distal peripheral airway structure, known as the terminal respiratory unit, and includes small bronchi and bronchioles. These tumors are believed to be derived from the Clara cells or alveolar type II epithelial cells, which can divide and form type II and type I alveolar pneumocytes. Squamous cell carcinoma (SCC) is the second major histological subtype of NSCLC that is derived from the epithelia of the major bronchi of the more central airways. The minor histological subtype of NSCLC is large cell carcinomas that are speculated to originate in stem cells of the larger airways [77]. The causative association between cigarette smoking and the development of NSCLC has been well established and widely known since the 1950s. The relationship with cigarette smoking was also distinctly associated with a greater proportion of the NSCLC being of the histological subtype, SCC, or the neuroendocrine-derived small cell carcinoma. However, over the last several decades, epidemiologic surveys have revealed accelerated increases in AC, but less rapid increases in SCC in both western and Asian countries despite relatively little change in the number of people who smoked [78]. The recognition that cigarette smoking is a major cause of NSCLC has served as an impetus to change the composition of the cigarette to decrease the concentrations of carcinogenic N-nitroso compounds and polynuclear aromatic hydrocarbons, as well as a reduction in tar and nicotine delivery via the addition of filter tips to the cigarette. The change in NSCLC type is thought to correspond with changes in the smoker, who adjusts his/her smoke intake to satisfy a conditioned need for nicotine, so that the volume of smoke inhaled delivers far more carcinogens to the peripheral lung parenchyma. Nicotine-compensating smoking patterns include an increased frequency of puff drawing and a stronger depth of inhalation. A trend over the recent decades is that the AC incidence continues to rise in a significant proportion of patients who are considered never-smokers. The Asian countries have the highest percentage of patients with never-smoker AC, which has reportedly reached as high as 40 %, whereas incidence data from Brazil and the US report that 34 % and 10 % of population are affected, respectively [79–81]. Currently, never-smoker lung cancer is regarded as a distinct disease entity with a unique tumorigenic pattern, clinicopathologic features, and natural history [82, 83]. Several studies have documented that among the never-smoker population, the majority of affected patients are women [78, 81, 82, 84, 85]. Actually, in never-smokers, the main causes are unknown. To date, the hypothesized risk factors associated with this gender propensity include a role for estrogen in lung carcinogenesis, susceptibility genes, prior lung disease, as well as exposure to environmental tobacco smoke at home, residential radon, and indoor air pollutants such as cooking oil vapors, and indoor coal and wood smoke [85].
The ErbB family has been extensively studied in lung cancer and this has produced some insights into the role of these receptors in disease progression. Various immunohistochemical studies have shown immunoreactivity for EGFR in ~65 to 90 % of NSCLC, and overexpression of the protein has been observed in up to 62 % of cases [86–89]. SCC is more frequently associated with high levels of expression of EGFR [86, 87]; however, receptor expression levels have been a weak prognostic indicator for NSCLC [88, 89]. EGFR kinase domain mutants are observed in ~48 % AC cases found in those that never smoked, which are mutually exclusive of ALK rearrangements and KRAS mutations also found in this tumor group [90]. ErbB2 mutations of the tyrosine kinase domain have also been associated with never-smoker AC and are found to be mutually exclusive of EGFR mutations [91]. The presence of these AC mutations in people who never smoked is associated with responsiveness to the EGFR TKIs, erlotinib and gefitinib [42, 91]. Retrospective studies suggest that kinase domain mutations are relatively rare in NSCLC in smokers [42, 91]. EGFRvIII mutants have been reported in 5 % of SCC, but kinase domain mutants are not observed in this histologic type [36, 42]. Among the EGFR ligands, EREG, TGFα, and AREG are frequently expressed (65–93 % of tumors), whereas EGF and HB-EGF are infrequently detected in tumors [87, 92]. The relationship between ligand expression and prognosis has become controversial, as indicators of sensitivity to EGFR inhibitors are sought. TGFα expression, whether detected by immunohistochemistry or in the serum of patients, appears to predict poor prognosis and lack of responsiveness to EGFR TKIs [93, 94]. AREG expression in NSCLC may be associated both with responsiveness [95, 96] and unresponsiveness to EGFR TKIs [93, 94]. These contradictory results maybe related to differences in the sensitivity of the methods used for detection of the ligand. With the exception of AC, where survival is driven by EGFR kinase domain mutations, it appears that despite high levels of the EGFR and ErbB ligand expression, most NSCLC are refractory to the currently available EGFR-targeted therapeutics.
Head and neck squamous carcinoma
These cancers arise from the stratified squamous epithelia of the mucosal surfaces within the head and neck, with the majority of them having the histologic designation of SCC. The use of smoking and/or chewing tobacco and distilled alcohol are the primary risk factors associated with the development of head and neck squamous cell carcinomas (HNSCC). EGFR expression has been reported in 90–100 % of HNSCC and overexpression of the protein as determined by immunohistochemistry occurs in 80 % of the tumors [97–99]. Increased EGFR copy number and EGFR phosphorylation have been associated with poor prognosis [97, 100]. EGFRvIII mutants are fairly prevalent in these tumors ranging from 17 to 42 % of HNSCC samples and these tumors are refractory to EGFR-targeted therapeutics [101, 102]. EGF, TGFα, and AREG were reported as expressed in ~65, 90, and 45 % of HNSCC tumor samples, respectively [98, 102–104]. The expression of both AREG and TGFα by HNSCC has been correlated with poor prognosis and advanced disease stage [98, 102]. AREG expression also has been associated with insensitivity to cetuximab combined with docetaxel treatment [102]. Similar to NSCLC, a minority of HNSCC tumors respond positively to EGFR TKI and the long-term benefits are modest [105].
Prostate cancer
In contrast to the frequent expression of EGFR found in groups of NSCLC and HNSCC, only a small subset of prostate cancers express ErbB receptors. Each member of the ErbB receptor family has been detected by IHC in approximately 15–30 % of prostate adenocarcinomas [106–108]. However, expression of EGFR was more prevalent (~40 %) in androgen-independent tumors, with up to 16 % of this subset demonstrating EGFR amplification [107, 109, 110]. TGFα and AREG proteins are observed in the majority (~70 %) of prostate adenocarcinomas, but EGF expression levels are decreased in the tumor relative to the prostate epithelia [108, 111–113]. Given the restricted subset of tumors that express EGFR, receptor-targeted therapeutics will not be considered as a first-line therapeutic for prostate cancer. However, since over 90 % of prostate cancer metastasizes to bone, EGFR-targeted agents could be used as part of a strategy to interrupt supportive microenvironment signaling.
Breast cancer
The development of microarray platforms capable of simultaneously evaluating gene expression from a large portion of the genome has led to a major breakthrough in the classification of breast cancer tumors. Gene expression profiles that classify breast cancers into novel molecular subtypes offer a method to view the contribution of ErbB family members to disease progression. The subclasses are: normal breast-like, ErbB2 amplified, luminal A, luminal B, and basal [114–116]. The ErbB2 amplified, basal, and luminal B subtypes have substantially worse prognosis than the normal breast-like and luminal A [114–116].
The basal molecular subtype of breast cancer has the highest incidence of EGFR expression. Originally, basal cancers were considered tumors with a phenotype characteristic of that found in cells that contact the basement membrane. Mammary basal cell epithelial markers include the expression of keratin five and 14 (basal keratins), P cadherin as well as troponin found in myoepithelial cells [114–116]. Recently, a subgroup of basal tumors enriched with epithelial to mesenchymal transition markers have been identified and are now called claudin-low tumors [117, 118]. Both basal and claudin-low tumors generally express low to non-detectable immunoreactive levels of estrogen receptor, progesterone receptor, and ErbB2. As such, they have been termed “triple receptor-negative,” a classification that is associated with a poor prognosis [119]. Triple receptor-negative cancers are correlated with poor survival and high rates of distant metastasis, and are generally high-grade, large tumors. Immunophenotyping studies indicated that 50–70 % of these cancers expressed high levels of EGFR immunoreactivity [120]. Low levels of EGFR expression in these tumors is correlated with reduced numbers of distant metastasis [121]. Basal tumors frequently expressed high levels of EGFR mRNA and this expression was correlated with TGFα and ADAM-17 [122]. Thus, a sizable fraction of basal or claudin-low cancers would likely exhibit autocrine TGFα-EGFR signaling and these have poor prognosis. A subset of triple-negative tumors do metastasize to bone [123].
Luminal A tumors express ER along with GATA binding protein 3, X-box binding protein 1, trefoil factor 3, and other estrogen-regulated genes in addition to high levels of the luminal keratins K8 and 18 [114–116]. Luminal B tumors tend to express the above markers at slightly reduced levels, but have an upregulated cassette of genes, including proliferation-related genes such as myb and components involved in DNA replication [114–116]. There is no specific ErbB family member gene signature recognized in the luminal A or B tumors. Increased AREG mRNA expression was observed in the luminal A subclass [122], but ADAM-17 levels were low in this class of tumors relative to other subtypes. These observations suggest that although most ERα+ luminal A breast cancers express AREG, lack of EGFR negates the possibility of autocrine EGFR signaling. Current evidence neither supports nor refutes whether AREG participates in paracrine signaling, as luminal tumor types typically express low levels of ADAM-17. Despite having an overall better prognosis than ErbB2 amplified and basal tumors, ER+ tumors are more likely to metastasize to bone than to other organs [124].
