Abstract
Species of Broussonetia have been essential in the development of papermaking technology. In Japan and Korea, a hybrid between B. monoica and B. papyrifera (= B. × kazinoki) known as kōzo and daknamu is still the major source of raw materials for making traditional paper washi and hanji, respectively. Despite their cultural and practical significance, however, the origin and taxonomy of kōzo and daknamu remain controversial. Additionally, the long-held generic concept of Broussonetia s.l., which included Sect. Allaeanthus and Sect. Broussonetia, was challenged as phylogenetic analyses showed Malaisia is sister to the latter section. To re-examine the taxonomic proposition that recognizes Allaeanthus, Broussonetia, and Malaisia (i.e., Broussonetia alliance), plastome and nuclear ribosomal DNA (nrDNA) sequences of six species of the alliance were assembled. Characterized by the canonical quadripartite structure, genome alignments and contents of the six plastomes (160,121–162,594 bp) are highly conserved, except for the pseudogenization and/or loss of the rpl22 gene. Relationships of the Broussonetia alliance are identical between plastome and nrDNA trees, supporting the maintenance of Malaisia and the resurrection of Allaeanthus. The phylogenomic relationships also indicate that the monoecy in B. monoica is a derived state, possibly resulting from hybridization between the dioecious B. kaempferi (♀) and B. papyrifera (♂). Based on the hypervariable ndhF-rpl32 intergenic spacer selected by sliding window analysis, phylogeographic analysis indicates that B. monoica is the sole maternal parent of B. × kazinoki and that daknamu carries multiple haplotypes, while only one haplotype was detected in kōzo. Because hybridizations between B. monoica and B. papyrifera are unidirectional and have occurred rarely in nature, our data suggest that daknamu might have originated via deliberate hybrid breeding selected for making hanji in Korea. On the contrary, kōzo appears to have a single origin and the possibility of a Korean origin cannot be ruled out.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paper is one of the greatest inventions of all time (Hunter 1978). Amongst a variety of raw materials used for making paper, the moraceous tree species paper mulberry (Broussonetia papyrifera) was essential in the development of the technology in ancient China (Barker 2002). Although the importance of B. papyrifera in the modern paper industry has decreased, hybrid paper mulberry trees known as kōzo and daknamu remain essential in the production of washi and hanji, traditional paper products from Japan and Korea, respectively (Mizumura et al. 2017; Won 2019). Despite the cultural and practical importance of washi and hanji, however, relationships between kōzo and daknamu and their origin(s) have been puzzling (Ohba and Akiyama 2014; Yun and Kim 2009). To settle controversies surrounding kōzo and daknamu, the taxonomy of Broussonetia first needs to be clarified. However, Corner (1962)’s long-held generic concept of Broussonetia (i.e., Broussonetia s.l.) that comprised seven species in two sections (i.e., Sect. Allaeanthus and Sect. Broussonetia) was recently challenged (Chung et al. 2017). This article addresses these issues using phylogenomic and phylogeographic approaches.
Corner (1962)’s Broussonetia L’Hér. ex Vent. sect. Allaeanthus (Thwaites) Corner included four species distributed in Madagascar [B. greveana (Baillon) C.C.Berg], Sri Lanka [B. zeylanica (Thwaites) Corner], Indochina [B. kurzii (Hook.f.) Corner; Fig. 1a, b], and the Philippines [B. luzonica (Blanco) Bureau; Fig. 1c, d]. All four species have economic and cultural importance, with the tough-fibered B. zeylanica historically used for papermaking in Sri Lanka (Thwaites 1854), B. luzonica and B. kurzii used as culinary ingredients (LaFrankie 2010; Tangkanakul et al. 2006), and B. greveana sought after for its valuable wood (Louppe 2008). Broussonetia sect. Broussonetia comprised three species, B. kaempferi Siebold (Fig. 1 h, i), B. monoica Hance (Fig. 1j), and B. papyrifera (L.) L’Hér. ex Vent. (Fig. 1k–m) distributed in East Asia and northern Indochina (Chung et al. 2017). All three species contain long fibers suitable for papermaking (Chûjô 1950; Mizumura et al. 2017). In addition to papermaking, B. papyrifera is also the prime material for making bark cloth by the Austronesian-speaking peoples in the Pacific (Chang et al. 2015; Penailillo et al. 2016) and known for its medicinal and nutritional properties and fast-growing habit (Peng and Shen 2018). Consequently, this fibrous tree has been widely translocated since prehistorical times (Chang et al. 2015) and become an invasive tree species worldwide (Chung et al. 2017).
Morphology of Allaeanthus, Malaisia, and Broussonetia: (a) A. kurzii, pistillate inflorescences, (b) A. kurzii, staminate inflorescences, (c) A. luzonicus, pistillate inflorescences, (d) A. luzonicus, staminate inflorescences, (e) M. scandens, pistillate inflorescences, (f) M. scandens, staminate inflorescences, (g) M. scandens, syncarps, (h) B. kaempferi, pistillate inflorescences, (i) B. kaempferi, staminate inflorescences, (j) B. monoica, pistillate (left two) and staminate (right two) inflorescences, (k) B. papyrifera, pistillate inflorescences, (l) B. papyrifera, staminate inflorescence, (m) B. papyrifera, syncarps, (n) kōzo (B. × kazinoki) plantation in Shiroishi, Miyagi, Japan. a, b: courtesy of Preecha Karaket; c, i: courtesy of Danilo N. Tandang; d: photo by Forest & Kim Starr/CC BY 3.0 US; e, f: courtesy of Pi-Fong Lu; j, k, l, m, n: photos by K.-F. Chung; i: Alan Kwok/Ada Tai/CC BY-NC 4.0
Despite being a small genus, the extensive intraspecific morphological variation (Fig. 2), lack of type designation (Akiyama et al. 2013; Chung et al. 2017), and long history of cultivation, selection and hybrid breeding (Peng and Shen 2018; Peng et al. 2014) has led to much confusion and controversy in the taxonomy of the multi-purpose Broussonetia s.l.. The rationale used by Corner (1962) for reducing Allaeanthus Thwaites to a section of Broussonetia was: “There are no major differences between these sections, which are not generically distinct.” Although Sections Allaeanthus and Broussonetia appear to be similar in their capitate, globose pistillate inflorescences and racemose to spicate staminate inflorescences (Corner 1962), the former is a tropical group, while the latter is mainly distributed in temperate to subtropical regions (Chung et al. 2017). Morphologically, reproductive organs of the two sections are also different. The slender catkins characterized by Sect. Allaeanthus (Fig. 1b, d) can be distinguished from the cylindric (Fig. 1 h, m) or globose (Fig. 1j) staminate inflorescences of Sect. Broussonetia. Styles of Sect. Allaeanthus are white (Fig. 1a), while those in Sect. Broussonetia are pinkish to purple when mature (Fig. 1h, j, k). Corner (1962) also noted that drupes of Sect. Broussonetia are stipitate within the sessile perianth, while drupes of Sect. Allaeanthus are sessile. The two sections can also be distinguished by the fruit color, with green to yellowish in Sect. Allaeanthus (Fig. 1c) and orange-red in Sect. Broussonetia (Fig. 1 m).
