Abstract
Multiple myeloma (MM), one of the most intractable malignancies, is characterized by the infiltration and growth of plasma cells, the most differentiated cells in the B-cell lineage, in the bone marrow. Despite the introduction of novel therapeutic agents, including proteasome inhibitors and immunomodulatory drugs, the prognosis of patients with MM is still worse than that of most hematological malignancies. A better understanding of the molecular pathogenesis of the disease is essential to achieve any improvement of treatment outcome of MM patients. All MM cases pass through the phase of asymptomatic expansion of clonal plasma cells, referred to as monoclonal gammopathy of undetermined significance (MGUS). It has long been believed that MM evolves linearly from MGUS to terminal phases, such as extramedullary tumors and plasma cell leukemia via the accumulation of novel mutations. However, recent studies using next-generation sequencing have disclosed the complex genomic architecture of the disease. At each step of progression, the acquisition of novel mutations is accompanied by subclonal evolution from reservoir clones with branching patterns. Each subclone may carry novel mutations and distinct phenotypes, including drug sensitivity. In addition, minor clones already exist at the MGUS stage, which could expand later in the clinical course, resulting in relapse and/or leukemic conversion. The ultimate goal of treatment is to eradicate all clones, including subclonal populations with distinct biological characteristics. This goal could be achieved by further improving treatment strategies that reflect the genomic landscape of the disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple myeloma (MM), one of the most intractable malignancies, is characterized by the infiltration and growth of malignant plasma cells in the bone marrow [1–3]. Despite the introduction of high-dose chemotherapy with stem cell support and novel therapeutic agents, including proteasome inhibitors and immunomodulatory drugs (IMiDs), the prognosis of patients with MM is still worse than that of other hematological malignancies, such as acute myeloid leukemia and diffuse large B-cell lymphoma [4–7]. A better understanding of the molecular pathogenesis of the disease is essential to achieve any improvement of treatment outcome of MM patients.
It is now widely believed that all MM cases pass through the phase of asymptomatic expansion of clonal plasma cells, referred to as monoclonal gammopathy of undetermined significance (MGUS) [8, 9]. When the proportion of malignant plasma cells, which can be distinguished by the surface marker combination of CD19−/CD56+/CD45−/CD38+ from their normal counterparts (CD19+/CD56−/CD45+/CD38+), is <10 % of all bone marrow mononuclear cells, the disease is diagnosed as MGUS by definition (Fig. 1). When the proportion of malignant plasma cells exceeds 10 %, the disease condition is defined as MM, which is classified into two groups, namely, smoldering (asymptomatic) MM and symptomatic MM, according to the absence or presence of so-called CRAB symptoms (hyperCalcemia, Renal failure, Anemia and lytic Bone disease). It is estimated that MGUS progresses to MM at the rate of 1–2 % per year. The growth and survival of MM cells are strongly supported by bone marrow stroma cells via direct contact and/or stroma-derived soluble factors, such as interleukin-6 [10–13]; however, upon disease progression, MM cells acquire the ability of stroma-independent growth and extend outside the bone marrow to form extra-medullary lesions (plasmacytoma) or become leukemic (plasma cell leukemia) at terminal stages.
