Abstract
We report results of karyotype analyses using nine species of Maianthemum from China. The species, except M. atropurpureum (with 2n=72), had 2n=36, and the results support the earlier suggestion that Maianthemum has x=18 with 2n=36 in most species. The species examined, however, showed marked differences in karyotype, particularly in the numbers of metacentric, submetacentric, and acrocentric chromosomes as well as in the number of satellites. In addition, we distinguished three different modes based on the number of clear gaps in chromosome length variation: unimodal, bimodal, and trimodal. The unimodal variation (with no gap) was found in M. dahuricum and M. atropurpureum, the bimodal variation (with one gap) in M. tatsienense, and the trimodal variation (with two gaps) in M. bifolium, M. forrestii, M. japonicum, M. henryi, M. purpureum, and M. lichiangense. In the trimodal variation, the positions of the two gaps may differ from species to species. In addition, the frequency of acrocentric chromosomes per complement was generally higher in the trimodal variation than in the unimodal and bimodal variations. Results of our analyses, which had not been clearly presented prior to this, may provide a better understanding of species evolution in the tribe Polygonatae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maianthemum Wiggers sensu lato (including Smilacina), a genus of the tribe Polygonatae of Ruscaceae (Rudall et al. 2000; APGII 2003), comprises about 35 species distributed in eastern Asia and North America, with one species M. bifolium extending its distribution range from Asia to Europe (Wu and Raven 2000). In the Old World, the genus is represented by 19 species, which occur mainly in the Hengduan Mountains, southwest China (Li 1990). Recent morphological and molecular studies do not clearly show relationships of Maianthemum with other genera in Polygonatae (Rudall et al. 2000; Yamashita and Tamura 2000; Yamashita and Tamura 2004). Like other genera of Ruscaceae (including Convallariaceae), Maianthemum has been relatively well studied cytologically. Till now, 18 of the 35 species have been studied with respect to chromosome features (Table 1). Those cytological data show that there is a variation in karyotype of somatic chromosomes in Maianthemum, and that evaluation is needed as to which karyotype is basic or derived in the genus.
This paper provides results of our study on karyotype analyses of somatic chromosomes in nine Old World species of Maianthemum, and taken together with results of earlier studies, discusses how the genus is likely to be characterized according to chromosome characters. We will also discuss how we can use chromosome data in a future extensive study of Maianthemum and Polygonatae.
Materials and methods
Nine species of Maianthemumthat were collected from China were examined in this study (Table 2). Voucher specimens are deposited in the Herbarium of the Kunming Institute of Botany, the Chinese Academy of Sciences (KUN). Individuals of those species were collected from their native habitats, transplanted in clay pots, and cultivated in the Botanical Garden of the Kunming Institute of Botany.
Root tips were collected from each individual and pretreated with a solution of 0.1% aqueous colchicines for 4–5 h at 20–21°C. After fixation by Carnoy solution (ethanol:acetic acid=3:1) at 4°C, the root tips were dissociated in a mixture of 1 N HCl and 45% acetic acid (1:1) for 30 s at 60°C, stained by 1% acetic orcein for 2–3 h, and squashed on a glass slide. Observations were made using nuclei at interphase, prophase and metaphase, and measurements of chromosome arms were based on at least five cells per individual.
Terminology of chromosome morphology based on the position of a centromere followed Levan et al. (1964). For comparison among different karyotypes at mitotic metaphase, a karyotype formula like 2n=36=12m+10sm+14st(2sat) was used following Kim et al. (2003). In this example, the chromosomes comprise 12 metacentric chromosomes, 10 submetacentric chromosomes, and 14 acrocentric chromosomes (including two chromosomes with a satellite).