Although Corner (1962)’s circumscription of Broussonetia has been widely followed (e.g., Berg et al. 2006; Chang et al. 1998; LaFrankie 2010; Rohwer 1993; Wunderlin 1997; Yun and Kim 2009; Zhou and Gilbert 2003), whether the genus should be placed in Tribe Artocarpeae (Corner 1962), Tribe Broussonetieae (Chang et al. 1998), or Tribe Moreae (Berg et al. 2006; Rohwer 1993) has been disputed. Based on the plastid ndhF and nuclear ribosomal 26 S subunit sequences, Clement and Weiblen (2009) showed that molecular data did not support previous morphology-based classifications of Moraceae. In their revised tribal classification, Broussonetia was placed in Tribe Dorstenieae (Clement and Weiblen 2009). Within Tribe Dorstenieae, molecular data also showed that B. papyrifera is sister to the lianaceous Trophis scandens (Lour.) Hook. & Arn. [Trophis sect. Malaisia (Blanco) C.C.Berg], while other sampled species of Trophis form a clade sister to Morus L. (Clement and Weiblen 2009). To rectify the polyphyletic Trophis, Clement and Weiblen (2009) reinstated the generic status of Malaisia Blanco [i.e., Malaisia scandens (Lour.) Planch.; Fig. 1e–g]. Both the tribal classification of Broussonetia in Dorstenieae and its sister group relationship with Malaisia are also consistent with recent studies using Hyb-Seq data (Gardner et al. 2021; Zerega and Gardner 2019). Meanwhile, based on a nearly complete taxon sampling, Chung et al. (2017) further showed that, while both Sect. Allaeanthus and Sect. Broussonetia are monophyletic, Broussonetia s.l. is paraphyletic, with Sect. Broussonetia sister to M. scandens before the former joining Sect. Allaeanthus to form a monophyletic Broussonetia s.l.. To rectify the paraphyletic Broussonetia s.l., Chung et al. (2017) reinstated Allaeanthus to maintain the generic status of Malaisia, avoiding nomenclatural changes that would have also generated an expanded Broussonetia without obvious diagnostic characters. The three genera can be identified by the following key (Berg et al. 2006; Corner 1962):
-
1. Syncarps globose, thickly set with slender stalked bracts of various shapes more or less covering the drupes; seeds 2–3 mm long; endocarps crustaceous to ligneous; cotyledons equal, conduplicate to plane…2
-
1. Syncarps with few strongly projecting drupes each invested by the utricular perianth, the bracts short; seeds 6–7 mm long; endocarps membranous; cotyledons very unequal, the large one thickly fleshy and folded…Malaisia
-
2. Styles white; exocarps green to yellowish, sessile; seeds compressed, smooth, ligneous, the keel not double…Allaeanthus
-
2. Style pinkish to purple; exocarps orange-red, stipitate within the sessile perianth; seeds slightly compressed, papillate-asperate, crustaceous, the keel double at the base…Broussonetia
Although Malaisia has never been synonymized under Broussonetia, Corner (1962) placed the two genera in the same couplet of the taxonomic key to the Asian genera of Artocarpeae, implicitly suggesting their morphological similarity. Nevertheless, the female florets of M. scandens form contracted capitate to short-spicate inflorescences (Fig. 1e) which has caused much difficulty in placing it within Moraceae (Corner 1962). Geographically, M. scandens is distributed from Taiwan and southern China to throughout SE Asia and the western Pacific islands (Berg et al. 2006), partially overlapping with the Southeast Asian Allaeanthus and the mainly East Asian Broussonetia (Chung et al. 2017). The unique morphology of Malaisia and its overlapping geographic distribution with Allaeanthus and Broussonetia stimulated the current study to further test Chung et al. (2017)’s recent taxonomic treatment using phylogenomic data.
By resolving phylogenetic relationships and taxonomic controversies of Allaeanthus, Broussonetia, and Malaisia (i.e., the Broussonetia alliance), this study also aims to track the origin of the hybrid paper mulberry trees kōzo and daknamu used for traditional papermaking in Japan and Korea, respectively. Papermaking technology was introduced from China to Korea during the 3rd to 6th century (Song and Munn 2004; Yun and Kim 2009), primarily using the native Broussonetia papyrifera (Jeong 2015). According to Nihon Shoki (Chronicles of Japan), papermaking, as well as B. papyrifera, was introduced from Korea to Japan in 610 AD by the Korean monk Dam Jing (Kitamura and Murata 1980; Mizumura et al. 2017; Song and Munn 2004), though there is documentation that papermaking was introduced earlier to Japan by the Japanese Buddhist monk Dōkyō (Hunter 1978).
In Japan, Broussonetia is called kōzo zoku (zoku means ‘genus’ in Japanese) and the name ‘kōzo’ has long been applied to all Broussonetia species used by craftspeople for making washi and growers of these trees (Chûjô 1950). Taxonomically, early Japanese botanists regarded B. kazinoki Siebold as the correct scientific name for kōzo (Chûjô 1950; Kitamura and Okamoto 1962; Ohwi 1965). However, such definition was challenged after Kitamura and Murata (1980) studied the name hime-kōzo (‘hime’ means small in Japanese) recorded in two Japanese classic herbal books, Honzō Kōmoku Keimō (Dictated Compendium of Materia Medica) by Ranzan Ono and Sōmoku-Zusetsu (An Iconography of Herbaceous and Woody Plants of Japan) by Yokusai Iinuma. Kitamura and Murata (1980) concluded that B. kazinoki should be the correct name for hime-kōzo, a monoecious (Fig. 1j) shrub with leaning branches (Chung et al. 2017) and variable leaf shapes (Fig. 2b) distributed in Japan (Okamoto 2006), Korea (Yun and Kim 2009), China (Zhou and Gilbert 2003), and Taiwan (Liao 1996). In ancient Japan, hime-kōzo was simply called kōzo and used for papermaking (Kitamura and Okamoto 1962). By the Edo Period (1603–1868 AD), however, a hybrid became the preferred material for making washi and gradually the name kōzo (Fig. 1n) was applied specifically to the hybrid (Kitamura and Murata 1980), inevitably resulting in subsequent and widespread confusion regarding its definition. By distinguishing hime-kōzo from the broadly defined kōzo (sensu Chûjô 1950, Kitamura and Okamoto 1962; Ohwi 1965), Kitamura and Murata (1980) applied kōzo specifically to those plants cultivated for making washi and regarded kōzo as the hybrid between B. kazinoki and B. papyrifera, though no evidence was provided.
Although Kitamura and Murata (1980)’s concept of Japanese Broussonetia had been immediately and widely followed (e.g., Mizumura et al. 2017; Okamoto 2006; Yamazaki 1989), the identity of B. kazinoki remained confusing. In Japan, ‘kazinoki’ has been the common name for B. papyrifera since ancient times, and rightfully cited in Siebold (1830) when the name B. kazinoki was first proposed (though the name was not validly published until 1846; Akiyama et al. 2013). However, because the modern type concept had not yet been developed in the 19th Century (Fosberg 1992), neither Siebold (1830) nor the subsequent work (i.e., Siebold and Zuccarini 1846) specified specimens that were essential to resolve the puzzling identity of B. kazinoki. To stabilize the taxonomy of Japanese plant names, “Siebold collection of Japanese plants” was investigated and names described by Siebold (and Zuccarini) were lectotypified (Akiyama et al. 2013). As a result, a specimen from Siebold’s collection at Botanische Staatssammlung München (i.e., M0120984) was selected as the lectotype of Broussonetia kazinoki Siebold, with the comment that “This specimen is not the real Japanese “Kazinoki”, i.e., Broussonetia papyrifera (L.) Vent., but Japanese kōzo” (Akiyama et al. 2013). Based on their lectotypification, Ohba and Akiyama (2014) revised Broussonetia of Japan. Because B. kazinoki was taken by the nothospecies kōzo, B. monoica, a name long synonymized under B. kazinoki (e.g., Zhou and Gilbert 2003), succeeded as the earliest validly published and correct name for hime-kōzo (Ohba and Akiyama 2014). Subsequently, the multiplication sign was added (i.e., B. × kazinoki) by Chung et al. (2017) according to Article H.3 of the Code (McNeill et al. 2012).