The expression pattern of surface markers during B-cell differentiation and transformation to myeloma. Germinal center B cells (GC cells) lose and acquire the surface expression of CD10 and CD27, respectively, when they differentiate into post-GC cells concomitant with the acquisition of the capability to produce high-affinity immunoglobulins through somatic hypermutations (SHM), receptor editing (Receptor Edit) and class switch recombination (CSR). Normal plasma cells (PC) change to express the hallmark protein CD38 upon differentiation from CD38-negative post-GC cells, but they retain the ability to express CD19. In striking contrast, myeloma cells (malignant PC) lack the ability to express CD19. In addition, the expression levels of CD56 and CD45 increase and decrease, respectively, during malignant transformation. When the proportion of malignant PC (M) is <10 % of bone marrow mononuclear cells, the disease condition is considered to be monoclonal gammopathy of undetermined significance (MGUS) by definition. The diagnosis of multiple myeloma (MM) is made when the proportion of malignant PC exceeds 10 % in the bone marrow
The molecular abnormalities underlying each step of myeloma development have been elucidated as: (1) transformation of normal plasma cells to MGUS; (2) progression of MGUS to MM; (3) final evolution to extra-medullary diseases. Although it was long believed that MM evolves linearly from MGUS through MM to extra-medullary diseases, recent detailed analyses with refined technology, including comparative genomic hybridization, single nucleotide polymorphism arrays, and next-generation sequencing, have unveiled the complex genomic landscape of the disease. Such studies have greatly increased our understanding of MM, but raised additional important questions which have also to be resolved.
Molecular mechanisms underlying the progression of multiple myeloma
Plasma cells are terminally differentiated cells of the B-cell lineage, which have experienced somatic hypermutation and class switch recombination (Fig. 1) and, consequently, acquired the ability to produce specific immunoglobulins upon exposure to cognate antigens [14, 15]. They arrest in the G0/G1 phase of the cell cycle as long-lived plasma cells in the bone marrow or memory B-cells in the lymph nodes until the next antigen challenge. Therefore, the initial step in the development of myeloma should be the acquisition of a growth advantage by dormant plasma cells or memory B-cells. This process is accomplished through deregulation of a critical regulator of the G1/S transition, the cyclin D family, via two mechanisms in most but not all cases (Fig. 2a). The first mechanism is the generation of recurrent chromosomal translocations involving the immunoglobulin heavy chain (IgH) locus at 14q32; this takes place in approximately 50 % of cases during the somatic hypermutation or class switch recombination process—with a few exceptions [16]. The most frequent recurrent chromosomal translocation is t(11;14)(q13;q32), observed in 15–20 % of cases [17], followed by t(4;14)(p16;q32), with a 12–15 % prevalence [18, 19]. Other translocations, including t(14;16)(q32;q23), t(14;20)(q32;q11), and t(6;14)(p21;q32), are more rarely detected in MM patients (<5 %) [20]. In myeloma cells harboring t(11;14)(q13;q32), the cyclin D1 gene, which is located at 11q13, is aberrantly driven by the heavy chain enhancer that is in close proximity due to chromosomal translocation [21]. Similarly, the cyclin D3 gene at 6p21 is overexpressed in MM cells carrying t(6;14)(p21;q32) [22]. The t(4;14)(p16;q32) translocation results in deregulation of the histone methyltransferase MMSET (also known as WHSC1/NSD2/KMT3G), which causes a decrease in the H3K27me3 level and an increase in the H3K36me2 level along the entire genome, resulting in the derangement of several genes, including cyclin D2 [23, 24]. It has been reported that MMSET overexpression also impairs DNA repair via the modulation of H4K20 methylation [25]. Translocations t(14;16)(q32;q23) and t(14;20)(q32;q11) upregulate Maf family transcription factors c-Maf and MafB, the target genes of which also include cyclin D2 [26, 27]. The second mechanism underlying the malignant transformation of plasma cells to MGUS is hyperdiploidy, which is observed in up to 55 % of MM patients, including 10 % with overlap with 14q translocations [28–31]. For unknown reasons, the odd-numbered chromosomes 3, 5, 7, 9, 11, 15, 19, and 21 show a gain. The precise mechanisms by which hyperdiploidy causes plasma cell transformation is not understood at present; however, the most prevalent hyperdiploidy (approx. 30 %), namely, trisomy 11, may cause cyclin D1 overexpression due to an increase in gene dosage.