Results
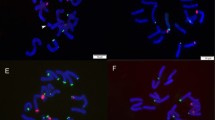
In all nine species examined, the nucleus had many chromatin threads and chromomeres at interphase (Fig. 1), and they were darkly, densely stained throughout the whole chromosome at prophase (Fig. 2). Chromosomes at metaphase were counted 2n=36 or 72 (Fig1, 3–11). They were diverse in shape, having a centromere at the median, submedian, subterminal, or terminal position. Their sizes also varied and ranged from 21.5–2.4 μm. A satellite may be present on one, two or more chromosomes. Based on their shape and size, the chromosomes at metaphase showed a unimodal, bimodal, or trimodal arrangement. Descriptions of the metaphase chromosomes for individual species are given below.
Maianthemum bifolium
The chromosome number of Maianthemum bifolium was 2n=36, with the karyotype formula of 12m+10sm+14st(2sat). Satellites were observed in the proximal regions of short arms in the 11th pair of chromosomes. The chromosome length ranged from about 11.6 μm (the 1st and longest pair) to 3.2 μm (the 18th and shortest pair). However, there were clear gaps in chromosome length between the 1st pair and the subsequent 2nd to 10th pairs (about 9.2–6.3 μm long) and further between the 10th pair and the subsequent 11th to 18th pairs (about 4.2–3.2 μm long) (Fig. 2). Thus the chromosomes showed a trimodal variation with respect to length. The chromosome features were similar to those reported by Kawano et al. (1967). In addition to 2n=36, 42, and 54 have also been reported for the species (Table 1). These numbers may be erroneous.
Maianthemum dahuricum
The chromosome number of Maianthemum dahuricum was 2n=36, with the karyotype formula of 14m+16sm+6st(2sat). Satellites were observed in the proximal regions of short arms in the 9th pair of chromosomes. The chromosome length gradually changed from about 9.7 to 2.9 μm (Fig. 2, 13). Since there was no clear gap in length between the chromosomes, the chromosomes were unimodal in variation. The karyotype differed from the one (2n=36=20m+12sm+2st+2t[2sat]) reported by Hong and Zu (1987). More individuals must be examined for reconfirmation.
Maianthemum forrestii
Maianthemum forrestii was investigated for the first time. The chromosome number was 2n=36, with the karyotype formula of 14m+16sm(1sat)+6st. A satellite was observed in the interstitial region of the short arm of one of the 10th pair of chromosomes. The chromosomes showed a trimodal variation in length (Fig. 2, 14). The chromosome length ranged from about 20.5 (to 14.8) μm (the 1st and longest pair) to 4.2 μm (the 18th and shortest pair). There were clear gaps in chromosome length between the 1st pair and the subsequent 2nd to 10th pairs (about 13.2–7.2 μm long) and further between the 10th pair and the subsequent 11th to 18th pairs (about 6.2–4.2 μm long).
Maianthemum tatsienense
The chromosome number of Maianthemum tatsienense was 2n=36, with the karyotype formula of 18m+10sm+8st. Satellites were not observed. The chromosome length ranged from about 15.8 (to 14.5) μm long (the 1st and longest pair) to 3.7 μm (18th and shortest pair), showing a bimodal variation. The first pair was much longer than the others. The chromosomes from the 2nd pair to the 18th shortest pair gradually varied in length from about 12.1to 3.7 μm (Fig. 2, 15). The karyotype of this species differed from those reported earlier (i.e., 2n=36=16m+10sm+10st[2sat] [Wang et al. 1990], and 2n=36=22m+2sm+12st[2sat] [Wang et al. 1993]).
Maianthemum japonicum
The chromosome number of Maianthemum japonicum was 2n=36, with the karyotype formula of 12m(2sat)+14sm+10st. Satellites were observed in the interstitial regions of short arms in the 10th pair of chromosomes. The chromosomes showed a trimodal variation in length. The chromosome length ranged from about 13.2 μm (the 1st and longest pair) to 2.4 μm (the 18th and shortest pair). There were clear gaps in chromosome length between the 1st pair and the subsequent 2nd through 8th pairs (about 8.4–5.3 μm long) and further between the 8th pair and the subsequent 9th through 18th pairs (about 4.0–2.4 μm long) (Fig. 2, 16). The karyotype we reported here differed from those reported earlier (i.e., 2n=36=20m+6sm+10st[2sat] [Hong and Zu 1987] and 2n=36=20m[2sat]+6sm+10st [Hong and Wilhelm 1990]).