In Korea, daknamu, also regarded as a hybrid species (Kim et al. 1992; Yun and Kim 2009), has been the favored material and clonally propagated for making hanji for centuries (Won 2019). Although widely cultivated in Korea since ancient times, daknamu was not botanically described until Yun and Kim (2009) published the name Broussonetia × hanjiana M.Kim. However, while kōzo has long been considered as a cultivated plant in Japan, Yun and Kim (2009) regarded B. × hanjiana a natural hybrid because its type specimen was collected in the pristine evergreen broadleaved forest of Gageo Island, the only place in Korea where both B. monoica and B. papyrifera co-occur naturally with B. × hanjiana. Because kōzo and daknamu are both the hybrid between B. monoica and B. papyrifera, Chung et al. (2017) synonymized B. × hanjiana under B. × kazinoki. Before the introduction of machine-made paper, both kōzo and daknamu were extensively cultivated and clonally propagated for a wide range of paper products in Japan (Mizumura et al. 2017) and Korea (Yun and Kim 2009). If kōzo had originated in Japan, the hybridization must have occurred after the introduction of Broussonetia papyrifera in the 7th Century (Won 2019). Alternatively, kōzo might derive from daknamu, given that both B. monoica and B. papyrifera are native to Korea (Yun and Kim 2009). Using plastid and nuclear markers, Won (2019) confirmed that daknamu is indeed a hybrid between B. monoica (♀) and B. papyrifera (♂). Additionally, a likely incidence of back-cross and introgression of daknamu (♂) to B. monoica (♀) was also detected (Won 2019). However, no studies have yet investigated these two hybrids jointly.
This study aims to settle taxonomic disputes of the Broussonetia alliance and track origins of kōzo and daknamu. We assembled full plastid genome (plastome) sequences of the alliance which so far are only available for B. papyrifera (KX828844), “B. kazinoki” (MH223642 and MW465960), and the synthetic hybrid paper mulberry “B. kazinoki × B. papyrifera” (Xu et al. 2018). Additionally, nuclear ribosomal DNA sequences were assembled to test for the congruence between plastid and nuclear genomes. Based on phylogenomic relationships, Chung et al. (2017)’s taxonomic proposition is tested and character (e.g., gene loss and reproductive syndrome) state evolution is discussed. Based on the hypervariable region of plastome sequences identified in this study, we also address the following questions: Do kōzo and daknamu each have single or multiple origins? Do they share haplotypes? Have kōzo and daknamu experienced bottlenecks from the hybridization? Does kōzo or daknamu have higher haplotype diversity? If kōzo was introduced from Korea to Japan, as historical documents, had kōzo experienced a second bottleneck and showed reduced haplotype diversity?
Materials and methods
Plastome and nrDNA assembly
Based on Chung et al. (2017), we sampled six species of the Broussonetia alliance, including all three species of Broussonetia, two of the four species of Allaeanthus, and the monotypic Malaisia scandens. All voucher specimens were deposited at the Herbarium, Biodiversity Research Center, Academia Sinica, Taipei (HAST). Genomic DNA was extracted using CTAB method (Doyle 1991) and further purified by Monarch® PCR & DNA Cleanup Kit. Purified genomic DNAs were sent to the Genomic Core Lab of Institute of Molecular Biology, Academia Sinica for library preparation using TruSeq® Nano DNA Library Prep Kit (Illumina Inc., San Diego, CA, USA). The DNA was fragmented into target 350 bp by M220 Focused-ultrasonicator™ (Covaris, Woburn, MA, USA) and ligated to TruSeq Single Index adapters. Sequencing was done by MiSeq (Illumina Inc., San Diego, CA, USA) using Illumina Reagent Kit v3 (600 cycle) with pair-end mode, read length = 2 × 300 bp, and insert size = 350–550 bp (determined by Bioanalyzer High Sensitivity DNA Analysis). The samples were multiplexed with other Broussonetia samples not included in the current study. Plastomes were assembled using Geneious software version 11 (Kearse et al. 2012) with the following procedure: The adapter and barcode sequences were first removed. Then, we removed the low-quality bases at the start and the end of the reads by the modified Mott algorithm in Geneious (Error probability limit = 0.05). Then, trimmed reads were mapped to the plastome of Morus notabilis C.K.Schneid. (KP939360) using the “Map to Reference” function in Geneious with “Medium-Low Sensitivity” and default settings. Regions that contained indel(s) between the focal sample and the reference genome might not be correctly assembled by the reference-based method and could exhibit conflicts in the mapping results. Those regions were corrected by extending the reliably assembled regions into problematic parts and ‘iteratively mapping’ the reads to the contigs to obviate the effect of the reference genome. To obtain a complete plastome, the remaining gaps and uncertain regions that could not be resolved by iterative mapping were filled by Sanger sequencing using the primers designed from the reference genome and PCR condition listed in Table S1. Meanwhile, junctions between large single copy (LSC), small single copy (SSC), and inverted repeats (IRs) were verified by PCR. Nuclear ribosomal DNA (nrDNA) sequences, spanning across the partial external transcribed spacer (ETS), 18 S rRNA gene, internal transcribed spacer (ITS) 1, 5.8 S rRNA gene, ITS2, 26 S rRNA gene, and partial nontranscribed spacer (NTS) of the six species of Broussonetia alliance were assembled by retrieving the raw reads and mapping to the published nrDNA sequences of Ficus tikoua Bureau. (JF317367, JF317386, and EU091641) using the “Map to Reference” function of Geneious with “Medium-Low Sensitivity” and default settings.
Genome annotation and comparison
The newly assembled plastomes were annotated based on seven published plastomes of urticalean rosids (i.e., Sytsma et al. 2002), including Moraceae [Broussonetia papyrifera (KX828844), Ficus carica L. (NC035237; Rabah et al. 2017), and Morus notabilis (KP939360; Chen et al. 2016)], Urticaceae [Debregeasia saeneb (Forssk.) Hepper & Wood (KY419997; Zhang et al. 2017)], Cannabaceae [Humulus lupulus L. (KT266264; Vergara et al. 2016) and Cannabis sativa (KY084475; Vergara et al. 2016)], and Ulmaceae [Ulmus pumila L. (KY244086; Zuo et al. 2017)], using the transfer annotation feature in Geneious (similarity = 65%). To further verify the identified tRNAs, the structures and anti-codons of putative tRNAs were checked in tRNAscan-SE 2.0 (Lowe and Chan 2016). The genome maps were visualized by OrganellarGenomeDraw (Lohse et al. 2013). The numbers of variable sites were calculated using PAUP* version 4.0 (Swofford 2002). To detect genome rearrangement, multiple genome alignment of the 13 urticalean rosids plastomes was conducted using Mauve version 1.1.3 (Darling et al. 2004) launched in Geneious.
Sliding window analysis of plastomes
To detect evolutionary hotspots in the plastomes for phylogeographic and phylogenetic studies, the nucleotide diversity (π) values were calculated based on two datasets using sliding window analysis. The first dataset comprised the six newly assembled plastomes of the Broussonetia alliance, aiming to find high divergence hotspots for recent speciation and phylogeographic studies. The second dataset comprised 13 plastomes of the urticalean rosids for family level phylogenetic studies. Plastome sequences were aligned using MAFFT (Katoh and Standley 2013). The sliding window analysis was performed using PopGenome R package (Pfeifer et al. 2014) to calculate π (Nei and Li 1979) for the window length of 800 bp and step size of 400 bp.
Phylogenomic analyses
A preliminary phylogenomic analysis of the urticalean rosids were conducted using 16 plastomes (Fig. S1). Because MW465960 (“Broussonetia kazinoki”), MF496038 (“Broussonetia kazinoki × Broussonetia papyrifera”), and MH223642 (“Broussonetia kazinoki”) are almost identical and appear to be conspecific with B. monoica (MH189567) that was newly assembled in the current study (Fig. S1), the former three plastomes were not included in our final analysis. Phylogenomic relationships of the remaining 13 plastomes and eight nrDNA sequences of urticalean rosids were reconstructed using maximum likelihood (ML) and Bayesian inference (BI) methods. Plastome sequences were aligned using MAFFT version 7 with default parameters (Katoh and Standley 2013). For plastome sequences, phylogenomic relationships were reconstructed based on LSC, SSC and a single IR. The nucleotide substitution rates and distribution shapes were estimated by bModeltest (Bouckaert and Drummond 2017), with the model GTR+Г selected for all partitions and all analyses. The concatenated plastid data were divided into four partitions: protein-coding sequences, tRNAs, rRNAs, and other non-coding sequences. Model parameters were unlinked across partitions. The nrDNA phylogeny was reconstructed based on ITS1, ITS2, and rRNA (18 S, 5.8 S, and 26 S) regions. Two partitions (i.e., ITS regions and rRNA regions) with unlinked model parameters were applied. ML analyses were performed using RAxML version 8 (Stamatakis 2014). Twenty heuristic searches with 1,000 bootstrap replicates were conducted, and the congruence was checked manually. BI tree was reconstructed by BEAST2 (Bouckaert et al. 2014) with 100,000,000 generations and sampling every 5,000 generations. The effective sample sizes (ESS) of final posterior probability were confirmed in Tracer version 1.7 (Rambaut et al. 2018). If the BI tree topology was congruous with the ML tree, the posterior probabilities greater than 0.80 were mapped to the backbone of the ML tree at corresponding nodes.