a The overexpression of cyclin D is commonly observed during the initial step of the development of MM. Cyclin D1 and D3 are directly transactivated by the immunoglobulin heavy chain (IgH) enhancer in myeloma cells carrying t(11;14) and t(6;14), respectively. The histone methyltransferase MMSET, which is driven by the IgH enhancer, induces cyclin D2 overexpression in myeloma cells carrying t(4;14). Similarly, Maf family transcription factors activate the cyclin D2 gene in myeloma cells carrying t(14;16) and t(14;20). In addition, trisomy of chromosome 11 results in the enhanced expression of cyclin D1 due to an increase in gene dosage. Overexpressed cyclin Ds confer a growth advantage to plasma cells and also induce genomic instability to trigger secondary genetic abnormalities. b Overview of genetic abnormalities related to the initiation and progression of multiple myeloma. Chromosomal translocations involving 14q and hyperdiploidy are observed during the initial step of the malignant transformation of normal plasma cells (Normal PC). The progression from monoclonal gammopathy of undetermined significance (MGUS) to myeloma is associated with Ras point mutations, c-Myc overexpression, DNA hypomethylation, and the deletion of chromosome 13. The final step, i.e., the development of extramedullary diseases, is accompanied by the activation of mutations of components of the NF-κB pathway, chromosomal translocation involving the c-myc gene, 1q gain, and the loss of 1p and 17p
A further increase in the growth potential is required for MGUS to progress to MM because this process is quantitatively defined by an increase (>10 %) in the number of malignant plasma cells in the bone marrow. The acquisition of this additional growth advantage is believed to be mediated through the activation of two major oncogenes, Ras and Myc, at this stage (Fig. 2b). The frequency of the point mutations of K-Ras and N-Ras increases from approximately 7 % in MGUS to 24–27 % in MM [32, 33]. Little is currently known about the mechanisms underlying the acquisition of non-synonymous point mutations of Ras family oncogenes by MM cells, but the involvement of APOBEC3 cytidine deaminase is suggested by a recent comprehensive analysis of the mutation signature of multiple cancers (Fig. 3) [34, 35]. During the progression of MGUS to MM, c-Myc overexpression occurs at a frequency of 15 %, probably via superenhancer-mediated hyperactivation of transcription. This offers the possibility of therapeutic intervention with small molecular inhibitors of the superenhancer-binding transactivator BRD4, collectively called BET inhibitors [36, 37]. The deletion of chromosome 13 is often observed at this stage, especially in t(11;14)-positive myeloma, and this deletion contributes to disease progression via haploinsufficiency of the RB tumor suppressor gene at 13q14 [38]. Chromosome 13 deletion can also be an early event in MMSET- and Maf-deregulated myeloma, in which a small number of driver translocations sometimes exist in the entire clonal population, and may function as an initiating event instead of—or in addition to—cyclin D overexpression [38]. The progression of MGUS to MM is often accompanied with DNA hypomethylation, which may be caused by aging-linked spontaneous deamination of methylated cytidine residues (Fig. 3) [34, 35]. DNA hypomethylation causes genomic instability, thereby further accelerating the progression of the disease.
Two mutation signatures observed in multiple myeloma. a Fraction of contribution of each mutation type in each context for the two mutation patterns identified by whole-exon sequencing. The major components contributing to each signature are highlighted with arrows (adapted from Bolli et al. [35] with permission). b The mechanisms by which each mutation signature is formed in myeloma cells. The signature 5 mechanism is generated by 5-oxidation and spontaneous deamination of methylated cytidine residues and is closely related to aging. The signature 2 mechanism is generated by APOBEC3-mediated C to T conversion
The terminal stage of myeloma progression is characterized by the ability for stroma-independent growth, which results in extramedullary diseases and plasma cell leukemia (PCL). The main driver of stroma-independent growth of MM cells is the constitutive activation of NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells): the interaction with bone marrow stroma is mediated by NF-κB-dependent expression of adhesion molecules, such as VLA-4, which is sustained by direct contact and/or stroma-derived cytokines. The homozygous deletion of the genes encoding inhibitors of the NF-κB pathways is one mechanism of the constitutive activation of NF-κB; such deletions are often detected in MM cells at this stage, including BIRC2/3 on chromosome 11 (approx. 7 %), TRAF3 on chromosome 14 (approx. 3 %), and CYLD on chromosome 16 (approx. 3 %) (Fig. 4) [39, 40]. Terminal-stage myeloma also shows massive structural chromosomal abnormalities, such as complex translocations involving the 8q24 locus, in which the c-Myc gene is located [41–43], duplication of the long arm of chromosome 1 (1q gain), and deletions of 1p32 or 17p13 [44–46]. The loss of 17p results in the inactivation of p53, as has been observed at the end stage of many cancers, including chronic myeloid leukemia (CML).
Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling pathway mutations in multiple myeloma. Two pathways, namely, the canonical and non-canonical pathways both lead to the activation of NF-κB and target gene expression. Arrows and bars indicate activating and inhibitory steps, respectively. Negative regulators of NF-κB signaling that undergo loss-of-function mutations in MM are designated in red (cIAP1/2 is transcribed from the BIRC2/3 genes). Positive regulators and effectors of NF-κB signaling that undergo gain-of-function mutations in MM are designated in sky blue. In both cases, these mutations lead to constitutive activation of NF-κB, as evidenced by nuclear translocation of p65/p50 in the canonical pathway and of p52/RelB in the non-canonical pathway, and increased target gene expression in MM
Intra-clonal heterogeneity in multiple myeloma
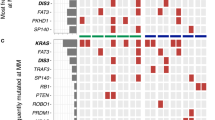
It has widely been believed that cancer is initiated in a single clone (cancer stem cell) and then evolves via the acquisition of genetic events that change the biology of the cells toward a more malignant and drug-resistant state in a relatively stepwise manner (Fig. 5a). This view is best exemplified by the clinical course of CML. Hematopoietic stem cells harboring the BCR-ABL fusion gene self-renew and propagate differentiated offspring that constitute large tumor populations as CML stem cells or CML-initiating cells. These cells respond well to ABL inhibitors but change to become drug resistant following acquisition of point mutations in the BCR-ABL gene (accelerated phase). When devastating events such as the loss of p53 take place, a CML clone finally transforms to acute leukemias (blastic crisis). The progression of MM has long been considered to follow the linear evolution pattern. In support of this view, a study using whole-exon sequencing demonstrated that the numbers of non-synonymous point mutations are linearly increased from MGUS, smoldering MM, symptomatic MM to PCL (median numbers of non-synonymous point mutations are 13, 28, 31 and 59, respectively) [47]. However, three seminal reports on the detailed time-course analyses of primary samples revealed the existence of a more complex genomic architecture of the disease [48–50]. In these studies, the comparison of copy number abnormalities or point mutations revealed a certain degree of heterogeneity within tumor clones of the same patients; for example, the relapsed clones would carry only a part of the sets of mutations observed at diagnosis with or without additional genetic abnormalities (Fig. 6). Moreover, clones harboring novel mutations often emerge at the time of relapse or at the end of the disease, especially at the PCL phase. These observations strongly suggest that minor subclones derived from an initiating clone exist at the time of diagnosis at undetectable frequencies and are ultimately responsible for relapse. This pattern is the most frequent one observed, in up to 50 % of cases. In approximately 30 % of cases, the relapsed clones possess novel genetic changes in addition to the original mutations and, therefore, represent classical clonal evolution (Fig. 7) [51]. The Kernel density plot analyses of clonal distribution using whole-exome data revealed that intra-clonal heterogeneity is already present at the premalignant phases of MM, namely MGUS and smoldering MM [47]. In addition, a study focusing on patients with MM carrying t(4;14) found that this driver mutation of myelomagenesis was sometimes detected only in a minor subclone at diagnosis, becoming dominant later or at relapse [52]. These results indicate that clonal diversity arises at very early phases of the disease and that clonal competition may affect the rate of disease progression. The progression of MM appears to be driven via branching evolutionary patterns according to the Darwinian model of evolution, which was originally proposed to explain the origin of the species, rather than following a linear multistep process (Fig. 5b). In other words, there are multiple clones with the variable ability to propagate descendants at each step of disease progression, and an increase in these reservoir clones may strongly affect the biological behavior of the entire disease, including malignant phenotype and drug sensitivity. In this concept, high-risk abnormalities, such as deregulation of the Maf family, may not be a simple consequence of disease progression, but a driving force of disease progression via accelerating clonal heterogeneity, ultimately leading to a clonal dominance of the selected ones [49, 53]. The feature of intra-clonal heterogeneity has a significant impact on how MM should be treated and on how therapeutic effects should be considered within the concept implicit in evolutionary biology.