Maianthemum henryi
The chromosome number of Maianthemum henryi was 2n=36, with the karyotype formula of 10m+14sm+12st(2sat). Satellites were observed in the interstitial regions of short arms in 10th pair of chromosomes. The chromosomes showed a trimodal variation in length. The chromosome length ranged from about 13.2 μm (the 1st and longest pair) to 3.2 μm (the 18th and shortest pair). There were clear gaps in chromosome length between the 1st pair and the subsequent 2nd to 10th pairs (about 10.3–6.3 μm long) and further between the 10th pair and the subsequent 11th to 18th pairs (about 4.2–3.2 μm long) (Fig. 3, 17). The karyotype we reported here differed from those reported earlier (i.e., 2n=36=16m+10sm+10st[2sat] [Hong and Zu 1987] and 2n=36=12m+16sm+6st+2t[2sat] [Wang et al. 1993]).
Maianthemum purpureum
The chromosome number of Maianthemum purpureum was 2n=36, with the karyotype formula of 12m+10sm+14st(2sat). Satellites were observed in the interstitial regions of short arms in the ninth pair of chromosomes. The chromosomes showed a trimodal variation in length. The chromosome length ranged from about 12.6 μm (the 1st and longest pair) to 3.4 μm (the 18th and shortest pair). There were clear gaps in chromosome length between the 1st pair and the subsequent 2nd to 10th pairs (about 8.9–5.8 μm long) and further between the 10th pair and the subsequent 11th to 18th pairs (about 4.7–3.4 μm long) (Fig. 3, 18). The karyotype we reported here agrees with the one reported previously by Mehra and Sachdeva (1976).
Maianthemum atropurpureum
The chromosome number of Maianthemum atropurpureum was 2n=72, which is the first record for the species. The karyotype formula is presented as 24m+30sm+18st. Satellites were not observed. The chromosome length gradually changed from about 13.2 to 3.2 μm (Fig. 3, 19). Since there was no clear gap in length among the chromosomes, the chromosomes were unimodal in variation.
Maianthemum lichiangense
The species Maianthemum lichiangense was investigated for the first time. The chromosome number was 2n=36, with the karyotype formula of 14m+12sm+10st. Satellites were not observed. The chromosomes showed a trimodal variation in length. The chromosome length ranged from about 21.5 μm (the 1st and longest pair) to 4.7 μm (the 18th and shortest pair). There were clear gaps in chromosome length between the 1st to 7th pairs (about 21.5–12.4 μm long) and the subsequent 8th to 10th pairs (about 9.2–8.4 μm long), and further between the 8th to 10th pairs and the subsequent 11th to 18th pairs (about 7.1–4.7 μm long) (Fig. 3, 20).
Discussion
As presented above, eight of the nine species of Maianthemum investigated had 2n=36. Only M. atropurpureum had 2n=72, but the species is known to have 2n=36 also (Wang et al. 1993). By putting these data together with information reported in earlier studies (see Table 1), we now have chromosome data from a total of about 20 species in Maianthemum. There is no doubt that Maianthemum has x=18 with 2n=36 in most species as already suggested (Tamura 1995). The species examined, however, showed marked differences in karyotype, particularly in the numbers of metacentric, submetacentric, and acrocentric chromosomes as well as in the number of satellites (Table 2). In addition, our karyotype data were often different from those reported in the earlier studies. There is a possibility that variation exists in karyotype among different individuals of the same species. This must be confirmed in future studies using more individuals from the same population as well as from different populations. Until it is confirmed whether a species has a consistent karyotype or not, the available karyotype data do not allow us to critically compare between species.