Phylogeographic analyses
Based on the sliding window analysis, matK-rps16, rps16-psbK, rps4-ndhJ, ndhF-rpl32, rpl32-ccsA, and ycf1 were identified as the most variable regions suitable for phylogeographic analyses (see Results). However, because our samples included substantial herbarium materials (Table S2) with compromised DNA quality, we first tested the suitability of the six potential markers for easiness of PCR amplification and Sanger sequencing. Consequently, ndhF-rpl32 intergenic spacer (IGS), which was also adopted by Won (2019) based on Chang et al. (2015), was chosen for phylogeographic analyses.
In combination with Won (2019)’s samples, a total of 81 samples were sequenced, including 9 kōzo from 9 washi workshops of 3 prefectures of Japan, 9 daknamu from 7 provinces of South Korea, and 63 B. monoica from China (14 samples from 7 provinces), Japan (32 samples from 17 prefectures), South Korea (4 samples from 3 provinces), and Taiwan (13 samples from 7 counties). Table S2 details the collecting information of the 81 accessions.
Genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) protocol. Conditions for PCR amplification and sequencing were detailed in Won (2019) using primers listed in Table S3. Sequences were assembled in Geneious software version 11 (Kearse et al. 2012). The assembled sequences were aligned by MAFFT version 7 (Katoh and Standley 2013) with default parameters. The haplotype network was reconstructed and visualized using the R package pegas (Paradis 2010). Because pegas cannot handle indel and single nucleotide polymorphism (SNP) at the same time, two long indels (9-bp and 77-bp) were replaced with SNPs to maximize sequence information. Gaps, ambiguous nucleotides, mononucleotide repeats, and microsatellite-like sequence variation at the junctions of the 77-bp indel (Fig. S2) were ignored due to uncertainty of objective alignment. The most parsimonious links in the haplotype network were determined by the default settings in pegas following the algorithm of Templeton et al. (1992). The sequences of unique haplotypes are available in Online Resource 3. The distribution map of haplotypes was drawn using the R package ggplot2. The nucleotide diversity, haplotype diversity, analysis of molecular variance (AMOVA), and pairwise GST were calculated in R package pegas and mmod (Winter 2012). The source code is available in Online Resource 4.
Results
Characteristics of plastomes
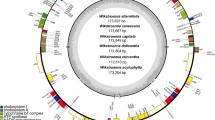
The six newly assembled plastomes display the typical circular, quadripartite structure of angiosperms (Kwon et al. 2020; Ruhlman and Jansen 2021), composed of an LSC, an SSC, and a pair of IRs (Fig. 3). For assembly quality (Table 1), 1,379,002 (Broussonetia papyrifera) to 2,208,733 (Allaeanthus kurzii) trimmed reads were mapped to the final plastomes, with the mean average coverage for each base pair ranging between 171.4 ± 58.7 (Malaisia scandens) and 566.5 ± 153.9 (A. kurzii). The plastomes of Broussonetia (B. kaempferi: 160,625 bp; B. monoica: 160,777 bp; B. papyrifera: 160,121 bp) are slightly smaller than the plastomes of Malaisia (M. scandens: 161,313 bp) and Allaeanthus (A. kurzii: 162,170 bp; A. luzonicus: 162,594 bp). Gene contents of the six plastomes of the Broussonetia alliance are identical, composed of 111 unique genes including 4 rRNA genes, 30 tRNA genes, and 77 protein-coding genes (Table S4). Among the 30 unique tRNA genes, the sequence of the trnM-CAU gene in the LSC differs from the trnM-CAU gene in IRs and thus they are regarded as different. Introns were found in 18 genes, including 12 protein-coding genes and 6 tRNA genes. Among the 111 unique genes, 17 genes are completely duplicated in IRs. In addition, the trans-splicing rps12 gene is partially duplicated (2nd and 3rd exon) in IRs. Based on plastome sequence alignment using Mauve, rearrangements were not detected among the six plastomes nor in the other urticalean rosids sampled in the present study (Fig. S3). Comparison of 12 plastomes of urticalean rosids also shows that the IR boundaries are highly conserved within the Broussonetia alliance, with a slight IR contraction in the LSC-IRA boundary in Malaisia and an expansion of ca. 600 bp in the ycf1 gene in (A) luzonicus in the SSC-IRB boundary (Fig. 4). However, as shown in Fig. 5, a premature stop codon was identified in the rpl22 gene of plastomes of (B) papyrifera, Malaisia scandens, Allaeanthus kurzii, and (A) luzonicus, indicating that the gene has pseudogenized. Additionally, this locus was completely lost in (B) monoica and B. kaempferi.
Evolutionary hotspots in plastomes
The Broussonetia alliance dataset shows that the nucleotide diversity (π) ranges from 0 to 0.04132 with a mean ± standard deviation (SD) of 0.01287 ± 0.00906. Six highly variable regions (π > 0.03) including matK-rps16, rps16-psbK, rps4-ndhJ, ndhF-rpl32, rpl32-ccsA, and ycf1 were identified (Fig. S4a). Three of these regions lie in the LSC region and three in the SSC region. Five out of six regions are non-coding regions located within intergenic spacers. In the urticalean rosids dataset, the nucleotide diversity ranges from 0.00116 to 0.06988 with a mean ± SD of 0.02345 ± 0.01328. The identified hotspot regions [i.e., matK, rpoC2, ndhF, and ycf1 (π > 0.045)] are all protein-coding genes (Fig. S4b). In both datasets, π is much lower in IR regions than LSC and SSC.
Phylogenomic analyses
The alignment of the 13 plastomes of urticalean rosids is 151,354 bp (LSC + SSC + a single IR), with 12,424 parsimony-informative sites (8.21%), 16,913 parsimony-uninformative sites (11.17%), and 122,017 constant sites (80.62%). Phylogenetic analyses using both ML and BI methods resolved Moraceae, Urticaceae (Debregeasia saeneb), Cannabaceae (Cannabis sativa and Humulus lupulus), and Ulmaceae (Ulmus pumila) forming successively sister and increasingly more inclusive monophyletic groups with full support values (Fig. 6a), congruent with previous studies using Sanger sequencing (Sytsma et al. 2002; Zhang et al. 2011). Within Moraceae, the Broussonetia alliance and Ficus form a clade sister to Morus, also congruent with early studies (Chung et al. 2017; Clement and Weiblen 2009). Within the Broussonetia alliance, Broussonetia and Malaisia form a clade sister to Allaeanthus, identical to relationships revealed in Chung et al. (2017). Within Broussonetia, B. kaempferi and B. monoica form a clade sister to B. papyrifera, as revealed in Chung et al. (2017).
Comparison of plastome and nrDNA phylogenies. (a) Plastome phylogeny of 13 urticalean rosids. Numbers at each node are bootstrap support value/posterior probability. Nodes without number indicates full support (100/1.0). Premature stop codons in genes are indicate as yellow bars. Gene deletions are indicated as red bars. Within the Broussonetia alliance, all species are dioecious (blue) except for B. monoica (green). (b) nrDNA phylogeny of the Broussonetia alliance and the out groups. Numbers at each node are bootstrap support value/posterior probability. Nodes without number indicates full support (100/1.0)
The alignment of the eight nrDNA is 5,899 bp, with 200 parsimony-informative sites (3.39%), 336 parsimony-uninformative sites (5.70%), and 5,363 constant sites (90.91%). The topology of the nrDNA phylogeny (Fig. 6b) is completely congruent with that of the plastome phylogeny (Fig. 6a), though supporting values at the Broussonetia + Malaisia clade is low (BS = 65%, PP = 0.53).