EGFR and activation of PTHrP gene expression
The connection between EGFR and PTHrP gene expression originally stemmed from attempts to understand the physiological role of the peptide in epithelial tissues. After the PTHrP gene was cloned, RNA peptide expression surveys indicated that PTHrP was expressed in various cultured primary epithelial cells [125–129]. In some cases, the epithelial cells were grown in serum-free media systems where various growth factors could be added individually. EGF was used in these media systems to promote proliferation of the primary epithelial cells. The inclusion of EGF in the media of primary keratinocytes, mammary, or prostate epithelial cells resulted in substantial increases in PTHrP mRNA and protein secreted into the media. These findings were interpreted as evidence that PTHrP gene expression was stimulated in connection with proliferation of epithelial cells and it was concluded that EGFR was a likely regulator of this calcitropic factor in epithelial-derived cancers [125–129].
The development of TKIs specific for EGFR provided an opportunity to expand the connection between the receptor and PTHrP gene expression in epithelial cells cultured under basal conditions without the addition of exogenous ligands. Treatment of primary keratinocytes with EGFR TKI, PD153035, resulted in a ~90 % decrease in PTHrP mRNA expression [130]. This compound was used to treat a series of breast cancer and lung SCC cells and these treatments resulted in a 50–75 % reduction in PTHrP mRNA levels [131, 132]. Ectopic expression of EGFR in a receptor-null lung SCC and breast cancer cell lines resulted in a substantial increase in PTHrP mRNA [132, 133]. Knockdown of EGFR was also able to reduce PTHrP mRNA expression in the cancer cell lines [133, 134]. The findings described are consistent with the concept that autocrine stimulation of EGFR is responsible for a substantial fraction of basal PTHrP gene expression of epithelial cancer cells in culture.
The next question that arose was whether a particular type of ligand regulated expression. Under primary culture conditions, keratinocytes dramatically upregulated AREG, but they also expressed several other ligands, including TGFα, HB-EGF, and EPGN [135, 136]. Blockade of AREG reduced PTHrP mRNA by ~70 % in primary keratinocytes, suggesting that this was the predominant ligand regulating expression [130]. Relative to other ligands, AREG levels were the highest in the majority of breast cancer and lung SCC lines evaluated and blockade by siRNA or antibodies efficiently reduced basal PTHrP mRNA levels [131–133]. The precise reason for high levels of AREG produced by these various cell types grown in culture is not clear, but this gene is activated by a multitude of pathways that are upregulated in cancers including the hippo pathway, WNTs, estrogen receptor, and cAMP/PKA [137–140]. Nevertheless, exogenous treatment with EGF, TGFα, and HB-EGF was capable of increasing PTHrP mRNA in keratinocytes and breast cancer and lung SCC lines, suggesting that activation of PTHrP can occur with ligands that induce rapid EGFR turnover, as well as those that do not [130, 132, 133, 141].
There has been some progress in understanding the signaling pathways that regulate PTHrP gene expression downstream of EGFR. For example, it is well established that PTHrP gene expression is activated by the ERK and p38 pathways [142, 143]. The use of various inhibitors and dominant negative constructs to the ERK and P38 pathways efficiently repress PTHrP gene expression in cell lines that exhibit autocrine EGFR signaling [130, 132, 133, 141, 144]. Although multiple tyrosine phosphorylation sites on EGFR serve as docking sites for signal transduction proteins that activate the MAPK pathway, the PLC/PKC pathway may be particularly important to the activation of PTHrP gene expression by EGFR in breast epithelial and cancer cells [132]. In fact, EGFR stimulates basal PTHrP gene expression in MDA-MB-231 cells in the presence of constitutively activated ras-protein [134]. However, the signaling downstream of the receptor has not been carefully investigated in other cell types where EGFR is responsible for basal PTHrP gene expression.
The influence of EGFR signaling on transcriptional regulation of the PTHrP gene has been studied, but is not completely understood. The PTHrP gene has three promoters (two tata boxes, P1 and P3, and a GC-rich region, P2) distributed over 2500 bp of the upstream regulatory region [145]. The P3 TATA promoter is used in most cell types and its regulation is fairly well understood. Transient transfection and EMSA have identified functional ETS, SMAD, and SPI binding sequences within the core promoter 50 bases upstream of the P3 TATA box. The P3 promoter is activated by the EGFR in all cells studied. Among the identified cis-acting sequences, only the ETS binding site is required for EGFR activation of PTHrP-luciferase constructs in keratinocytes and breast cancer lines (see Fig. 3) [130, 132]. In addition, EGFR signaling also activates the P1 promoter in lung SCC lines, which probably contributes to the high levels of PTHrP expression in this cell type.
The identity of the ETS proteins that mediate EGFR activation of PTHrP gene expression in the various cell types studied is not known. However, a recent chromatin immunoprecipitation/next-generation sequencing (ChIP-seq) study in prostate epithelial and cancer cell lines may shed some light on the relationship between ETS factors and PTHrP gene expression [146]. Normal prostate cells express more than 15 ETS family transcription factors, but most prostate cancers aberrantly express one additional ETS factor (ERG, ETV1, ETV4, or ETV5) due to a chromosomal rearrangement [147]. These overexpressed ETS genes appear to be oncogenic because they induce invasive behaviors in prostate epithelial cells [148–150]. ChIP-seq studies comparing the genomic binding sites of oncogenic ETS members (ERG, ETV1, and ETV4) to binding sites of ETS proteins normally expressed in prostate cells (ETS1 and GABPA) found that both types bound the PTHrP-P3 core promoter region (see Fig. 4) [146]. The sequence of the PTHrP P3-ETS binding site CCGGAAGC is similar to the consensus ETS binding site identified in the core promoter of most housekeeping genes [151]. These core promoter ETS sites are bound by many different ETS proteins in vivo and, thus, are not proposed to have specificity within the ETS family [152]. However, the prostate studies identified an additional ETS-bound region 54 KB upstream of the P3 promoter [146]. This potential enhancer was occupied by the oncogenic ETS proteins, ETV1, ETV4 and ERG, but not the non-oncogenic ETS1 and GABPA (Fig. 4). Like other enhancers bound specifically by the oncogenic ETS proteins, this region had binding sites for both ETS and AP-1 transcription factors, and antibodies to the AP-1 subunit, JUND, pulled down the same sequence. Since PTHrP is the closest gene, this upstream sequence may well serve as an enhancer for the calcitropic factor. Neighboring ETS and AP-1 binding sites have been shown to respond to RAS/MAPK signaling initiated by EGFR [153]. Therefore, we speculate that PTHrP gene expression induced by EGFR signaling may be controlled by interactions between the core promoter (including the ETS sequence there) and the ETS and AP1 sequences 54 kb upstream. It is worth noting that mapping an enhancer histone mark, H3K4 monomethylation, in the genomic region preceding the PTHrP gene [154] identifies many potential enhancers that vary by cell type, indicating that the transcription factors that regulate PTHrP expression may also vary in different biological contexts.
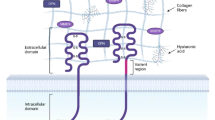
ETS factor binding in the region of PTHrP gene. ChIP-seq data using antibodies to the indicated factors are shown aligned to a genomic region (hg18, chr12:27,968,000–28,098,000) surrounding the PTHLH (PTHrP) gene [145, 153]. Antibodies to endogenous ETS1 and GABPA assayed these ETS proteins in a cell line derived from normal prostate (RWPE-1). An anti-FLAG antibody detected FLAG-ERG and FLAG-ETV1 exogenously expressed in RWPE-1 cells. Antibodies recognizing endogenous ETV4 (oncogenic ETS) and JUND (AP-1 subunit) were used for ChIP in PC3 prostate cancer cells (derived from a bone metastasis). H3K4Me1 and H3K4Me3 are histone marks that correlate with enhancer and promoter regions, respectively [202]. Histone methylation data are shown as an overlay from multiple cell types and is from the ENCODE Project, displayed using the UCSC genome browser [203]. A dashed gray line represents the PTHrP P3 core promoter and a solid gray line represents the potential −54 kb enhancer
EGFR and PTHrP signaling in cancer progression
In the years since its discovery, there has been an extensive study of the role of PTHrP and its contribution to the progression of neoplastic disease. The upregulation of PTHrP in response to signaling by EGFR raises the question as to how its activation might contribute to or antagonize tumor progression. As the story of PTHrP unfolds, it is clear that the answer to that question depends on the context. Several IHC surveys of PTHrP expression in primary breast and NSCLC suggest that expression of the protein is associated with better prognosis [155, 156]. In contrast, PTHrP expression by metastatic tumors within the bone microenvironment facilitates tumor growth and osteolysis. Thus, PTHrP is probably like many other secreted signaling molecules, exhibiting a microenvironment-dependent inhibitory or stimulatory effect on tumor progression.
EGFR-stimulated PTHrP as an autocrine growth factor
Although there has been much work published on non-PTH receptor (PTHR)-mediated effects of PTHrP on cells, there has not been sufficient evidence to establish a well-defined model by which the PTHrP mid-region and c-terminus and nuclear PTHrP would contribute to the progression of most cancer cell types. However, a role for PTHrP signaling mediated by the PTHR in the progression of cancer has been firmly established.
Cells of mesodermal origin such as osteoblasts, chondrocytes, and kidney are generally associated with high levels of PTHR expression [157, 158]. In contrast, most epithelia originating from the ectoderm or endoderm express very low levels of PTHR, but are often sources for PTHrP. Since most cancers originate in epithelia derived from ectoderm or endoderm, it is not surprising that high levels of PTHR are not typically observed in most human carcinomas, which do in fact express high levels of EGFR. Given this reciprocal relationship, it is not expected that activation of PTHrP gene expression by EGFR signaling would necessarily result in autocrine PTHR signaling. Nevertheless, a recent report indicated that treatment with TKI AG1478 decreased PTHrP production in a set of HNSCC lines that expressed both EGFR and PTHR [159]. Treatment of the cells with either EGFR TKI or siRNA to PTHrP inhibited cell growth in vitro and decreased motility and migration [159]. These findings indicate that EGFR-upregulated PTHrP could induce increased proliferation and invasive behaviors in epithelial cancer cells that express relatively low levels of PTHR.