Two models of disease progression of multiple myeloma. a In general, tumor cells are considered to be derived from a single initiating cell (D) and then to linearly evolve through the acquisition and accumulation of somatic mutations with sequential selections, leading to total domination by the fittest clone (clonal evolution). b Recent approaches with whole-genome sequencing suggest that the molecular events underlying myeloma development and progression are not attained in a linear fashion but rather through branching, nonlinear pathways that are typical of those proposed by Charles Darwin to explain the evolution of species (Darwinian branching model). See caption to Fig. 6 for definition of symbols
Clonal divergence revealed by the detection of variant alleles (a case reported by Egan et al. [48]). In this example, there are 15 variants common to all clones and shared by a common ancestor (filled circle). Six variants (filled star) are common to only the clones at diagnosis (clone 1.1) and the second relapse (clone 1.2), whereas no variants are common to the clones at the first relapse (clone 2) and leukemic phase (clone 3). The greatest divergence is observed between the first relapse and leukemic phase, with 7 unique variants detected in each sample (filled triangle and filled square, respectively). SVN Single-nucleotide variants
Three temporal types of disease progression were identified by analyzing the samples of relapsed cases: 1 the relapsing disease is identical to the diagnostic clone (vertical red arrow), which is genetically stable but resistant to chemotherapies, 2 linear evolution from the diagnostic clone with the acquisition of only a few novel abnormalities, 3 direct emergence of genetically identical clones from tumor-initiating cells or ancestor clones (marked D). The most frequently observed type of disease progress is the third type (approx. 50 %). See caption to Fig. 6 for definition of symbols
Clinical implications
Linear evolution of cancer is naturally linked to the notion of a risk-adapted treatment strategy. In the case of MM, the best example of such a strategy is the mSMART (Mayo Stratification for Myeloma And Risk-adapted Therapy) strategy, in which therapeutic options are divided into four intensities according to risk factors [54, 55]. The standard risk factors include the genetic alterations observed at the transformation of normal plasma cells to MGUS, with hyperdiploidy and chromosomal translocations directly accelerating the transactivation of cyclin D, namely t(11;14) and t(6;14). The high-risk factors are genetic abnormalities observed at the advanced stage of MM, such as del(17p13), and chromosomal translocations involving Maf family transcription factors. Indeed, several clinical trials have proved that genetic abnormalities observed at the advanced stage, such as del(17p13) and +1q21, act as independent worse prognostic factors in MM patients [56]. However, it is not surprising that the concept of intra-clonal heterogeneity substantially influences the treatment strategies of MM. As a proof of concept, Keats et al. [49] reported a case showing the inter-clonal difference in susceptibility to novel agents, thereby underscoring the importance of the comprehensive approach covering all clones (“one-size fits all”) so as not to allow selection of highly malignant and/or drug-resistant clones. For this purpose, the combination of highly active anti-myeloma drugs, such as proteasome inhibitors and IMiDs, is strongly recommended, although how to combine and order these to achieve an efficacious (chemo)therapeutic regimen is still controversial [57]. Given the existence of clonal heterogeneity at MGUS stages, early intervention is another tempting option, but also still a matter of debate [58, 59]. Early intervention would be successful if all clones are eradicated or significantly suppressed for a long time; otherwise, the result of this strategy would be the selection of drug-resistant clones and even more malignant conversion of the disease. Clinically, some cases are characterized by clones with slow growth and a stable genome that may remain indolent and stable for many years despite a suboptimal response to chemotherapy [60]. An approach that distinguishes indolent from more aggressive clones and tailored therapy accordingly is therefore of primary importance in clinical practice going forward even in light of the current concept of intra-clonal heterogeneity.