Phylogeographic analyses
The network depicting evolutionary relationships among the 16 haplotype is shown in Fig. 7. Based on the calculation of pegas, the 77-bp indel (Fig. S2) appears to have evolved independently between haplotypes cp-1 and cp-9, cp-2 and cp-4, and cp-10 and cp-12 (Fig. 7), likely triggered by the presence of the microsatellite-like sequences (Fig. S2). Of the 81 accessions of B. monoica and B. × kazinoki sampled (Table S2), 16 haplotypes were detected, with two major haplotypes (cp-1 and cp-2) carried by a majority (ca. 63%) of samples (Fig. 7). Of the 16 ndhF-rpl32 haplotypes, 15 were detected in B. monoica (N = 63), while only five haplotypes were found in B. × kazinoki (N = 18). Of the 18 accessions of B. × kazinoki sequenced, five haplotypes were detected in daknamu (N = 9), while kōzo (N = 9) carried only cp-1.
Overall, continental populations of China and Korea have higher nucleotide and haplotype diversities than their adjacent island populations of Taiwan and Japan (Table 2). According to AMOVA, haplotype compositions between regions (China, Korea, Japan and Taiwan) are highly differentiated (Table 3). However, the high within-region variance suggests the presence of population structure within regions, though a comprehensive sampling is required to give detailed information. Estimates of pairwise GST (Nei 1973) and G″ST (Hedrick 2005) also indicate the highest level of population differentiation between China and Taiwan, while the lowest values of both GST and G″ST are found between Korea and Japan (Table 4).
Discussion
Taxonomy of Broussonetia alliance
Using the genome skimming approach, complete plastome and nrDNA sequences of six species of the Broussonetia alliance were assembled and their phylogenomic relationships were reconstructed. Given the identical relationships between plastome and nrDNA trees among genera of the Broussonetia alliance (Fig. 6), our current data clearly show that Broussonetia s.l. is paraphyletic, results that are also congruent with those of Chung et al. (2017) and Gardner et al. (2021). To rectify the paraphyly of Broussonetia s.l., Malaisia can either be synonymized under Broussonetia s.l. or the generic status of Sect. Allaeanthus must be resurrected as proposed by Chung et al. (2017). To achieve an objective generic delimitation, Backlund and Bremer (1998), Linder et al. (2010), Heenan and Smissen (2013), and Hsieh et al. (2022) advocated five criteria: (1) prioritizing primary (i.e., family, genus, and species) over secondary ranks (i.e., subgenus, section, etc.), (2) maximizing phylogenetic information and reducing redundancy in a classification, (3) recognizing evolutionarily equivalent (i.e., clade age, phylogenetic distance, and morphology) groups as the same rank, (4) delimiting genus that is morphologically, ecologically, and geographically homogenous, and (5) taking into account the full taxonomic history of the group and minimizing name changes to maintain nomenclatural stability. Although our phylogenetic relationships of the Broussonetia alliance is also compatible with an expanded Broussonetia that synonymizes both Allaeanthus and Malaisia as sections, we support Chung et al. (2017)’s proposition because Malaisia is morphologically distinct (Corner 1962) and has never been subsumed under Broussonetia. Maintaining the generic status of Malaisia and resurrecting Allaeanthus also prioritizes the primary rank genus over the secondary rank section, maximizes our phylogenomic conclusions (Fig. 6), delimitates three genera that are morphologically (Fig. 1), ecologically, and geographically homogenous, reflects the full taxonomic histories of the three groups, and avoids an unnecessary name change.
Character evolution and origin of B. monoica
By mapping gene loss/deletion on the robust and congruent phylogenomic relationship, the rpl22 gene appears to have been pseudogenized prior to the diversification of the Broussonetia alliance and lost completely in the clade composed of B. kaempferi and B. monoica (Fig. 6). Additionally, our analyses also detected premature stop codons in the rps3 gene in Cannabaceae and the ndhF gene in Cannabis sativa (Fig. 6a) not previously known in the family (e.g., Zhang et al. 2018).
The robust and congruent phylogenomic relationships among species of the Broussonetia alliance also allow us to track the evolution of reproductive systems in the Broussonetia alliance. Except for B. monoica which is monoecious (Fig. 1j), all other species of the Broussonetia alliance are dioecious (Chung et al. 2017), indicating that the monoecy in B. monoica is a derived state (Fig. 6a) and suggesting that dioecy is not an evolutionary dead end (Schaefer and Renner 2010; Zhang et al. 2019) as commonly assumed (Heilbuth 2000). Volz and Renner (2008) surmised that transitions between monoecy and dioecy in Cucurbitaceae might correlate with polyploidy, resulted from hybridization between dioecious diploids and hermaphroditic polyploids. In hindsight, the vegetative morphology of the monoecious B. monoica, such as its shrubby habit (Chung et al. 2017), and the highly variable and oblique leaves (Fig. 2b) appear to be intermediate between the lianaceous and oblong-leaved B. kaempferi (Fig. 2a) and the arborescent and ovate-leaved (but often divided) B. papyrifera (Fig. 2c), suggesting that B. monoica could have resulted from hybridization of the former two dioecious species (Ďurkovič et al. 2012; Gil and Kim 2016; Liu et al. 2019; Tamaki et al. 2021). Cytologically, while chromosome numbers of both B. kaempferi (Narita and Yosinaga 1955) and B. papyrifera (Oginuma and Tobe 1995) are 2n = 26, Seki (1950) reported 2n = 26 and 39 for B. monoica (as B. kazinoki). If B. monoica did originate through hybridization between B. kaempferi and B. papyrifera, the cytological data suggest that B. monoica is either a homoploid hybrid or a triploid, supporting Volz and Renner (2008)’s speculation that changes in ploidy level could trigger the transition of sexual system. Under the hybridization scenario, the sister relationship between B. kaempferi and B. monoica in the plastome tree (Fig. 6a) would also indicate that the former should be the maternal parent of the latter species. The proposition on the hybrid origin of B. monoica is currently under investigation using genomic approaches.
Phylogeography of B. monoica and origin of B. × kazinoki
Although the main islands of Japan and Taiwan had been intermittently connected to the Asian Continent to various degrees by land bridges over recurrent glacial periods, the Korea Strait and Taiwan Strait have both been effective geographic barriers for gene flow since the end of last glacial maximum as revealed by phylogeographic studies (Huang 2011; Park et al. 2000; Qiu et al. 2009, 2011). However, while the current study reveals that B. monoica displays a marked phylogeographic pattern between populations of China and Taiwan, the low values of both GST and G″ST and largely overlapping haplotypes carried by B. monoica and B. × kazinoki of Korea and Japan indicate that the Korean Strait has been a porous barrier for the dispersal of the two species (Table 4; Fig. 7).
Because plastids are maternally inherited in Broussonetia papyrifera (Zhang et al. 2003), the hypervariable ndhF-rpl32 IGS (Fig. S4a) provides an ideal marker for investigating maternal origins of kōzo and daknamu (B. × kazinoki). In our analysis, all haplotypes (except for cp-15 which is one mutation away from cp-4) detected in B. × kazinoki are also carried by B. monoica (Won 2019). Although our sampling of both B. monoica and B. × kazinoki is far from comprehensive, the presence of five haplotypes in kōzo and daknamu clearly shows multiple origins of the hybrid (Londo et al. 2006; Miller and Schaal 2005) and an asymmetrical hybridization (Hamzeh et al. 2007; Zha et al. 2010) between B. monoica and B. papyrifera.