EGFR and PTHR as paracrine targets in the primary tumor microenvironment
An emerging role for EGFR signaling in cancer progression is its control over the expression of other cytokines and growth factors. The most well-developed example of this is the regulation of colony stimulating factor-1 (CSF-1) expression by EGFR in breast cancer cells. The production of CSF-1 differentiates monocytes to macrophages and enhances their survival in the tumor microenvironment. In response to CSF-1, macrophages produce EGF, which induces migration and invasive behaviors in breast cancer cells as well as increases CSF-1 production. Interactions between tumor cells and macrophages promote tumor progression and metastasis [160–162].
In the absence of PTHR on the tumor cell, EGFR-mediated activation of PTHrP may facilitate interactions with the tumor microenvironment. Among the cells present in the tumor stroma are various fibroblasts, resident cells of the particular organ, as well as derivatives from the bone marrow. Fibroblasts also express both EGFR and PTHR [163, 164]. It is well established that tumor-associated fibroblasts facilitate the growth of epithelial tumors. The fibroblasts within a specific tumor appear to be heterogeneous and may have distinct roles in tumor progression [165–167]. A subset of fibroblasts appear to take on a myofibroblastic phenotype that is characterized by the reorganization of the cytoskeleton in a manner similar to contractile cells accompanied by the expression of the smooth muscle actin [168]. Myofibroblasts appear to be a substantial source of growth factors such as stromal derived factor/CXCL12 and VEGF. These factors support carcinoma growth and angiogenesis. Myofibroblasts are responsible for the production of dense acellular type I collagen-rich extracellular matrix that endows the malignancies with a characteristic firmness when palpated [165–167]. EGFR is expressed in myofibroblasts and the activation of its receptor is associated with directed migration and the production of cytokines and growth factors that stimulate angiogenesis [169, 170]. Tissue fibroblasts are known to express modest levels of PTHR [163]. Recent evidence suggests that PTHR expression decreases as tissue fibroblasts undergo myofibroblast differentiation [171]. Activation of PTHR with exogenous PTHrP inhibited myofibroblast differentiation of lung fibroblasts [171]. At this point, the overall impact of either PTHR signaling or EGFR signaling on tumor-associated fibroblasts has not been comprehensively studied; however, these early findings are intriguing and suggest that increased PTHR signaling might inhibit the generation of myofibroblasts in the tumor microenvironment. This would be consistent with PTHrP serving as an inhibitor of growth in primary tumors.
EGFR and PTHrP in cancer-induced bone pathology
Osteolytic bone metastasis: a vicious cycle
A model called the vicious cycle has been developed to explain how cancer cells direct the resident cells of bone to uncouple the physiological linkage between bone matrix destruction and new bone formation in bone metastasis [172]. In the bone, tumor cells produce cytokines and growth factors that engage in paracrine signaling with osteoclasts, cells that breakdown bone matrix, and osteoblasts which are responsible for bone formation [172, 173]. Osteoclast formation is mediated mainly through RANK (receptor activator of nuclear factor β-ligand) and its agonist RANKL (RANK ligand), the latter of which is produced by osteoblasts and bone marrow stromal cells [172, 174]. Osteoblasts also produce a soluble antagonist of RANKL called osteoprotegerin (OPG) [174, 175]. Thus, osteoclast formation is regulated by the balance between RANKL and OPG in the bone microenvironment [172]. In various xenograft models, the neoplastic cells produce several growth factors and cytokines that perturb the RANKL/OPG ratio and increase the number of precursors that can be differentiated to osteoclasts [173, 176, 177]. The osteoclast-mediated resorption of bone causes the release of growth factors embedded in the bone matrix. These matrix-derived growth factors bind and stimulate their cognate receptors on the invasive cancer cell, resulting in increased tumor cell proliferation and production of cytokines that skew the RANKL/OPG ratio toward increased osteoclastogenesis, thereby propagating a vicious cycle of tumor cell proliferation and bone destruction [173, 178].
EGFR and bone
Studies on malignancy-associated hypercalcemia have long established that TGFα increases the formation of bone resorbing osteoclasts in bone marrow cultures and animal models [179, 180]. EGFR is expressed on both chondrocytes and cells of the osteoblast lineage in animals and humans [181]. Further studies related to bone turnover suggest additional roles for EGFR ligands in the pathogenesis of osteolytic lesions. Parathyroid hormone (PTH), the main serum calcium regulator, stimulates a 10- to 20-fold increase in AREG gene transcription and a modest increase in transcription of the TGFα and HB-EGF genes [140, 182]. PTHR, like other serpentine G-protein-coupled receptors, is coupled to proteases (such as ADAM-17) that cleave ErbB receptor ligand precursors and enable the release of mature, soluble ligands [183].
Exogenous EGFR ligands stimulate the proliferation of osteoblasts, inhibit their differentiation, and decrease their mineralization capacity [140]. Four-week-old transgenic mice lacking AREG expression exhibit less trabecular bone in the tibia than do wild-type littermates [140]. EGFR signaling may mediate the impact of PTH on the recruitment and expansion of cells committed to the osteoblast lineage, whereas excessive ligand signaling could prevent these cells from undergoing terminal differentiation and mineralized bone formation [140]. Thus, EGFR signaling regulates differentiation of osteoblasts and this could contribute to cancer-mediated diseases of bone by reducing matrix production.
EGFR ligands and osteolysis
There is growing evidence that EGFR signaling in osteoblasts directly contributes to osteolysis or bone resorption. EGF, TGFα, and MDA-MB-231 cells (which express various ErbB ligands) stimulate bone turnover and osteoclastogenesis in various model systems [179, 180, 184, 185]. This osteoclastogenesis is accompanied by decreased OPG expression and minimal change in RANKL expression by the bone cells [185]. EGFR TKI inhibit CSF-1 and RANKL production from human bone marrow stromal cells and osteoclast formation in vitro [186]. These studies clearly support the concept that EGFR signaling within the osteoblast promotes osteoclastogenesis through perturbation of the RANKL/OPG balance.
Recently, it was found that a bone-seeking clone of MDA-MB-231 cells that overexpress the proteases MMP1 and ADAMTS-1 dramatically increases AREG shedding. Conditioned medium from the MDA-MB-231/ADAMTS-1/MMP1 cells altered the RANKL/OPG balance in a primary mouse bone cell culture and enhanced osteoclastogenesis. This enhanced osteoclastogenesis could be inhibited by EGFR TKI, gefitinib, or by the anti-EGFR antibody, cetuximab. These agents (gefitinib 100 mg/kg daily or cetuximab 100 mg/kg weekly) prevented MDA-MB-231/ADAMTS-1/MMP1 cells from stimulating the formation of osteolytic lesions in the bone of immunocompromised mice containing these cells [187]. These findings suggest that the overexpression of EGFR ligands in the bone microenvironment could drive osteoclastogenesis.
EGFR and PTHrP in osteolysis
In the MDA-MB-231 model of breast cancer metastasis to bone, PTHR signaling is one of the key events in regulating the vicious cycle of breast cancer osteolysis and colonization [188]. MDA-MB-231 cells express PTHrP that stimulates RANKL expression and inhibits OPG expression in cells of the osteoblast lineage [188]. The pattern of PTHrP expression by breast cancers at various stages of progression resembles that displayed by metastasis virulence factors [189]. PTHrP expression is lower in primary breast cancers that ultimately metastasize to bone than in other primary breast tumors. However, PTHrP expression is very high among metastatic tumor cells within the bone microenvironment [156, 190–192]. PTHrP gene expression in these metastatic tumor cells appears to be stimulated by TGFβ released from the bone matrix via osteoclast activity [172, 178].
The signaling between the PTHrP and EGFR systems is not simply directed from the tumor cell to the microenvironment. As indicted above in many epithelial cancer types including the MDA-MB-231 line, autocrine EGFR is coupled to PTHrP gene expression [69, 130, 131, 145]. Knockdown of EGFR inhibited PTHrP gene expression and substantially reduced osteoclastogenesis in vitro as well as osteoltyic growth of MDA-MB-231 cells that were injected into the tibia [134]. A similar autocrine loop drives PTHrP gene expression in lung SCC, and reconstitution of that signaling by ectopic expression of the receptor in EGFR-null cells leads to aggressive osteoltyic lesions when placed in the bone [133]. The EGFR TKI, erlotinib, inhibited osteoltyic factors, PTHrP, IL-8, IL-11, and VEGF, in the pulmonary mucoepidermoid carcinoma cell line NCI-H292, and high doses of the compound (100 mg/kg) also reduced osteolytic growth of these cells after intratibial injection [193]. Blockade of autocrine EGFR stimulation in cancer cells and the accompanying repression of PTHrP gene expression may effectively restore the RANK/OPG balance on the osteoblast lineage and slow the growth of bone metastases.
The impact of EGFR-targeted therapeutics has been surprisingly effective in slowing or eliminating the growth of lung cancer and breast cancer cell line xenografts in the bone of immunocompromised mice. However, all of these studies used an exceedingly high dose (100 mg/kg, daily) of these compounds, and the effect of lower doses was not reported [187, 193]. Also, extensive evaluation of the impact of this high-dose treatment of bones from non-tumor bearing animals was not presented. In contrast, treatment of animals bearing MDA-MB-231 cells in their tibias with a moderate dose (10 mg/kg, daily) of an AREG blocking antibody enhanced tumor growth and produced an increase in osteoclasts in non-tumor bearing mice [134]. Addition of intermediate levels of the AREG antibody or gefitinib to bone marrow cultures containing cancer cells increased osteoclastogenesis [134]. Besides osteoblasts and osteoclasts, bone marrow is composed on many additional cell types that might be influenced by EGFR TKIs or their off-target effects. For example moderate 10 mg/kg doses of erlotinib have recently been reported to increase hematopoietic stem cell mobilization in response to G-CSF in mice [194]. Derivatives of hematopoietic stem cells include osteoclasts, monocytes, myeloid suppressor cells, and megakaryocytes, all of which could influence the growth of cancer cells in the bone [195]. These later findings argue that much more work has to be done to establish the precise effects of EGFR inhibitors on the bone and bone marrow. It is possible that review of data from the large number of lung cancer patients who have been treated with EGFR-targeted therapeutics could provide some insight as to what the long-term effect of pharmacological doses of these compounds is on human bone and bone marrow.