Conclusion and future perspective
In this review, we have surveyed our current understanding and knowledge of the molecular pathogenesis of MM and its clinical relevance. Substantial proportions of MM cases appear to progress through the classical pathway of linear clonal evolution, but are still associated with the early emergence of intra-clonal heterogeneity. At each step of progression, the acquisition of novel mutations is accompanied by subclonal evolution with branching patterns from reservoir clones. Each subclone may carry novel mutations and distinct phenotypes, including drug sensitivity. In addition, minor clones in the early pool could expand later in the clinical course, resulting in relapse and/or leukemic conversion in a considerable subset of cases. This complex genomic architecture makes it difficult to develop effective drugs targeting driver mutations. In MM, fewer than 30 genes are recurrently mutated in at least 5 % of patients and, notably, many of these are present in subclones or minor clones, which are supposed to act as drivers, such as Ras family oncogenes. The ultimate goal of treatment is to achieve long-term remission and cure by eradicating all clonal cells, including subclonal populations, with distinct biological characteristics. It seems difficult to achieve this goal, but a promising future is inferred from the mutation landscape: the median number of non-synonymous recurrent mutations is 35 per genome in MM, which is virtually identical to that of B-cell lymphomas (30 per genome) and falls between acute leukemias (12–13 per genome) and intractable solid tumors such as lung cancer and malignant melanoma (>500 per genome) [34].
References
Palumbo A, Anderson K (2011) Multiple myelomas. N Engl J Med 364:1046–1060
Mitsiades CS, Davies FE, Laubach JP et al (2011) Future directions of next-generation novel therapies, combination approaches, and the development of personalized medicine in myeloma. J Clin Oncol 29:1916–1923
Corre J, Munshi N, Avet-Loiseau H (2015) Genetics of multiple myeloma: another heterogeneity level? Blood 125:1870–1876
Gay F, Larocca A, Wijermans P et al (2011) Complete response correlates with long-term progression-free and overall survival in elderly myeloma treated with novel agents: analysis of 1175 patients. Blood 117:3025–3031
Bringhen S, Mateos MV, Zweegman S et al (2013) Age and organ damage correlate with poor survival in myeloma patients: meta-analysis of 1435 individual patient data from 4 randomized trials. Haematologica 98:980–987
Sonneveld P, Goldschmidt H, Rosiñol L et al (2013) Bortezomib-based versus nonbortezomib-based induction treatment before autologous stem-cell transplantation in patients with previously untreated multiple myeloma: a meta-analysis of phase III randomized, controlled trials. J Clin Oncol 31:3279–3287
Kumar SK, Dispenzieri A, Lacy MQ et al (2014) Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia 28:1122–1128
Weiss BM, Abadie J, Verma P et al (2009) A monoclonal precedes multiple myeloma in most patients. Blood 113:5418–5422
Agarwal A, Ghobrial IM (2013) Monoclonal gammopathy of undetermined significance and smoldering multiple myeloma: a review of the current understanding of epidemiology, biology, risk stratification, and management of myeloma precursor disease. Clin Cancer Res 19:985–994
Hideshima T, Mitsiades C, Tonon G et al (2007) Understanding multiple myeloma pathogenesis in the bone marrow to identify new therapeutic targets. Nat Rev Cancer 7:585–598
Meads MB, Hazlehurst LA, Dalton WS (2008) The bone marrow microenvironment as a tumor sanctuary and contributor to drug resistance. Clin Cancer Res 14:2519–2526
Noborio-Hatano K, Kikuchi J, Takatoku M et al (2009) Bortezomib overcomes cell-adhesion-mediated drug resistance through downregulation of VLA-4 expression in multiple myeloma. Oncogene 28:231–242