Nevertheless, our study also showed that there are three different modes in chromosome length variation: unimodal, bimodal, and trimodal. A few authors had also already noticed the presence of the gaps in chromosome length when chromosmes were serially arranged from long to short ones (Therman 1956; Kawano and Iltis 1966; Kawano et al. 1967; Sen 1974; LaFrankie 1986), but they did not distinguish the three variation modes as we did. The unimodal variation was found in Maianthemum dahuricum and M. atropurpureum, the bimodal variation in M. tatsienense, and the trimodal variation in M. bifolium, M. forrestii, M. japonicum, M. henryi, M. purpureum, and M. lichiangense. In both the bimodal and trimodal variations, the first pair of chromosomes was longer than the remainder, except in M. lichiangense, where the first gap existed between the 7th and 8th pairs. In the case of trimodal variation, another gap existed between the 10th and 11th pairs in M. bifolium, M. forrestii, M. henryi, M. purpureum, and M. lichiangense, but between the 8th and 9th pairs in M. japonicum. In addition, there was a general tendency that the frequency of acrocentric chromosomes per complement was higher in the trimodal variation (27–39% except in M. forrestii [with 22%]) than in the unimodal (11 and 25%) and bimodal variation (22%). Differences in the position of the first and second gaps in the trimodal variation as well as in the frequency of acrocentric chromosomes per complement may suggest the presence of reciprocal translocation among the chromosomes throughout the past species evolution.
Recently, Yamashita and Tamura (2004) discussed the discrimination of unimodal and trimodal variation in chromosome length in the tribe Convallarieae (with x=19 and 18). In the unimodal variation, as in Maianthemum, there was no clear gap in chromosome length when chromosomes were arranged serially from long to short ones. On the other hand, the trimodal variation was composed of one pair of long chromosomes, seven pairs of medium-sized chromosomes, and 11 pairs of short chromosomes. In light of the phylogenetic tree generated by molecular evidence, Yamashita and Tamura (2004) showed that unimodal variation was plesiomorphic in the tribe, from which the trimodal variation was derived. In the case of Maianthemum, tribe Polygonatae, or even all other Ruscaceae as well, it is very likely that species sharing the same mode of chromosome length variation have close affinities.
Thus, while more species of Maianthemum, based on exact determination associated with voucher specimens, should be investigated with respect to their respective karyotypes, molecular analyses are required to clarify species and generic relationships. Resultant phylogenetic relationships would clarify trends of chromosome evolution. Recognition of the three modes in chromosome length variation as well as of additional differences within the trimodal variation we reported in this study will provide us new insight into a better understanding of species relationships and evolution.
References
APG II (2003) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141:399–436
Chuang TI, Chao CY, Hu WL, Kwan SC (1963) Chromosome numbers of the vascular plants of Taiwan I. Taiwania 1:51–66
Gu ZJ, Sun H (1998) The chromosome report of some plants from Motuo, Xizang (Tibet). Acta Bot Yunnanica 20:207–210
Gu ZJ, Wang L, Li H (1992) Karyomorphological studies of some monocots in dulongjiang area. Acta Bot Yunnanica 5(Suppl):77–90
Hara H, Kurosawa S (1963) Cytotaxonomical studies on Japono-Himalayan elements (1). J Jpn Bot 38:7–10
Hara H, Kurosawa S (1964) Cytotaxonomical remarks on some eastern Himalayan and Japanese plants. Adv Front Plant Sci (India) 8:25–32