Humoral hypercalcemia of malignancy in lung cancer: a vicious cycle at the endocrine level
During the 1970s and 1980s, SCC of the lung was the most common cause of humoral hypercalcemia [196–198]. Among lung SCC lines capable of producing hypercalcemia in immunocompromised animals, all expressed the receptor and EGFR and TKI reduced PTHrP mRNA levels in vitro [133, 141, 199]. Ectopic expression of EGFR in a receptor-null lung SCC line that expressed ADAM-17 and AREG leads to hypercalcemia when grown on the flank of nude mice [133]. Treatment of mice carrying two different lung SCC lines with 3 days of moderate 10 mg/kg daily oral doses of EGFFR TKI gefitinib was able to reduce calcium levels to within normal levels, as well as substantially reducing circulating PTHrP [141]. These findings suggest that the autocrine EGFR activation of PTHrP in the cancer cell substantially contributes to humoral hypercalcemia of malignancy caused by lung SCC.
Among the lung SCC lines that produced hypercalcemia in mice, the HARA line produced much higher levels of PTHrP mRNA in tumors from hypercalcemic animals than when simply grown in vitro [141]. This observation suggested that an extrinsic factor present in the animals activated PTHrP gene expression. A possible explanation was that factors released from the bone could serve as a positive feedback loop to activate PTHrP gene expression. High levels of calcium were proposed as one of the factors contributing to PTHrP gene expression in breast cancer cells within the bone microenvironment [200]. Subsequently, it was found that in all lung SCC lines studied, PTHrP secretion was increased in response to elevated calcium concentrations [201]. The calcium-sensing receptor gene from the HARA line contained a single nucleotide polymorphism that reduced the receptor’s activation threshold and contributed to increased PTHrP secretion [201]. The calcium-sensing receptor variants may provide a positive feedback loop between PTHrP and calcium that drive hypercalcemia in some lung SCC patients [201]. Thus, in some cases, hypercalcemia may be driven by a vicious cycle, which is induced by PTHrP secretion from the primary tumor that is reinforced by factors like calcium released from the bone.
Final considerations
EGFR is a potent regulator of PTHrP production in cancer cell lines that produce hypercalcemia and osteolytic bone metastases in mice. Can this signaling relationship be exploited in the clinic by using existing EGFR inhibitors, genfitinib, erlotinib and cetuximab, as part of targeted therapies to manage PTHrP-driven humoral hypercalcemia of malignancy and osteolytic bone metastases? From our perspective, EGFR inhibitors appear to hold greater promise as second- or third-line agents for the management of humoral hypercalcemia induced by lung cancer or other squamous carcinomas that express both high levels of EGFR and PTHrP. Hypercalcemia treatments tend to be transient interventions provided as part of palliative care, which may provide opportunities for the use of current EGFR-targeted therapeutics. In considering bone metastases, many studies have indicated that there are multiple redundant pathways by which cancer cells stimulate cells of the osteoblast lineage to recruit and activate osteoclasts, indicating PTHrP may not be central to this pathogenesis for many cancers. Additionally, interventions for bone metastasis would be used over longer time frames where drug resistance can emerge and off-target effects can have devastating consequences. Given the recent findings that EGFR signaling appears to limit hematopoietic stem cell recruitment in mice, we believe much more basic research needs to be done to determine how long-term blockade of the pathway will impact bone physiology before agents like genfitinib, erlotinib, and cetuximab could be considered to target bone metastases. So, we conclude that even though the intersection of EGFR and PTHrP pathways suggests novel avenues for clinical interventions in the area of cancer-mediated bone pathologies, there remain warning signs on the road ahead.
References
Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–37.
Stern DF, Heffernan PA, Weinberg RA. p185, a product of the neu proto-oncogene, is a receptorlike protein associated with tyrosine kinase activity. Mol Cell Biol. 1986;6(5):1729–40.
Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci USA. 2010;107(17):7692–7.
Wilson KJ, Gilmore JL, Foley J, Lemmon MA, Riese DJ 2nd. Functional selectivity of EGF family peptide growth factors: implications for cancer. Pharmacol Ther. 2009;122(1):1–8.
Daub H, Weiss FU, Wallasch C, Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379(6565):557–60.
Iwamoto R, Mekada E. Heparin-binding EGF-like growth factor: a juxtacrine growth factor. Cytokine Growth Factor Rev. 2000;11(4):335–44.
Prince RN, Schreiter ER, Zou P, Wiley HS, Ting AY, Lee RT, Lauffenburger DA. The heparin-binding domain of HB-EGF mediates localization to sites of cell–cell contact and prevents HB-EGF proteolytic release. J Cell Sci. 2010;123(Pt 13):2308–18.
Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164(5):769–79.
Tousseyn T, Jorissen E, Reiss K, Hartmann D. (Make) stick and cut loose—disintegrin metalloproteases in development and disease. Birth Defects Res C Embryo Today. 2006;78(1):24–46.
Chokki M, Eguchi H, Hamamura I, Mitsuhashi H, Kamimura T. Human airway trypsin-like protease induces amphiregulin release through a mechanism involving protease-activated receptor-2-mediated ERK activation and TNF alpha-converting enzyme activity in airway epithelial cells. Febs J. 2005;272(24):6387–99.
Edwin F, Wiepz GJ, Singh R, Peet CR, Chaturvedi D, Bertics PJ, Patel TB. A historical perspective of the EGF receptor and related systems. Methods Mol Biol. 2006;327:1–24.
Harris RC, Chung E, Coffey RJ. EGF receptor ligands. Exp Cell Res. 2003;284(1):2–13.
Riese DJ 2nd, Stern DF. Specificity within the EGF family/ErbB receptor family signaling network. BioEssays. 1998;20(1):41–8.
Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315(4):638–48.
Riese DJ 2nd, Gallo RM, Settleman J. Mutational activation of ErbB family receptor tyrosine kinases: insights into mechanisms of signal transduction and tumorigenesis. BioEssays. 2007;29(6):558–65.
Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;2005(1):1–13.
Roepstorff K, Grandal MV, Henriksen L, Knudsen SL, Lerdrup M, Grovdal L, Willumsen BM, van Deurs B. Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic. 2009;10(8):1115–27.
Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12(2):104–17.
Bublil EM, Yarden Y. The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr Opin Cell Biol. 2007;19(2):124–34.
Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7(7):505–16.
Earp HS, Dawson TL, Li X, Yu H. Heterodimerization and functional interaction between EGF receptor family members: a new signaling paradigm with implications for breast cancer research. Breast Cancer Res Treat. 1995;35(1):115–32.
Wada T, Qian XL, Greene MI. Intermolecular association of the p185neu protein and EGF receptor modulates EGF receptor function. Cell. 1990;61(7):1339–47.
Wang Z, Zhang L, Yeung TK, Chen X. Endocytosis deficiency of epidermal growth factor (EGF) receptor-ErbB2 heterodimers in response to EGF stimulation. Mol Biol Cell. 1999;10(5):1621–36.
Hendriks BS, Wiley HS, Lauffenburger D. HER2-mediated effects on EGFR endosomal sorting: analysis of biophysical mechanisms. Biophys J. 2003;85(4):2732–45.
Jones JT, Akita RW, Sliwkowski MX. Binding specificities and affinities of EGF domains for ErbB receptors. FEBS Lett. 1999;447(2–3):227–31.
Liu W, Volpe MA, Zscheppang K, Nielsen HC, Dammann CE. ErbB4 regulates surfactant synthesis and proliferation in adult rat pulmonary epithelial cells. Exp Lung Res. 2009;35(1):29–47.
Zscheppang K, Korenbaum E, Bueter W, Ramadurai SM, Nielsen HC, Dammann CE. ErbB receptor dimerization, localization, and co-localization in mouse lung type II epithelial cells. Pediatr Pulmonol. 2006;41(12):1205–12.
Hatakeyama M, Yumoto N, Yu X, Shirouzu M, Yokoyama S, Konagaya A. Transformation potency of ErbB heterodimer signaling is determined by B-Raf kinase. Oncogene. 2004;23(29):5023–31.
Frolov A, Schuller K, Tzeng CW, Cannon EE, Ku BC, Howard JH, Vickers SM, Heslin MJ, Buchsbaum DJ, Arnoletti JP. ErbB3 expression and dimerization with EGFR influence pancreatic cancer cell sensitivity to erlotinib. Cancer Biol Ther. 2007;6(4):548–54.
Liles JS, Arnoletti JP, Tzeng CW, Howard JH, Kossenkov AV, Kulesza P, Heslin MJ, Frolov A. ErbB3 expression promotes tumorigenesis in pancreatic adenocarcinoma. Cancer Biol Ther. 2010;10(6):555–63.
Boerner JL, Danielsen A, Maihle NJ. Ligand-independent oncogenic signaling by the epidermal growth factor receptor: v-ErbB as a paradigm. Exp Cell Res. 2003;284(1):111–21.
Downward J, Yarden Y, Mayes E, Scrace G, Totty N, Stockwell P, Ullrich A, Schlessinger J, Waterfield MD. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307(5951):521–7.
Kris RM, Lax I, Gullick W, Waterfield MD, Ullrich A, Fridkin M, Schlessinger J. Antibodies against a synthetic peptide as a probe for the kinase activity of the avian EGF receptor and v-erbB protein. Cell. 1985;40(3):619–25.