Despite the limited geographic sampling of both B. monoica and B. × kazinoki in Korea, high nucleotide and haplotype diversities comparable to the Chinese populations were detected (Table 2). Additionally, daknamu carries five haplotypes (Fig. 7), indicating recurrent and multiple origins of the hybrid. On the contrary, hybridization between B. monoica and B. papyrifera has never been reported in China or Taiwan where B. monoica and B. papyrifera are both common and often sympatrically distributed, except for the synthetic ‘hybrid paper mulberry’ (i.e., B. kazinoki × B. papyrifera) bred for the poverty alleviation project in China (Ni et al. 2020; Peng et al. 2014). Even in this ‘hybrid paper mulberry’, B. monoica is the maternal parent (Fig. S1), corroborating our inference that hybridization between B. monoica and B. papyrifera is possible only when the former species serves as the maternal parent. In Korea, B. papyrifera is restricted to islands and coastal areas while B. monoica is generally an inland species (Won 2019). Although the type specimen of B. × hanjiana was collected from the natural broadleaved forest of Gageo Island and likely originated naturally (Yun and Kim 2009), the largely non-overlapping distribution, the rarity of natural hybridization between B. monoica and B. papyrifera elsewhere, and asymmetrical hybridization between the two species suggests that the multi-origin daknamu might not have originated naturally but instead have been the result of deliberate and artificial hybridizations, possibly generated and selected for making hanji in Korea.
On the contrary, the proposition of a single origin of kōzo in Japan cannot be rejected given that all sequenced individuals of kōzo carry cp-1 (Fig. 7). Although our current sampling of kōzo is restricted to three prefectures (Miyagi, Nagano, and Tochigi) of Honshu, kōzo used by the washi workshop of Shiroishi, Miyagi was introduced from Uwajima, Ehime of Shikoku during the early 17th Century by Hidemune Date (Kahoku News, 25 February 2018) for encouraging washi production, indicating that kōzo carrying cp-1 might have a much wider distribution. The ca. 900 km translocation of kōzo from Ehime to Miyagi during the Edo period also indicates that the current distribution of kōzo cannot reflect the precise geographic origin of this economically important fiber crop in ancient Japan. Nevertheless, because B. monoica carrying cp-1 is also distributed in Japan (Fig. 7), our sampled kōzo could have originated indigenously, either naturally or artificially. On the other hand, the relatively low population differentiation between Japan and Korea (Table 4) and absence of genetic variation of kōzo suggests that a Korean origin of kōzo along with the introduction of the papermaking technique (Mizumura et al. 2017; Song and Munn 2004; Yun and Kim 2009) cannot be ruled out completely. Future studies using genomic approaches and collaborations with traditional washi and hanji makers are required to fully understand the origins of the intriguing kōzo and daknamu.
Given the importance of kōzo for washi and daknamu for hanji, our study not only addresses important evolutionary questions but also has profound value for the manufacture of traditional Korean and Japanese paper, which are also highly relevant to the understanding of the historical paper material and paper conservation. Because of the short histories of the introduction of papermaking to Korea and Japan, kōzo and daknamu also represent a unique study system to understand how cultural and societal developments and biological process of hybridization (Diamond 2002; Zeder 2015) could have shaped the genetic diversity of a hybrid domesticated crop species.
Change history
12 March 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10265-022-01382-z
References
Akiyama S, Thijsse G, Esser H-J, Ohba H (2013) Siebold and Zuccarini’s type specimens and original materials from Japan, Part 2. Angiosperms, Dicotyledoneae 1. J Jap Bot 88:346–377
Backlund A, Bremer K (1998) To be or not to be—principles of classification and monotypic plant families. Taxon 47:391–400. https://doi.org/10.2307/1223768
Barker C (2002) Plate 432. Broussonetia papyrifera. Bot Mag 19:8–18. https://doi.org/10.1111/1467-8748.00324
Berg CC, Corner EJH, Jarrett FM (2006) Moraceae, genera other than Ficus. In: Nooteboom HP (ed) Flora Malesiana, Series I, vol 17/Part 1. National Herbarium Nederland, Leiden, pp 1–152
Bouckaert RR, Drummond AJ (2017) bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol 17:e42. https://doi.org/10.1186/s12862-017-0890-6
Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537. https://doi.org/10.1371/journal.pcbi.1003537
Chang S, Wu C, Cao Z (1998) Moraceae. In: Chang S, Wu C (eds) Flora Reipublicae Popularis Sinicae, Tomus 23(1). Science Press, Beijing, pp 1–219 (In Chinese)
Chang C-S, Liu H-L, Moncada X, Seelenfreund A, Seelenfreund D, Chung K-F (2015) A holistic picture of Austronesian migrations revealed by phylogeography of Pacific paper mulberry. Proc Natl Acad Sci USA 112:13537–13542. https://doi.org/10.1073/pnas.1503205112
Chen C, Zhou W, Huang Y, Wang Z-Z (2016) The complete chloroplast genome sequence of the mulberry Morus notabilis (Moreae). Mitochondrial DNA A 27:2856–2857. https://doi.org/10.3109/19401736.2015.1053127
Chûjô K (1950) Notes on Broussonetia papyrifera Vent (Kazinoki), B. kazinoki Sieb (Kôzo) and B. kaempferi Sieb (Turu-Kôzo). J Jap For Soc 32:329–334 (In Japanese with English abstract)
Chung K-F, Kuo W-H, Hsu Y-H, Li Y-H, Rubite RR, Xu W-B (2017) Molecular recircumscription of Broussonetia (Moraceae) and the identity and taxonomic status of B. kaempferi var. australis. Bot Stud 58:e11. https://doi.org/10.1186/s40529-017-0165-y
Clement WL, Weiblen GD (2009) Morphological evolution in the mulberry family (Moraceae). Syst Bot 34:530–552. https://doi.org/10.1600/036364409789271155
Corner EJH (1962) The classification of Moraceae. Gard Bull Singapore 19:187–252
Darling ACE, Mau B, Blattner FR, Perna NT (2004) Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. https://doi.org/10.1101/gr.2289704
Diamond J (2002) Evolution, consequences and future of plant and animal domestication. Nature 418:700–707. https://doi.org/10.1038/nature01019
Doyle J (1991) DNA protocols for plants. In: Hewitt GM, Johnston AWB, Young JPW (eds) Molecular techniques in taxonomy. Springer, Berlin, pp 283–293
Ďurkovič J, Kardošová M, Čaňová I, Lagaňa R, Priwitzer T, Chorvát D, Cicák A, Pichler V (2012) Leaf traits in parental and hybrid species of Sorbus (Rosaceae). Am J Bot 99:1489–1500. https://doi.org/10.3732/ajb.1100593
Fosberg FR (1992) An essay on lectotypification. Taxon 41:321–323. https://doi.org/10.2307/1222339
Gardner EM, Audi L, Zhang Q, Sauquet H, Monro AK, Zerega NJC (2021) Phylogenomics of Brosimum (Moraceae) and allied genera, including a revised subgeneric system. Taxon 70:778–792. https://doi.org/10.1002/tax.12503
Gil H-Y, Kim S-C (2016) Viola woosanensis, a recurrent spontaneous hybrid between V. ulleungdoensis and V. chaerophylloides (Violaceae) endemic to Ulleung Island. Korea. J Plant Res 129:807–822. https://doi.org/10.1007/s10265-016-0830-3