Kikuchi J, Koyama D, Mukai HY et al (2014) Suitable drug combination with bortezomib for multiple myeloma under stroma-free conditions and in contact with fibronectin or bone marrow stromal cells. Int J Hematol 99:726–736
Nutt SL, Taubenheim N, Hasbold J et al (2011) The genetic network controlling plasma cell differentiation. Semin Immunol 23:341–349
Pieper K, Grimbacher B, Eibel H (2013) B-cell biology and development. J Allergy Clin Immunol 131:959–971
Walker BA, Wardell CP, Johnson DC et al (2013) Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood 121:3413–3419
González D, van der Burg M, García-Sanz R et al (2007) Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma. Blood 110:3112–3121
Chesi M, Nardini E, Brents LA et al (1997) Frequent translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet 16:260–264
Chesi M, Nardini E, Lim RSC et al (1998) The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92:3025–3034
Iida S, Rao PH, Butler M et al (1997) Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet 17:226–230
Chesi M, Bergsagel PL, Brents LA et al (1996) Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood 88:674–681
Bergsagel PL, Kuehl WM, Zhan F et al (2005) Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 106:296–303
Lauring J, Abukhdeir AM, Konishi H et al (2008) The multiple myeloma-associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood 111:856–864
Martinez-Garcia E, Popovic R, Min D-J et al (2011) The MMSET histone methyltransferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 117:211–220
Pei H, Zhang L, Luo K et al (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470:124–128
Hurt EM, Wiestner A, Rosenwald A et al (2004) Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell 5:191–199
Chesi M, Bergsagel PL, Shonukan OO et al (1998) Frequent dysregulation of the c-maf protooncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood 91:4457–4463
Sawyer JR, Waldron JA, Jagannath S et al (1995) Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet 182:41–49
Laï JL, Zandecki M, Mary JY et al (1995) Improved cytogenetics in multiple myeloma: a study of 151 patients including 117 patients at diagnosis. Blood 85:2490–2497
Calasanz MJ, Cigudosa JC, Odero MD et al (1997) Cytogenetic analysis of 280 patients with multiple myeloma and related disorders: primary breakpoints and clinical correlations. Genes Chromosom Cancer 18:84–93
Smadja NV, Bastard C, Brigaudeau C et al (2001) Hypodiploidy is a major prognostic factor in multiple myeloma. Blood 98:2229–2238
Chapman MA, Lawrence MS, Keats JJ et al (2011) Initial genome sequencing and analysis of multiple myeloma. Nature 471:467–472
Lohr JG, Stojanov P, Carter LS et al (2014) Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell 25:91–101
Alexandrov LB, Nik-Zainal S, Wedge DC et al (2013) Signatures of mutational processes in human cancer. Nature 500:415–421
Bolli N, Avet-Loiseau H, Wedge DC et al (2014) Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun 5:2997. doi:10.1038/ncomms3997
Delmore JE, Issa GC, Lemieux ME et al (2011) BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146:904–917
Loven J, Hoke HA, Lin CY et al (2013) Selective inhibition of tumor oncogenesis by disruption of super-enhances. Cell 153:320–334
Chiecchio L, Dagrada GP, Ibrahim AH et al (2009) Timing of acquisition of deletion 13 in plasma cell dyscrasias is dependent on genetic context. Haematologica 94:1708–1713
Annunziata CM, Davis RE, Demchenko Y et al (2007) Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12:115–130
Demchenko YN, Glebov OK, Zingone A et al (2010) Classical and/or alternative NF-κB pathway activation in multiple myeloma. Blood 115:3541–3552