Ekstrand AJ, Longo N, Hamid ML, Olson JJ, Liu L, Collins VP, James CD. Functional characterization of an EGF receptor with a truncated extracellular domain expressed in glioblastomas with EGFR gene amplification. Oncogene. 1994;9(8):2313–20.
Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91(16):7727–31.
Ji H, Zhao X, Yuza Y, Shimamura T, Li D, Protopopov A, Jung BL, McNamara K, Xia H, Glatt KA, Thomas RK, Sasaki H, Horner JW, Eck M, Mitchell A, Sun Y, Al-Hashem R, Bronson RT, Rabindran SK, Discafani CM, Maher E, Shapiro GI, Meyerson M, Wong KK. Epidermal growth factor receptor variant III mutations in lung tumorigenesis and sensitivity to tyrosine kinase inhibitors. Proc Natl Acad Sci USA. 2006;103(20):7817–22.
Ding H, Shannon P, Lau N, Wu X, Roncari L, Baldwin RL, Takebayashi H, Nagy A, Gutmann DH, Guha A. Oligodendrogliomas result from the expression of an activated mutant epidermal growth factor receptor in a RAS transgenic mouse astrocytoma model. Cancer Res. 2003;63(5):1106–13.
Huang PH, Miraldi ER, Xu AM, Kundukulam VA, Del Rosario AM, Flynn RA, Cavenee WK, Furnari FB, White FM. Phosphotyrosine signaling analysis of site-specific mutations on EGFRvIII identifies determinants governing glioblastoma cell growth. Mol BioSyst. 2010;6(7):1227–37.
Narita Y, Nagane M, Mishima K, Huang HJ, Furnari FB, Cavenee WK. Mutant epidermal growth factor receptor signaling down-regulates p27 through activation of the phosphatidylinositol 3-kinase/Akt pathway in glioblastomas. Cancer Res. 2002;62(22):6764–9.
Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305(5687):1163–7.
Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, Kupfer D, Wilson R, Kris M, Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101(36):13306–11.
Sharma SV, Settleman J. ErbBs in lung cancer. Exp Cell Res. 2009;315(4):557–71.
Sharma SV, Fischbach MA, Haber DA, Settleman J. “Oncogenic shock”: explaining oncogene addiction through differential signal attenuation. Clin Cancer Res. 2006;12(14 Pt 2):4392s–5s.
Choi SH, Mendrola JM, Lemmon MA. EGF-independent activation of cell-surface EGF receptors harboring mutations found in gefitinib-sensitive lung cancer. Oncogene. 2007;26(11):1567–76.
Filosto S, Becker CR, Goldkorn T. Cigarette smoke induces aberrant EGF receptor activation that mediates lung cancer development and resistance to tyrosine kinase inhibitors. Mol Cancer Ther. 2012;11(4):795–804.
Khan EM, Lanir R, Danielson AR, Goldkorn T. Epidermal growth factor receptor exposed to cigarette smoke is aberrantly activated and undergoes perinuclear trafficking. FASEB J. 2008;22(3):910–7.
Filosto S, Khan EM, Tognon E, Becker C, Ashfaq M, Ravid T, Goldkorn T. EGF receptor exposed to oxidative stress acquires abnormal phosphorylation and aberrant activated conformation that impairs canonical dimerization. PLoS ONE. 2011;6(8):e23240.
Khan EM, Heidinger JM, Levy M, Lisanti MP, Ravid T, Goldkorn T. Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking. J Biol Chem. 2006;281(20):14486–93.
Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2008;314(17):3093–106.
Ceresa BP. Regulation of EGFR endocytic trafficking by rab proteins. Histol Histopathol. 2006;21(9):987–93.
Eden ER, White IJ, Futter CE. Down-regulation of epidermal growth factor receptor signalling within multivesicular bodies. Biochem Soc Trans. 2009;37(Pt 1):173–7.
Wiley HS. Trafficking of the ErbB receptors and its influence on signaling. Exp Cell Res. 2003;284(1):78–88.
Elenius K, Paul S, Allison G, Sun J, Klagsbrun M. Activation of HER4 by heparin-binding EGF-like growth factor stimulates chemotaxis but not proliferation. Embo J. 1997;16(6):1268–78.
Shing Y, Christofori G, Hanahan D, Ono Y, Sasada R, Igarashi K, Folkman J. Betacellulin: a mitogen from pancreatic beta cell tumors. Science. 1993;259(5101):1604–7.
Higashiyama S, Abraham JA, Miller J, Fiddes JC, Klagsbrun M. A heparin-binding growth factor secreted by macrophage-like cells that is related to EGF. Science. 1991;251(4996):936–9.
Sweeney C, Lai C, Riese DJ 2nd, Diamonti AJ, Cantley LC, Carraway KL 3rd. Ligand discrimination in signaling through an ErbB4 receptor homodimer. J Biol Chem. 2000;275(26):19803–7.
Waterman H, Sabanai I, Geiger B, Yarden Y. Alternative intracellular routing of ErbB receptors may determine signaling potency. J Biol Chem. 1998;273(22):13819–27.
French AR, Sudlow GP, Wiley HS, Lauffenburger DA. Postendocytic trafficking of epidermal growth factor–receptor complexes is mediated through saturable and specific endosomal interactions. J Biol Chem. 1994;269(22):15749–55.
Reddy CC, Niyogi SK, Wells A, Wiley HS, Lauffenburger DA. Engineering epidermal growth factor for enhanced mitogenic potency. Nat Biotechnol. 1996;14(13):1696–9.
Kochupurakkal BS, Harari D, Di-Segni A, Maik-Rachline G, Lyass L, Gur G, Kerber G, Citri A, Lavi S, Eilam R, Chalifa-Caspi V, Eshhar Z, Pikarsky E, Pinkas-Kramarski R, Bacus SS, Yarden Y. Epigen, the last ligand of ErbB receptors, reveals intricate relationships between affinity and mitogenicity. J Biol Chem. 2005;280(9):8503–12.
Strachan L, Murison JG, Prestidge RL, Sleeman MA, Watson JD, Kumble KD. Cloning and biological activity of epigen, a novel member of the epidermal growth factor superfamily. J Biol Chem. 2001;276(21):18265–71.
Sasaki E, Arakawa T, Fujiwara Y, Kawada N, Fukuda T, Higuchi K, Komurasaki T, Kobayashi K. Epiregulin stimulates proliferation of rabbit gastric cells in primary culture through autophosphorylation of the epidermal growth factor receptor. Eur J Pharmacol. 1997;338(3):253–8.
Shelly M, Pinkas-Kramarski R, Guarino BC, Waterman H, Wang LM, Lyass L, Alimandi M, Kuo A, Bacus SS, Pierce JH, Andrews GC, Yarden Y. Epiregulin is a potent pan-ErbB ligand that preferentially activates heterodimeric receptor complexes. J Biol Chem. 1998;273(17):10496–505.
Shoyab M, McDonald VL, Bradley JG, Todaro GJ. Amphiregulin: a bifunctional growth-modulating glycoprotein produced by the phorbol 12-myristate 13-acetate-treated human breast adenocarcinoma cell line MCF-7. Proc Natl Acad Sci USA. 1988;85(17):6528–32.
Barnard JA, Graves-Deal R, Pittelkow MR, DuBois R, Cook P, Ramsey GW, Bishop PR, Damstrup L, Coffey RJ. Auto- and cross-induction within the mammalian epidermal growth factor-related peptide family. J Biol Chem. 1994;269(36):22817–22.
Cook PW, Mattox PA, Keeble WW, Pittelkow MR, Plowman GD, Shoyab M, Adelman JP, Shipley GD. A heparin sulfate-regulated human keratinocyte autocrine factor is similar or identical to amphiregulin. Mol Cell Biol. 1991;11(5):2547–57.
Neelam B, Richter A, Chamberlin SG, Puddicombe SM, Wood L, Murray MB, Nandagopal K, Niyogi SK, Davies DE. Structure–function studies of ligand-induced epidermal growth factor receptor dimerization. Biochemistry. 1998;37(14):4884–91.
Adam R, Drummond DR, Solic N, Holt SJ, Sharma RP, Chamberlin SG, Davies DE. Modulation of the receptor binding affinity of amphiregulin by modification of its carboxyl terminal tail. Biochim Biophys Acta. 1995;1266(1):83–90.
Gilmore JL, Scott JA, Bouizar Z, Robling A, Pitfield SE, Riese DJ, 2nd, Foley J. Amphiregulin-EGFR signaling regulates PTHrP gene expression in breast cancer cells. Breast Cancer Res Treat. 2008;110(3):493–505.
Stern KA, Place TL, Lill NL. EGF and amphiregulin differentially regulate Cbl recruitment to endosomes and EGF receptor fate. Biochem J. 2008;410(3):585–94.
Willmarth NE, Baillo A, Dziubinski ML, Wilson K, Riese DJ 2nd, Ethier SP. Altered EGFR localization and degradation in human breast cancer cells with an amphiregulin/EGFR autocrine loop. Cell Signal. 2009;21(2):212–9.
Willmarth NE, Ethier SP. Autocrine and juxtacrine effects of amphiregulin on the proliferative, invasive, and migratory properties of normal and neoplastic human mammary epithelial cells. J Biol Chem. 2006;281(49):37728–37.
Chung E, Graves-Deal R, Franklin JL, Coffey RJ. Differential effects of amphiregulin and TGF-alpha on the morphology of MDCK cells. Exp Cell Res. 2005;309(1):149–60.
Baldys A, Gooz M, Morinelli TA, Lee MH, Raymond JR Jr, Luttrell LM, Raymond JR Sr. Essential role of c-Cbl in amphiregulin-induced recycling and signaling of the endogenous epidermal growth factor receptor. Biochemistry. 2009;48(7):1462–73.