Hamzeh M, Sawchyn C, Perinet P, Dayanandan S (2007) Asymmetrical natural hybridization between Populus deltoides and P. balsamifera (Salicaceae). Can J Bot 85:1227–1232. https://doi.org/10.1139/B07-105
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638. https://doi.org/10.1111/j.0014-3820.2005.tb01814.x
Heenan PB, Smissen RD (2013) Revised circumscription of Nothofagus and recognition of the segregate genera Fuscospora, Lophozonia, and Trisyngyne (Nothofagaceae). Phytotaxa 146:1–31. https://doi.org/10.11646/phytotaxa.146.1.1
Heilbuth JC (2000) Lower species richness in dioecious clades. Am Nat 156:221–241. https://doi.org/10.1086/303389
Hsieh C-L, Yu C-C, Huang Y-L, Chung K-F (2022) Mahonia vs. Berberis unloaded: Generic delimitation and infrafamilial classification of Berberidaceae based on plastid phylogenomics. Front Plant Sci 12:e720171. https://doi.org/10.3389/fpls.2021.720171
Huang S-F (2011) Historical biogeography of the flora of Taiwan. J Natl Taiwan Mus 64:33–63 (In Chinese with English abstract)
Hunter D (1978) Papermaking. Dover Publications, New York
Jeong SH (2015) A study on manufacturing technologies and excellence of Korean traditional paper. Korean J Cult Herit Stud 48:96–131 (In Korean with English abstract)
Katoh K, Standley DM (2013) MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kim M, Kim T-J, Lee S (1992) A taxonomic study of the Korea Broussonetia (Moraceae) by multivariate analysis. Korean J Pl Taxon 22:241–254 (In Korean with English abstract)
Kitamura S, Murata G (1980) Coloured illustrations of woody plants of Japan, vol II. Hoikusha Publishing Co., Osaka (In Japanese)
Kitamura S, Okamoto S (1962) Coloured illustrations of trees and shrubs of Japan. Hoikusha Publishers, Osaka (In Japanese)
Kwon E-C, Kim J-H, Kim N-S (2020) Comprehensive genomic analyses with 115 plastomes from algae to seed plants: structure, gene contents, GC contents, and introns. Genes Genom 42:553–570. https://doi.org/10.1007/s13258-020-00923-x
LaFrankie JVJ (2010) Trees of Tropical Asia: An Illustrated Guide to Diversity. Black Tree Publication, Inc., Manila
Liao J-C (1996) Moraceae. In: Editorial Committee of the Flora of Taiwan (ed) Flora of Taiwan, vol 2, 2nd edn. Department of Botany, National Taiwan University, Taipei, Taiwan, pp 136–195
Linder HP, Baeza M, Barker NP, Galley C, Humphreys AM, Lloyd KM, Orlouch DA, Purie MD, Simon BK, Walsh N, Verboom GA (2010) A generic classification of the Danthonioideae (Poaceae). Ann MO Bot Gard 97:306–364. https://doi.org/10.3417/2009006
Liu S-H, Tseng Y-H, Zure D, Rubite RR, Balangcod TD, Peng C-I, Chung K-F (2019) Begonia balangcodiae sp. nov. from northern Luzon, the Philippines and its natural hybrid with B. crispipila, B. × kapangan nothosp. nov. Phytotaxa 407:5–21. https://doi.org/10.11646/phytotaxa.407.1.3
Lohse M, Drechsel O, Kahlau S, Bock R (2013) OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:W575–W581. https://doi.org/10.1093/nar/gkt289
Londo JP, Chiang Y-C, Hung K-H, Chiang T-Y, Schaal BA (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA 103:9578–9583. https://doi.org/10.1073/pnas.0603152103
Louppe D (2008) Broussonetia greveana (Baill.) C.C.Berg. In: Louppe D, Oteng-Amoako AA, Brink M (eds) Prota 7(1): Timbers/Bois d’oeuvre 1 [CD-Rom]. PROTA, Wageningen
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. https://doi.org/10.1093/nar/gkw413
McNeill J, Barrie FR, Buck WR, Demoulin V, Greuter W, Hawksworth DL, Herendeen PS, Knapp S, Marhold K, Prado J, van Prud’homme WF, Smith GF, Wiersema JH, Turland NJ (2012) International Code of Nomenclature for algae, fungi and plants (Melbourne Code) adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. Regnum Veg 154:1–240
Miller A, Schaal B (2005) Domestication of a Mesoamerican cultivated fruit tree, Spondias purpurea. Proc Natl Acad Sci USA 102:12801–12806. https://doi.org/10.1073/pnas.0505447102
Mizumura M, Kubo T, Moriki T (2017) Japanese paper: history, development and use in Western paper conservation. In: Whymark F (ed) Adapt & evolve 2015: East Asian materials and techniques in western conservation. The Institute of Conservation, London, pp 43–59
Narita Y, Yosinaga K (1955) Studies on the breeding of paper mulberry (Broussonetia). III. On the chromosome number of Broussonetia kaempferi Sieb. (TURU-KÔZO). J Jap Forest Soc 37:52 (In Japanese with English abstract)
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323. https://doi.org/10.1073/pnas.70.12.3321
Nei M, Li W-H (1979) Mathematical-model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273. https://doi.org/10.1073/pnas.76.10.5269
Ni JW, Su S, Li H, Geng YH, Zhou HJ, Feng YZ, Xu XQ (2020) Distinct physiological and transcriptional responses of leaves of paper mulberry (Broussonetia kazinoki × B. papyrifera) under different nitrogen supply levels. Tree Physiol 40:667–682. https://doi.org/10.1093/treephys/tpaa021
Oginuma K, Tobe H (1995) Karyomorphology of some Moraceae and Cecropiaceae (Urticales). J Plant Res 108:313–326. https://doi.org/10.1007/Bf02344357
Ohba H, Akiyama S (2014) Broussonetia (Moraceae) in Japan. J Jap Bot 89:123–128 (In Japanese with English abstract)
Ohwi J (1965) Flora of Japan. Smithsonian Institute, Washington, D.C.
Okamoto M (2006) Moraceae. In: Iwatsuki K, Boufford DE, Ohba H (eds) Flora of Japan, vol IIa. Kodansha Ltd., Tokyo, pp 67–77
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420. https://doi.org/10.1093/bioinformatics/btp696
Park S-C, Yoo D-G, Lee C-W, Lee E-I (2000) Last glacial sea-level changes and paleogeography of Korea (Tsushima) Strait. Geo-Mar Lett 20:64–71. https://doi.org/10.1007/s003670000039
Penailillo J, Olivares G, Moncada X, Payacan C, Chang C-S, Chung K-F, Matthews PJ, Seelenfreund A, Seelenfreund D (2016) Sex distribution of paper mulberry (Broussonetia papyrifera) in the Pacific. PLoS ONE 11:e0161148. https://doi.org/10.1371/journal.pone.0161148
Peng X, Shen S (2018) The paper mulberry: a novel model system for woody plant research. Chin Bull Bot 53:372–381. https://doi.org/10.11983/CBB17106 (In Chinese with English abstract)
Peng X, Teng L, Wang X, Wang Y, Shen S (2014) De novo assembly of expressed transcripts and global transcriptomic analysis from seedlings of the paper mulberry (Broussonetia kazinoki × Broussonetia papyifera). PLoS ONE 9:e97487. https://doi.org/10.1371/journal.pone.0097487
Pfeifer B, Wittelsburger U, Ramos-Onsins SE, Lercher MJ (2014) PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol 31:1929–1936. https://doi.org/10.1093/molbev/msu136
Qiu Y-X, Sun Y, Zhang X-P, Lee J, Fu C-X, Comes HP (2009) Molecular phylogeography of East Asian Kirengeshoma (Hydrangeaceae) in relation to Quaternary climate change and landbridge configurations. New Phytol 183:480–495. https://doi.org/10.1111/j.1469-8137.2009.02876.x
Qiu Y-X, Fu C-X, Comes HP (2011) Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol 59:225–244. https://doi.org/10.1016/j.ympev.2011.01.012
Rabah SO, Lee C, Hajrah NH, Makki RM, Alharby HF, Alhebshi AM, Sabir JSM, Jansen RK, Ruhlman TA (2017) Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in cashew. Plant Genome-Us 10:e3. https://doi.org/10.3835/plantgenome2017.03.0020