Sawyer JR, Tian E, Heuck CJ et al (2014) Jumping translocations of 1q12 in multiple myeloma: a novel mechanism for deletion of 17p in cytogenetically defined high-risk disease. Blood 123:2504–2512
Affer M, Chesi M, Chen WD et al (2014) Promiscuous MYC locus rearrangements hijack enhancers but mostly super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 28:1725–1735
Walker BA, Wargell CP, Brioli A et al (2014) Translocations at 8q24 juxtapose MYC with genes that harbor superenhancers resulting in overexpression and poor prognosis in myeloma patients. Blood Cancer J 4:e191
Avet-Loiseau H, Attal M, Campion L et al (2012) Long-term analysis of the IFM 99 trials for myeloma: cytogenetic abnormalities [t(4;14), del(17p), 1q gains] play a major role in defining long-term survival. J Clin Oncol 30:1949–1952
Rajkumar SV, Gupta V, Fonseca R et al (2013) Impact of primary molecular cytogenetic abnormalities and risk of progression in smoldering multiple myeloma. Leukemia 27:1738–1744
Hebraud B, Leleu X, Lauwers-Cances V et al (2014) Deletion of the 1p32 region is a major independent prognostic factor in young patients with myeloma: the IFM experience on 1195 patients. Leukemia 28:675–679
Walker BA, Wardell CP, Melchor L et al (2014) Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia 28:384–390
Egan JB, Shi C-X, Tembe W et al (2012) Whole-genome sequencing of multiple myeloma from diagnosis to plasma cell leukemia reveals genomic initiating events, evolution, and clonal tides. Blood 120:1060–1066
Keats JJ, Chesi M, Egan JB et al (2012) Clonal competition with alternating dominance in multiple myeloma. Blood 120:1067–1076
Walker BA, Wardell CP, Melchor L et al (2012) Intraclonal heterogeneity and distinct molecular mechanisms characterize the development of t(4;14) and t(11;14) myeloma. Blood 120:1077–1086
Bahlis NJ (2012) Darwinian evolution and tiding clones in multiple myeloma. Blood 120:927–928
Hébraud B, Caillot D, Corre J et al (2013) The translocation t(4;14) can be present only in minor subclones in multiple myeloma. Clin Cancer Res 19:4634–4637
Magrangeas F, Avet-Loiseau H, Gouraud W et al (2013) Minor clone provides a reservoir for relapse in multiple myeloma. Leukemia 27:473–481
Rajkumar SV (2012) Multiple myeloma: 2012 update on diagnosis, risk-stratification, and management. Am J Hematol 87:79–88
Mikhael JR, Dingli D, Roy V et al (2013) Management of newly diagnosed symptomatic multiple myeloma: updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin Proc 88:360–376
Boyd KD, Ross FM, Chiecchio L et al (2012) A novel prognostic model in myeloma based on co-segregation adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia 26:349–355
Mateos MV, Martinez-Lopez J, Hernandez MT et al (2014) Comparison of sequential vs alternating administration of bortezomib, melphalan, prednisone (VMP) and lenalidomide plus dexamethasone (Rd) in elderly patients with newly diagnosed multiple myeloma (MM): GEM2010MAS65 trial. In: Burns LJ (ed) 56th ASH Annual Meeting. American Society of Hematology, Washington DC, Abstract #178
Mateos MV, Hernandez MT, Giraldo P et al (2013) Lenalidomide plus dexamethasone for high-risk smoldering multiple myeloma. N Engl J Med 369:438–481
Ghobrial IM, Landgren O (2014) How I treat smoldering multiple myeloma. Blood 124:3380–3388
Pineda-Roman M, Bolejack V, Arzoumanian V et al (2007) Complete response in myeloma extends survival without, but not with history of prior monoclonal gammopathy of undetermined significance or smouldering disease. Br J Haematol 136:393–399
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Furukawa, Y., Kikuchi, J. Molecular pathogenesis of multiple myeloma. Int J Clin Oncol 20, 413–422 (2015). https://doi.org/10.1007/s10147-015-0837-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-015-0837-0