Nishi E, Klagsbrun M. Heparin-binding epidermal growth factor-like growth factor (HB-EGF) is a mediator of multiple physiological and pathological pathways. Growth Factors. 2004;22(4):253–60.
Shoyab M, Plowman GD, McDonald VL, Bradley JG, Todaro GJ. Structure and function of human amphiregulin: a member of the epidermal growth factor family. Science. 1989;243(4894 Pt 1):1074–6.
Petersen I. The morphological and molecular diagnosis of lung cancer. Dtsch Arztebl Int. 2011;108(31–32):525–31.
Bryant A, Cerfolio RJ. Differences in epidemiology, histology, and survival between cigarette smokers and never-smokers who develop non-small cell lung cancer. Chest (Comparative Study). 2007;132(1):185–92.
Ahn MJ, Lee J, Park YH, Ahn JS, Ziogas A, Zell JA, Park K, Ou SH. Korean ethnicity as compared with white ethnicity is an independent favorable prognostic factor for overall survival in non-small cell lung cancer before and after the oral epidermal growth factor receptor tyrosine kinase inhibitor era. J Thoracic Oncol Off Publ Int Assoc Study Lung Cancer (Comparative Study). 2010;5(8):1185–96.
Toh CK, Gao F, Lim WT, Leong SS, Fong KW, Yap SP, Hsu AA, Eng P, Koong HN, Thirugnanam A, Tan EH. Never-smokers with lung cancer: epidemiologic evidence of a distinct disease entity. J Clin Oncol Off J Am Soc Clin Oncol (Multicenter Study). 2006;24(15):2245–51.
Santoro IL, Ramos RP, Franceschini J, Jamnik S, Fernandes AL. Non-small cell lung cancer in never smokers: a clinical entity to be identified. Clinics (Sao Paulo). 2011;66(11):1873–7.
Subramanian J, Govindan R. Lung cancer in ‘Never-smokers’: a unique entity. Oncology (Williston Park). 2010;24(1):29–35 (Review).
Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat Rev Cancer. 2007;7(10):778–90. (Review).
Wakelee HA, Chang ET, Gomez SL, Keegan TH, Feskanich D, Clarke CA, Holmberg L, Yong LC, Kolonel LN, Gould MK, West DW. Lung cancer incidence in never smokers. J Clin Oncol. 2007;25(5):472–8.
Clement-Duchene C, Vignaud JM, Stoufflet A, Bertrand O, Gislard A, Thiberville L, Grosdidier G, Martinet Y, Benichou J, Hainaut P, Paris C. Characteristics of never smoker lung cancer including environmental and occupational risk factors. Lung Cancer (Research Support, Non-U.S. Gov’t). 2010;67(2):144–50.
Hirsch FR, Varella-Garcia M, Bunn PA Jr, Di Maria MV, Veve R, Bremmes RM, Baron AE, Zeng C, Franklin WA. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21(20):3798–807.
Rusch V, Baselga J, Cordon-Cardo C, Orazem J, Zaman M, Hoda S, McIntosh J, Kurie J, Dmitrovsky E. Differential expression of the epidermal growth factor receptor and its ligands in primary non-small cell lung cancers and adjacent benign lung. Cancer Res. 1993;53(10 Suppl):2379–85.
Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–15.
Nicholson RI, McClelland RA, Gee JM, Manning DL, Cannon P, Robertson JF, Ellis IO, Blamey RW. Epidermal growth factor receptor expression in breast cancer: association with response to endocrine therapy. Breast Cancer Res Treat. 1994;29(1):117–25.
Kim HR, Shim HS, Chung JH, Lee YJ, Hong YK, Rha SY, Kim SH, Ha SJ, Kim SK, Chung KY, Soo R, Kim JH, Cho BC. Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer (Research Support, Non-U.S. Gov’t). 2012;118(3):729–39.
Rosell R, Taron M, Reguart N, Isla D, Moran T. Epidermal growth factor receptor activation: how exon 19 and 21 mutations changed our understanding of the pathway. Clin Cancer Res. 2006;12(24):7222–31.
Zhang J, Iwanaga K, Choi KC, Wislez M, Raso MG, Wei W, Wistuba II, Kurie JM. Intratumoral epiregulin is a marker of advanced disease in non-small cell lung cancer patients and confers invasive properties on EGFR-mutant cells. Cancer Prev Res (Phila). 2008;1(3):201–7.
Masago K, Fujita S, Hatachi Y, Fukuhara A, Sakuma K, Ichikawa M, Kim YH, Mio T, Mishima M. Clinical significance of pretreatment serum amphiregulin and transforming growth factor-alpha, and an epidermal growth factor receptor somatic mutation in patients with advanced non-squamous, non-small cell lung cancer. Cancer Sci. 2008;99(11):2295–301.
Kakiuchi S, Daigo Y, Ishikawa N, Furukawa C, Tsunoda T, Yano S, Nakagawa K, Tsuruo T, Kohno N, Fukuoka M, Sone S, Nakamura Y. Prediction of sensitivity of advanced non-small cell lung cancers to gefitinib (Iressa, ZD1839). Hum Mol Genet. 2004;13(24):3029–43.
Yonesaka K, Zejnullahu K, Lindeman N, Homes AJ, Jackman DM, Zhao F, Rogers AM, Johnson BE, Janne PA. Autocrine production of amphiregulin predicts sensitivity to both gefitinib and cetuximab in EGFR wild-type cancers. Clin Cancer Res. 2008;14(21):6963–73.
Vollebergh MA, Kappers I, Klomp HM, Buning-Kager JC, Korse CM, Hauptmann M, de Visser KE, van den Heuvel MM, Linn SC. Ligands of epidermal growth factor receptor and the insulin-like growth factor family as serum biomarkers for response to epidermal growth factor receptor inhibitors in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2010;5(12):1939–48.
Szabo B, Nelhubel GA, Karpati A, Kenessey I, Jori B, Szekely C, Petak I, Lotz G, Hegedus Z, Hegedus B, Fule T, Dome B, Timar J, Tovari J. Clinical significance of genetic alterations and expression of epidermal growth factor receptor (EGFR) in head and neck squamous cell carcinomas. Oral Oncol. 2011;47(6):487–96.
Rubin Grandis J, Melhem MF, Gooding WE, Day R, Holst VA, Wagener MM, Drenning SD, Tweardy DJ. Levels of TGF-alpha and EGFR protein in head and neck squamous cell carcinoma and patient survival. J Natl Cancer Inst. 1998;90(11):824–32.
Silva SD, Cunha IW, Younes RN, Soares FA, Kowalski LP, Graner E. ErbB receptors and fatty acid synthase expression in aggressive head and neck squamous cell carcinomas. Oral Dis. 2010;16(8):774–80.
Keller J, Shroyer KR, Batajoo SK, Zhao HL, Dong LM, Hayman MJ, Chan EL. Combination of phosphorylated and truncated EGFR correlates with higher tumor and nodal stage in head and neck cancer. Cancer Invest. 2010;28(10):1054–62.
Sok JC, Coppelli FM, Thomas SM, Lango MN, Xi S, Hunt JL, Freilino ML, Graner MW, Wikstrand CJ, Bigner DD, Gooding WE, Furnari FB, Grandis JR. Mutant epidermal growth factor receptor (EGFRvIII) contributes to head and neck cancer growth and resistance to EGFR targeting. Clin Cancer Res. 2006;12(17):5064–73.
Tinhofer I, Klinghammer K, Weichert W, Knodler M, Stenzinger A, Gauler T, Budach V, Keilholz U. Expression of amphiregulin and EGFRvIII affect outcome of patients with squamous cell carcinoma of the head and neck receiving cetuximab–docetaxel treatment. Clin Cancer Res. 2011;17(15):5197–204.
Christensen ME, Therkildsen MH, Poulsen SS, Bretlau P. Immunoreactive transforming growth factor alpha and epidermal growth factor in oral squamous cell carcinomas. J Pathol. 1993;169(3):323–8.
Tsai ST, Yang KY, Jin YT, Lin YC, Chang MT, Wu LW. Amphiregulin as a tumor promoter for oral squamous cell carcinoma: involvement of cyclooxygenase 2. Oral Oncol. 2006;42(4):381–90.
Zhang S, Chen J, Jiang H, Ma H, Yang B. Anti-epidermal growth factor receptor therapy for advanced head and neck squamous cell carcinoma: a meta-analysis. Eur J Clin Pharmacol. 2012;68(5):561–9.
Zellweger T, Ninck C, Bloch M, Mirlacher M, Koivisto PA, Helin HJ, Mihatsch MJ, Gasser TC, Bubendorf L. Expression patterns of potential therapeutic targets in prostate cancer. Int J Cancer. 2005;113(4):619–28.
Hernes E, Fossa SD, Berner A, Otnes B, Nesland JM. Expression of the epidermal growth factor receptor family in prostate carcinoma before and during androgen-independence. Br J Cancer. 2004;90(2):449–54.
Bostwick DG, Qian J, Maihle NJ. Amphiregulin expression in prostatic intraepithelial neoplasia and adenocarcinoma: a study of 93 cases. Prostate. 2004;58(2):164–8.
Bartlett JM, Brawley D, Grigor K, Munro AF, Dunne B, Edwards J. Type I receptor tyrosine kinases are associated with hormone escape in prostate cancer. J Pathol. 2005;205(4):522–9.
Di Lorenzo G, Tortora G, D’Armiento FP, De Rosa G, Staibano S, Autorino R, D’Armiento M, De Laurentiis M, De Placido S, Catalano G, Bianco AR, Ciardiello F. Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen-independence in human prostate cancer. Clin Cancer Res. 2002;8(11):3438–44.