Rambaut A, Drummond A, Xie D, Baele G, Suchard M (2018) Tracer v1.7, Available from http://beast.community/tracer
Rohwer JG (1993) Moraceae. In: Kubitzki K, Rohwer JG, Bittrich V (eds) The families and genera of vascular plants, vol 2. Springer-Verlag, Heidelberg, pp 438–453
Ruhlman TA, Jansen RK (2021) Plastid genomes of flowering plants: essential principles. In: Maliga P (ed) Chloroplast biotechnology: methods and protocols, methods in molecular biology, 2nd edn. Springer Science, New York, pp 3–47
Schaefer H, Renner SS (2010) A three-genome phylogeny of Momordica (Cucurbitaceae) suggests seven returns from dioecy to monoecy and recent long-distance dispersal to Asia. Mol Phylogenet Evol 54:553–560. https://doi.org/10.1016/j.ympev.2009.08.006
Seki H (1950) On the chromosome number of Broussonetia kazinoki Sieb. Jap J Genet 25:123–125. https://doi.org/10.1266/jjg.25.123 (In Japanese with English abstract)
Siebold PF (1830) Synopsis Plantarum Oeconomicarum Universi Regni Japonici. Verh Batav Genootsch Kunst 12:1–75
Siebold PF, Zuccarini JG (1846) Florae japonicae familiae naturales. Sectio altera. Abh Math-Phys Cl Königl Bayer Akad Wiss 4:123–240
Song M, Munn J (2004) Permanence, durability and unique properties of Hanji. Book Pap Gr Annu 23:127–136
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Swofford DL (2002) PAUP* 4.0: Phylogenetic Analysis Using Parsimony (* and other Methods), v. 4.0 Beta 10. Sinauer Associates, Sunderland
Sytsma KJ, Morawetz J, Pires JC, Nepokroeff M, Conti E, Zjhra M, Hall JC, Chase MW (2002) Urticalean rosids: circumscription, rosid ancestry, and phylogenetics based on rbcL, trnL-F, and ndhF sequences. Am J Bot 89:1531–1546. https://doi.org/10.1002/ajb2.1254
Tamaki I, Obora T, Ohsawa T, Matsumoto A, Saito Y, Ide Y (2021) Different population size change and migration histories created genetic diversity of three oaks in Tokai region, central Japan. J Plant Res 134:933–946. https://doi.org/10.1007/s10265-021-01323-2
Tangkanakul P, Trakoontivakorn G, Auttaviboonkul P, Niyomvit B, Wongkrajang K (2006) Antioxidant activity of northern and northeastern Thai foods containing indigenous vegetables. Agric Nat Resour 40:47–58
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633. https://doi.org/10.1093/genetics/132.2.619
Thwaites GHK (1854) Description of some new genera and species of Ceylon plants. Hooker’s J Bot Kew Gard Misc 6:298–304
Vergara D, White KH, Keepers KG, Kane NC (2016) The complete chloroplast genomes of Cannabis sativa and Humulus lupulus. Mitochondrial DNA A 27:3793–3794. https://doi.org/10.3109/19401736.2015.1079905
Volz SM, Renner SS (2008) Hybridization, polyploidy and evolutionary transitions between monoecy and dioecy in Bryonia (Cucurbitaceae). Am J Bot 95:1297–1306. https://doi.org/10.3732/ajb.0800187
Winter DJ (2012) MMOD: an R library for the calculation of population differentiation statistics. Mol Ecol Resour 12:1158–1160. https://doi.org/10.1111/j.1755-0998.2012.03174.x
Won H (2019) Test of the hybrid origin of Broussonetia × kazinoki (Moraceae) in Korea using molecular markers. Korean J Plant Taxon 49:282–293. https://doi.org/10.11110/kjpt.2019.49.4.282
Wunderlin RP (1997) Moraceae Link—Mulberry Family. In: Flora of North America Editorial Committee (ed) Flora of North America North of Mexico, vol 3. Oxford University Press, New York, pp 388–399
Xu Z, Yang G, Dong M, Wu L, Zhang W, Zhao Y (2018) The complete chloroplast genome of an economic and ecological plant, paper mulberry (Broussonetia kazinoki × Broussonetia papyifera). Mitochondrial DNA Part B 3:28–29. https://doi.org/10.1080/23802359.2017.1419088
Yamazaki T (1989) Moraceae. In: Satake Y, Hara H, Watari S, Tominari T (eds) Wild flowers of Japan: woody plants. Heibonsha Ltd., Tokyo, pp 85–93 (In Japanese)
Yun K-W, Kim M (2009) Taxnomic study of Broussonetia (Moraceae) in Korea. Korean J Pl Taxon 39:80–85
Zeder MA (2015) Core questions in domestication research. Proc Natl Acad Sci USA 112:3191–3198. https://doi.org/10.1073/pnas.1501711112
Zerega NJC, Gardner EM (2019) Delimitation of the new tribe Parartocarpeae (Moraceae) is supported by a 333-gene phylogeny and resolves tribal level Moraceae taxonomy. Phytotaxa 388:253–265. https://doi.org/10.11646/phytotaxa.388.4.1
Zha H-G, Milne RI, Sun H (2010) Asymmetric hybridization in Rhododendron agastum: a hybrid taxon comprising mainly F1s in Yunnan, China. Ann Bot 105:89–100. https://doi.org/10.1093/aob/mcp267
Zhang Q, Liu Y, Sodmergen (2003) Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol 44:941–951. https://doi.org/10.1093/pcp/pcg121
Zhang SD, Soltis DE, Yang Y, Li DZ, Yi TS (2011) Multi-gene analysis provides a well-supported phylogeny of Rosales. Mol Phylogenet Evol 60:21–28. https://doi.org/10.1016/j.ympev.2011.04.008
Zhang S-D, Jin J-J, Chen S-Y, Chase MW, Soltis DE, Li H-T, Yang J-B, Li D-Z, Yi T-S (2017) Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol 214:1355–1367. https://doi.org/10.1111/nph.14461
Zhang H-L, Jin J-J, Moore MJ, Yi T-S, Li D-Z (2018) Plastome characteristics of Cannabaceae. Plant Diversity 40:127–137. https://doi.org/10.1016/j.pld.2018.04.003
Zhang Q, Onstein RE, Little SA, Sauquet H (2019) Estimating divergence times and ancestral breeding systems in Ficus and Moraceae. Ann Bot 123:191–204. https://doi.org/10.1093/aob/mcy159
Zhou Z, Gilbert MG (2003) Moraceae. In: Wu ZY, Raven PH, Hong DY (eds) Flora of China, vol 5. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis, pp 21–73
Zuo L-H, Shang A-Q, Zhang S, Yu X-Y, Ren Y-C, Yang M-S, Wang J-M (2017) The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE 12:e0171264. https://doi.org/10.1371/journal.pone.0171264
Acknowledgements
The authors thank Harvard University Herbaria (HUH) and Herbarium of Biodiversity Research Center, Academia Sinica (HAST) for permission to sample the herbarium specimens and curators of the Herbaria of National Museum of Nature and Science (TNS) and Tohoku University (TUS) for assistances during the visit. The hospitality of Shiroishi City government, Kurafuto Shiroishi, and Endo Tadao’s Family are deeply appreciated. K.-F. Chung’s travel to HUH was supported by 2016 Sargent Award for Visiting Scholar, Arnold Arboretum, Harvard University. We also thank two anonymous reviewers for constructive suggestions for the manuscript, Trevor Padgett for improving the writing, and Preecha Karaket, Danilo N. Tandang, and Pi-Fong Lu for permission to use their photographs. This study was supported by the Thematic Research Program grant of Academia Sinica (AS-TP-107-L18 AS-ASCDC-107-304) and a grant of the Minister of Science and Technology (MOST 109-2621-B-001-005-MY2) to K.-F. Chung.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article is revised due to electronic supplementary files were incorrectly cross linked with different supplementary files and corrected in this version.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kuo, WH., Liu, SH., Chang, CC. et al. Plastome phylogenomics of Allaeanthus, Broussonetia and Malaisia (Dorstenieae, Moraceae) and the origin of B. × kazinoki. J Plant Res 135, 203–220 (2022). https://doi.org/10.1007/s10265-022-01369-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-022-01369-w