Hong DY, Wilhelm S (1990) Cytotaxonomical studies of the Liliaceae in the Changbai Mountains of northeastern China. Cathaya 2:151–164
Hong DY, Zu XY (1987) Cytotaxonomical of 10 species of 6 genera. Acta Phytotax Sinica 25:245–253
Kawano S, Iltis HH (1963) Cytotaxonomy of the genus Smilacina (Liliaceae): karyotype analysis of some eastern North American species. Chromosoma (Berl) 14:296–309
Kawano S, Iltis HH (1966) Cytotaxonomy of the genus Smilacina (Liliaceae): chromosome morphology and evolutionary consideration of New World species. Cytologia 31:12–28
Kawano S, Ihara M, Suzki M, Iltis HH (1967) Biosystematic studies on Maianthemum (Liliaceae-Polygonatae): somatic chromosome number and morphology. Bot Mag (Tokyo) 80:345–352
Kim JS, Pak JH, Seo BB, Tobe H (2003) Karyotypes of metaphase chromosomes in diploid populations of Dendranthema zawadskii and related species (Asteraceae) from Korea: diversity and evolutionary implications. J Plant Res 116:47–55
Laane MM, Lie T (1985) Fremstilling av kromosompreparater med enkle metoder. Blyttia 7–15
LaFrankie JV (1986) Morphology and taxonomy of the New World species of Maianthemum (Liliaceae). J Arnold Arbor 67:371–439
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centrometric position on chromosome. Hereditas 52:201–220
Li H (1990) Infrageneric system of the genus Maianthemum. Acta Bot Yunnanica Suppl 3:1–12
Mehra PN, Sachdeva SK (1976) Cytological observation on some W Himalayan Monocots: Smilacaceae, Liliaceae and Trilliaceae. Cytologia 41:5–22
Mehra PN, Sachdeva SK (1979) Cytological observations on some East-Himalayan monocots. Cytologia 44:233–240
Rudall PJ, Conran JG, Chase MW (2000) Systematics of Ruscaceae/Convallariaceae: a combined morphological and molecular investigation. Bot J Linn Soc 134:73–92
Sen S (1974) Chromosome evolution in Polygonateae. Bull Bot Soc Bengal 28:103–111
Sokolovskaya AP, Probatova NS (1985) Chromosome numbers in the vascular plants from the Primorye territory, Kamchatka region, Amur valley and Sakhalin. Bot Zurn SSSR 70:997–999
Tamura MN (1995) A karyological review of the orders Asparagales and Liliales (Monocotyledonae). Feddes Repert 106:83–111
Therman E (1956) Cytotaxonomy of the tribe Polygonatae. Am J Bot 43:134–142
Wang SF, Xu JM, Yu SH (1990) Report on karyotype of Smilacina tatsienensis and Ophiopogon japonicus. Acta Phytotax Sin 28:207–210
Wang L, Gu ZJ, Gong X, Xiao TJ (1993) A Cytological Study of fifteen species in six genera of Liliaceae from Yunnan. Acta Phytotax Sin 31:549–559
Wu ZY, Raven PH (eds) (2000) Flora of China, vol 24. Science Press, Beijing, and Missouri Botanical Garden Press, St Louis, MO, pp 217–222
Yamashita J, Tamura MN (2000) Molecular phylogeny of the Convallariaceae (Asparagales). In: Willson KL, Morrison DA (eds) Monocots: systematics and evolution. CSIRO, Melbourne, pp 387–400
Yamashita J, Tamura MN (2004) Phylogenetic analyses and chromosome evolution in Convallarieae (Ruscaceae sensu lato) with some taxonomic treatments. J Plant Res 117:363–370
Acknowledgments
We are grateful to Profs. Li Heng and Kazuo Oginuma for their assistance in completing the study. The study was partly supported by grants from Chinese Natural and Science Foundation (CNSF 40332021 to H. Sun and 30300023 to Z.-L. Nie), and the Chinese Academy of Sciences (KSCX2-SW-009 to Y. P. Yang).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Meng, Y., Nie, ZL., Yang, YP. et al. Karyomorphology of Maianthemum sensu lato (Polygonatae, Ruscaceae). J Plant Res 118, 155–162 (2005). https://doi.org/10.1007/s10265-005-0205-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-005-0205-7