DeHaan AM, Wolters NM, Keller ET, Ignatoski KM. EGFR ligand switch in late stage prostate cancer contributes to changes in cell signaling and bone remodeling. Prostate. 2009;69(5):528–37.
Scher HI, Sarkis A, Reuter V, Cohen D, Netto G, Petrylak D, Lianes P, Fuks Z, Mendelsohn J, Cordon-Cardo C. Changing pattern of expression of the epidermal growth factor receptor and transforming growth factor alpha in the progression of prostatic neoplasms. Clin Cancer Res. 1995;1(5):545–50.
Myers RB, Kudlow JE, Grizzle WE. Expression of transforming growth factor-alpha, epidermal growth factor and the epidermal growth factor receptor in adenocarcinoma of the prostate and benign prostatic hyperplasia. Mod Pathol. 1993;6(6):733–7.
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–74.
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6.
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009.
Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23.
Hennessy BT, Gonzalez-Angulo AM, Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J, Sahin A, Agarwal R, Joy C, Liu W, Stivers D, Baggerly K, Carey M, Lluch A, Monteagudo C, He X, Weigman V, Fan C, Palazzo J, Hortobagyi GN, Nolden LK, Wang NJ, Valero V, Gray JW, Perou CM, Mills GB. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69(10):4116–24.
Korsching E, Jeffrey SS, Meinerz W, Decker T, Boecker W, Buerger H. Basal carcinoma of the breast revisited: an old entity with new interpretations. J Clin Pathol. 2008;61(5):553–60.
Burness ML, Grushko TA, Olopade OI. Epidermal growth factor receptor in triple-negative and basal-like breast cancer: promising clinical target or only a marker? Cancer J. 2010;16(1):23–32.
Viale G, Rotmensz N, Maisonneuve P, Bottiglieri L, Montagna E, Luini A, Veronesi P, Intra M, Torrisi R, Cardillo A, Campagnoli E, Goldhirsch A, Colleoni M. Invasive ductal carcinoma of the breast with the “triple-negative” phenotype: prognostic implications of EGFR immunoreactivity. Breast Cancer Res Treat. 2009;116(2):317–28.
Kenny PA, Bissell MJ. Targeting TACE-dependent EGFR ligand shedding in breast cancer. J Clin Invest. 2007;117(2):337–45.
Foley J, Nickerson NK, Nam S, Allen KT, Gilmore JL, Nephew KP, Riese DJ 2nd. EGFR signaling in breast cancer: bad to the bone. Semin Cell Dev Biol. 2010;21(9):951–60.
Clark GM, Sledge GW Jr, Osborne CK, McGuire WL. Survival from first recurrence: relative importance of prognostic factors in 1,015 breast cancer patients. J Clin Oncol. 1987;5(1):55–61.
Cramer SD, Peehl DM, Edgar MG, Wong ST, Deftos LJ, Feldman D. Parathyroid hormone-related protein (PTHrP) is an epidermal growth factor-regulated secretory product of human prostatic epithelial cells. Prostate. 1996;29(1):20–9.
Sebag M, Henderson J, Goltzman D, Kremer R. Regulation of parathyroid hormone-related peptide production in normal human mammary epithelial cells in vitro. Am J Physiol. 1994;267(3 Pt 1):C723–30.
Ferrari SL, Rizzoli R, Bonjour JP. Effects of epidermal growth factor on parathyroid hormone-related protein production by mammary epithelial cells. J Bone Miner Res. 1994;9(5):639–44.
Ferrari SL, Rizzoli R, Bonjour JP. Parathyroid hormone-related protein production by primary cultures of mammary epithelial cells. J Cell Physiol. 1992;150(2):304–11.
Henderson JE, Kremer R, Rhim JS, Goltzman D. Identification and functional characterization of adenylate cyclase-linked receptors for parathyroid hormone-like peptides on immortalized human keratinocytes. Endocrinology. 1992;130(1):449–57.
Cho YM, Lewis DA, Koltz PF, Richard V, Gocken TA, Rosol TJ, Konger RL, Spandau DF, Foley J. Regulation of parathyroid hormone-related protein gene expression by epidermal growth factor-family ligands in primary human keratinocytes. J Endocrinol. 2004;181(1):179–90.
Lorch G, Gilmore JL, Koltz PF, Gonterman RM, Laughner R, Lewis DA, Konger RL, Nadella KS, Toribio RE, Rosol TJ, Foley J. Inhibition of epidermal growth factor receptor signalling reduces hypercalcaemia induced by human lung squamous-cell carcinoma in athymic mice. Br J Cancer. 2007;97(2):183–93.
Gilmore JL, Scott JA, Bouizar Z, Robling A, Pitfield SE, Riese DJ 2nd, Foley J. Amphiregulin-EGFR signaling regulates PTHrP gene expression in breast cancer cells. Breast Cancer Res Treat. 2008;110(3):493–505.
Gilmore JL, Gonterman RM, Menon K, Lorch G, Riese DJ 2nd, Robling A, Foley J. Reconstitution of amphiregulin-epidermal growth factor receptor signaling in lung squamous cell carcinomas activates PTHrP gene expression and contributes to cancer-mediated diseases of the bone. Mol Cancer Res. 2009;7(10):1714–28.
Nickerson NK, Mohammad KS, Gilmore JL, Crismore E, Bruzzaniti A, Guise TA, Foley J. Decreased autocrine EGFR signaling in metastatic breast cancer cells inhibits tumor growth in bone and mammary fat pad. PLoS ONE. 2012;7(1):e30255.
Piepkorn M, Pittelkow MR, Cook PW. Autocrine regulation of keratinocytes: the emerging role of heparin-binding, epidermal growth factor-related growth factors. J Invest Dermatol. 1998;111(5):715–21.
Pittelkow MR, Cook PW, Shipley GD, Derynck R, Coffey RJ Jr. Autonomous growth of human keratinocytes requires epidermal growth factor receptor occupancy. Cell Growth Differ. 1993;4(6):513–21.
Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11(12):1444–50.
Katoh Y, Katoh M. Canonical WNT signaling pathway and human AREG. Int J Mol Med. 2006;17(6):1163–6.
Ciarloni L, Mallepell S, Brisken C. Amphiregulin is an essential mediator of estrogen receptor alpha function in mammary gland development. Proc Natl Acad Sci USA. 2007;104(13):5455–60.
Qin L, Partridge NC. Stimulation of amphiregulin expression in osteoblastic cells by parathyroid hormone requires the protein kinase A and cAMP response element-binding protein signaling pathway. J Cell Biochem. 2005;96(3):632–40.
Lorch G, Gilmore JL, Koltz PF, Gonterman RM, Laughner R, Lewis DA, Konger RL, Nadella KS, Toribio RE, Rosol TJ, Foley J. Inhibition of epidermal growth factor receptor signaling reduces hypercalcemia induced by human lung squamous-cell carcinoma in athymic mice. Br J Cancer. 2007;97(2):183–93.
Aklilu F, Gladu J, Goltzman D, Rabbani SA. Role of mitogen-activated protein kinases in the induction of parathyroid hormone-related peptide. Cancer Res. 2000;60(6):1753–60.
Kakonen SM, Selander KS, Chirgwin JM, Yin JJ, Burns S, Rankin WA, Grubbs BG, Dallas M, Cui Y, Guise TA. Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J Biol Chem. 2002;277(27):24571–8.
Furugaki K, Moriya Y, Iwai T, Yorozu K, Yanagisawa M, Kondoh K, Fujimoto-Ohuchi K, Mori K. Erlotinib inhibits osteolytic bone invasion of human non-small-cell lung cancer cell line NCI-H292. Clin Exp Metastasis. 2011;28(7):649–59.
Richard V, Rosol TJ, Foley J. PTHrP gene expression in cancer: do all paths lead to Ets? Crit Rev Eukaryot Gene Expr. 2005;15(2):115–32.
Hollenhorst PC, Ferris MW, Hull MA, Chae H, Kim S, Graves BJ. Oncogenic ETS proteins mimic activated RAS/MAPK signaling in prostate cells. Genes Dev (Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t). 2011;25(20):2147–57.
Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8(7):497–511.
Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, Mehra R, Chinnaiyan AM. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68(1):73–80.
Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, Yu J, Wang L, Montie JE, Rubin MA, Pienta KJ, Roulston D, Shah RB, Varambally S, Mehra R, Chinnaiyan AM. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448(7153):595–9.
Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10(2):177–88.
Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21(15):1882–94.
Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Ann Rev Biochem. 2011;80:437–71.
Watabe T, Yoshida K, Shindoh M, Kaya M, Fujikawa K, Sato H, Seiki M, Ishii S, Fujinaga K. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int J Cancer (Journal international du cancer; Research Support, Non-U.S. Gov’t). 1998;77(1):128–37.
A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol (Research Support, N.I.H., Extramural). 2011;9(4):e1001046.
Montgrain PR, Deftos LJ, Arenberg D, Tipps A, Quintana R, Carskadon S, Hastings RH. Prognostic implications of parathyroid hormone-related protein in males and females with non-small-cell lung cancer. Clin Lung Cancer. 2011;12(3):197–205.
Henderson M, Danks J, Moseley J, Slavin J, Harris T, McKinlay M, Hopper J, Martin T. Parathyroid hormone-related protein production by breast cancers, improved survival, and reduced bone metastases. J Natl Cancer Inst. 2001;93(3):234–7.
Lee K, Deeds JD, Bond AT, Juppner H, Abou-Samra AB, Segre GV. In situ localization of PTH/PTHrP receptor mRNA in the bone of fetal and young rats. Bone. 1993;14(3):341–5.
Lee K, Deeds JD, Segre GV. Expression of parathyroid hormone-related peptide and its receptor messenger ribonucleic acids during fetal development of rats. Endocrinology. 1995;136(2):453–63.
Yamada T, Tsuda M, Ohba Y, Kawaguchi H, Totsuka Y, Shindoh M. PTHrP promotes malignancy of human oral cancer cell downstream of the EGFR signaling. Biochem Biophys Res Commun. 2008;368(3):575–81.
Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65(12):5278–83.
Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 2009;69(24):9498–506.
Leek RD, Harris AL. Tumor-associated macrophages in breast cancer. J Mammary Gland Biol Neoplasia. 2002;7(2):177–89.
Hanafin NM, Chen TC, Heinrich G, Segre GV, Holick MF. Cultured human fibroblasts and not cultured human keratinocytes express a PTH/PTHrP receptor mRNA. J Invest Dermatol. 1995;105(1):133–7.
Kawamoto T, Nishi M, Takahashi K, Nishiyama T, Sato JD, Taniguchi S. Stimulation by transforming growth factor-beta of epidermal growth factor-dependent growth of aged human fibro-blasts: recovery of high affinity EGE receptors and growth stimulation by EGF. In Vitro Cell Dev Biol. 1989;25(10):965–70.
Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, Wheeler M, Spitler J, Rowley DR. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res. 2003;9(13):4792–801.
Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432(7015):332–7.
Littlepage LE, Egeblad M, Werb Z. Coevolution of cancer and stromal cellular responses. Cancer Cell. 2005;7(6):499–500.
Martin M, Pujuguet P, Martin F. Role of stromal myofibroblasts infiltrating colon cancer in tumor invasion. Pathol Res Pract. 1996;192(7):712–7.
Yoo J, Perez CE, Nie W, Edwards RA, Sinnett-Smith J, Rozengurt E. TNF-alpha induces upregulation of EGFR expression and signaling in human colonic myofibroblasts. Am J Physiol Gastrointest Liver Physiol. 2012;302(8):G805–14.
Konstantinopoulos PA, Vandoros GP, Karamouzis MV, Gkermpesi M, Sotiropoulou-Bonikou G, Papavassiliou AG. EGF-R is expressed and AP-1 and NF-kappaB are activated in stromal myofibroblasts surrounding colon adenocarcinomas paralleling expression of COX-2 and VEGF. Cell Oncol. 2007;29(6):477–82.
Rehan VK, Sakurai R, Wang Y, Santos J, Huynh K, Torday JS. Reversal of nicotine-induced alveolar lipofibroblast-to-myofibroblast transdifferentiation by stimulants of parathyroid hormone-related protein signaling. Lung. 2007;185(3):151–9.
Guise TA, Mohammad KS, Clines G, Stebbins EG, Wong DH, Higgins LS, Vessella R, Corey E, Padalecki S, Suva L, Chirgwin JM. Basic mechanisms responsible for osteolytic and osteoblastic bone metastases. Clin Cancer Res. 2006;12(20 Pt 2):6213s–6s.
Guise TA. Breaking down bone: new insight into site-specific mechanisms of breast cancer osteolysis mediated by metalloproteinases. Genes Dev. 2009;23(18):2117–23.
Thomas RJ, Guise TA, Yin JJ, Elliott J, Horwood NJ, Martin TJ, Gillespie MT. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999;140(10):4451–8.
Guise TA, Mundy GR. Cancer and bone. Endocr Rev. 1998;19(1):18–54.
Bendre MS, Gaddy-Kurten D, Mon-Foote T, Akel NS, Skinner RA, Nicholas RW, Suva LJ. Expression of interleukin 8 and not parathyroid hormone-related protein by human breast cancer cells correlates with bone metastasis in vivo. Cancer Res. 2002;62(19):5571–9.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–49.
Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103(2):197–206.
Ibbotson KJ, D’Souza SM, Ng KW, Osborne CK, Niall M, Martin TJ, Mundy GR. Tumor-derived growth factor increases bone resorption in a tumor associated with humoral hypercalcemia of malignancy. Science. 1983;221(4617):1292–4.
Ibbotson KJ, D’Souza SM, Smith DD, Carpenter G, Mundy GR. EGF receptor antiserum inhibits bone resorbing activity produced by a rat Leydig cell tumor associated with the humoral hypercalcemia of malignancy. Endocrinology. 1985;116(1):469–71.
Shupnik MA, Ip NY, Tashjian AH Jr. Characterization and regulation of receptors for epidermal growth factor in mouse calvaria. Endocrinology. 1980;107(6):1738–46.
Qin L, Qiu P, Wang L, Li X, Swarthout JT, Soteropoulos P, Tolias P, Partridge NC. Gene expression profiles and transcription factors involved in parathyroid hormone signaling in osteoblasts revealed by microarray and bioinformatics. J Biol Chem. 2003;278(22):19723–31.
Cole JA. Parathyroid hormone activates mitogen-activated protein kinase in opossum kidney cells. Endocrinology. 1999;140(12):5771–9.
Guise TA, Yoneda T, Yates AJ, Mundy GR. The combined effect of tumor-produced parathyroid hormone-related protein and transforming growth factor-alpha enhance hypercalcemia in vivo and bone resorption in vitro. J Clin Endocrinol Metab. 1993;77(1):40–5.
Zhu J, Jia X, Xiao G, Kang Y, Partridge NC, Qin L. EGF-like ligands stimulate osteoclastogenesis by regulating expression of osteoclast regulatory factors by osteoblasts: implications for osteolytic bone metastases. J Biol Chem. 2007;282(37):26656–64.
Normanno N, De Luca A, Aldinucci D, Maiello MR, Mancino M, D’Antonio A, De Filippi R, Pinto A. Gefitinib inhibits the ability of human bone marrow stromal cells to induce osteoclast differentiation: implications for the pathogenesis and treatment of bone metastasis. Endocr Relat Cancer. 2005;12(2):471–82.
Lu X, Wang Q, Hu G, Van Poznak C, Fleisher M, Reiss M, Massague J, Kang Y. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009;23(16):1882–94.
Guise TA, Yin JJ, Taylor SD, Kumagai Y, Dallas M, Boyce BF, Yoneda T, Mundy GR. Evidence for a causal role of parathyroid hormone-related protein in the pathogenesis of human breast cancer-mediated osteolysis. J Clin Invest. 1996;98(7):1544–9.
Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–84.
Southby J, Kissin MW, Danks JA, Hayman JA, Moseley JM, Henderson MA, Bennett RC, Martin TJ. Immunohistochemical localization of parathyroid hormone-related protein in human breast cancer. Cancer Res. 1990;50(23):7710–6.
Powell GJ, Southby J, Danks JA, Stillwell RG, Hayman JA, Henderson MA, Bennett RC, Martin TJ. Localization of parathyroid hormone-related protein in breast cancer metastases: increased incidence in bone compared with other sites. Cancer Res. 1991;51(11):3059–61.
Vargas SJ, Gillespie MT, Powell GJ, Southby J, Danks JA, Moseley JM, Martin TJ. Localization of parathyroid hormone-related protein mRNA expression in breast cancer and metastatic lesions by in situ hybridization. J Bone Miner Res. 1992;7(8):971–9.
Furugaki K, Moriya Y, Iwai T, Yorozu K, Yanagisawa M, Kondoh K, Fujimoto-Ohuchi K, Mori K. Erlotinib inhibits osteolytic bone invasion of human non-small-cell lung cancer cell line NCI-H292. Clin Exp Metastasis. 2011;28(7):649–59.
Ryan MA, Nattamai KJ, Xing E, Schleimer D, Daria D, Sengupta A, Kohler A, Liu W, Gunzer M, Jansen M, Ratner N, Le Cras TD, Waterstrat A, Van Zant G, Cancelas JA, Zheng Y, Geiger H. Pharmacological inhibition of EGFR signaling enhances G-CSF-induced hematopoietic stem cell mobilization. Nat Med. 2010;16(10):1141–6.
Park SI, Soki FN, McCauley LK. Roles of bone marrow cells in skeletal metastases: no longer bystanders. Cancer Microenviron. 2011;4(3):237–46.
Bender RA, Hansen H. Hypercalcemia in bronchogenic carcinoma. A prospective study of 200 patients. Ann Intern Med. 1974;80(2):205–8.
Mundy GR, Martin TJ. The hypercalcemia of malignancy: pathogenesis and management. Metabolism. 1982;31(12):1247–77.
Lazaretti-Castro M, Kayath M, Jamnik S, Santoro IL, Tadokoru H, Vieira JG. Prevalence of hypercalcemia in patients with lung cancer. Rev Assoc Med Bras. 1993;39(2):83–7.
Wysolmerski JJ, Vasavada R, Foley J, Weir EC, Burtis WJ, Kukreja SC, Guise TA, Broadus AE, Philbrick WM. Transactivation of the PTHrP gene in squamous carcinomas predicts the occurrence of hypercalcemia in athymic mice. Cancer Res. 1996;56(5):1043–9.
Sanders JL, Chattopadhyay N, Kifor O, Yamaguchi T, Butters RR, Brown EM. Extracellular calcium-sensing receptor expression and its potential role in regulating parathyroid hormone-related peptide secretion in human breast cancer cell lines. Endocrinology. 2000;141(12):4357–64.
Lorch G, Viatchenko-Karpinski S, Ho HT, Dirksen WP, Toribio RE, Foley J, Gyorke S, Rosol TJ. The calcium-sensing receptor is necessary for the rapid development of hypercalcemia in human lung squamous cell carcinoma. Neoplasia. 2011;13(5):428–38.
Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet (Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t). 2007;39(3):311–8.
Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, Fujita PA, Learned K, Rhead B, Smith KE, Kuhn RM, Karolchik D, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC Genome Browser. Nucl Acids Res (Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t). 2010;38((Database issue)):D620–5.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Foley, J., Nickerson, N., Riese, D.J. et al. At the crossroads: EGFR and PTHrP signaling in cancer-mediated diseases of bone. Odontology 100, 109–129 (2012). https://doi.org/10.1007/s10266-012-0070-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10266-012-0070-5