Abstract
Clonal growth can increase not only floral display but also geitonogamy and may affect sexual reproduction both positively and negatively. A clonal woody species, Prunus ssiori, was partially self-incompatible according to a pollination experiment. Its main pollinators, bumble bees, were often observed to consecutively visit inflorescences within a tree. Clone identification revealed that its genets formed mutually exclusive patches. These features suggest frequent geitonogamous pollination. In a 6.24-ha plot, 212 trees belonged to 59 genets, and 42 genets consisted of a single tree, whereas the rest contained two or more clonal trees. The largest genet had 65 trees and occupied 0.4 of a hectare. Fruit set was measured in 127 inflorescences sampled from nine maternal trees at the center of the plot. Paternal genets of 107 of their 300 seeds were assigned in the plot using microsatellites. There were no selfed seeds. Male reproductive success (the probability that individual trees of each genet sired a seed) increased as tree size increased, as the distance between the trees and maternal trees decreased, when the genet did not contain the maternal trees, and when the genet consisted of a single tree. Female reproductive success (fruit set in individual inflorescences of each maternal tree) increased as the within-tree geitonogamy index, which reflected the frequency of pollination within the maternal tree, decreased. These results suggest that clonal growth reduces male reproductive success, at least, in P. ssiori, because of pollen discounting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most clonal plants can also reproduce sexually and regenerate through seedlings. In such clonal plants, a spatial clonal structure resulting from the clonal growth strategy affects the distribution of flowers in genets that consist of genetically identical ramets originating from individual seedlings. The clonal growth strategy has been categorized into two extreme types along a continuum of clonal growth forms (Lovett Doust 1981): in the phalanx type, ramets are connected with short internodes and closely spaced, and genets form a clumped distribution of discrete clonal patches; on the other hand, in the guerrilla type, ramets are connected with long internodes and widely dispersed, and genets form an intermingled pattern of fragmented clonal patches. The abundance and distribution of flowers within a genet and the spatial patterns of floral patches of different genets can influence mate availability and pollinator behavior (Charpentier 2001). Thus, the clonal growth strategy has a potential impact on mating patterns and may affect reproductive success both positively and negatively (Handel 1985).
A negative effect of clonal growth on reproductive success is geitonogamy, i.e., self-pollination between flowers within a genet (Eckert 2000). Geitonogamy may result in pollen and ovule discounting and can reduce male and female reproductive success, respectively, in both self-compatible and self-incompatible plants (Waser and Price 1991; Harder and Barrett 1995). In self-incompatible plants, geitonogamy may lead to the wastage of pollen deposited on incompatible stigmas and stigma clogging by incompatible pollen, resulting in reduced fertilization. In self-compatible plants, geitonogamy can increase the selfing rate and the risk of inbreeding depression. The phalanx form of clonal growth is more likely than the guerrilla form to suffer from geitonogamy (Charpentier 2001). On the other hand, a positive effect of clonal growth on reproductive success is floral display, which attracts pollinators and increases mating opportunities (Klinkhamer et al. 1989). Pollinators may recognize floral display in various spatial scales, which can correspond to spatial aggregations of flowers within inflorescences, ramets, and genets. If pollinators respond to floral abundance in a large spatial scale, as in genets (Thompson 2001), clonal growth will increase pollinator visits and reproductive success (Kato and Hiura 1999). Therefore, clonal growth potentially has both positive and negative effects on reproductive success owing to floral display and geitonogamy, respectively.
After Charpentier (2001), some empirical studies investigated the relationship between clonal growth and reproductive success (Reusch 2001; Routley et al. 2004; Wang et al. 2005; Wilson et al. 2005; Clark-Tapia et al. 2006; Trapnell and Hamrick 2006). These studies often demonstrated that female reproductive success decreased with clonal growth due to geitonogamy but rarely examined male reproductive success using molecular markers. In a self-compatible species of marine eelgrass, clonal dominance reduced the outcrossing rate (Reusch 2001). In self-incompatible species, paternity of seeds has been investigated. In a domestic apple species, the siring rate per genet did not increase linearly with the number of clonal trees, which supports the idea that male reproductive success of individual trees decreases due to clonal growth (Routley et al. 2004). In an epiphytic orchid species, both male and female reproductive success decreased in genets with too few flowering ramets, probably due to their small floral display, and in genets with too many flowering ramets, possibly due to geitonogamy (Trapnell and Hamrick 2006). These two studies, however, ignored some factors affecting male reproductive success, such as the distance from females to males and the size of male plants, which have been considered in pollination models recently (Smouse and Sork 2004).
Prunus ssiori Fr. Schm. (Rosaceae) is a woody clonal species that can reproduce both sexually by hermaphrodite flowers and asexually by sprouting from secondary roots (Ogawa et al. 1999). Genets of this species formed mutually exclusive patches that reached more than 50 m in diameter (Nagamitsu et al. 2004), indicating clonal growth in the typical phalanx form. In addition to the clonal growth form, the number of trees in individual genets varies from a single tree to more than 30 within a population in 2.3 ha (Nagamitsu et al. 2004). These features of clonal structure are suitable for examination of the effects of clonal growth on reproductive success. In the genus Prunus, many microsatellite markers can be used to estimate paternity with accuracy (Testolin et al. 2000; Dirlewanger et al. 2002). The microsatellites enable the evaluation of male reproductive success using paternity assignments.
Our objective in this study is to examine the effects of clonal growth on both male and female reproductive success. The reproductive success was measured in terms of the male fertility of individual trees and fruit set in individual inflorescences, which can be regarded as reproductive efficiency per plant module. To accomplish the objective, our study proceeded through four steps.
-
First, a pollination experiment and pollinator observations were carried out to confirm the mating and pollination systems, respectively.
-
Second, genets were identified using microsatellites, and the spatial distribution of clones was described in a plot.
-
Third, paternity assignments of seeds and measurements of fruit set were conducted to estimate male and female reproductive success, respectively.
-
Finally, the effects of clonal growth on reproductive success were examined using statistical models including some factors in pollination process.
Materials and methods
Pollination experiment
In 2003, a pollination experiment was conducted in the arboretum of the Forestry and Forest Products Research Institute in Sapporo (43º0′N, 141º24′E, 140 m altitude). Twelve trees of four genets were planted in the arboretum. Among these, four trees were selected from the different genets. Bag, self, and outcross treatments and a control were assigned to four inflorescences sampled from each tree selected. In late May, the number of flower buds was counted in each of the 64 inflorescences sampled, and then these inflorescences were enclosed in fine-mesh nylon bags to prevent any pollinator visits to their flowers. After the flowers began to open, the bags were removed in the control during the flowering period in early June. In the self and outcross treatments, flowers were pollinated using a soft brush with pollen obtained from the same tree and from the three other trees, respectively, on 2–11 June. In these treatments, the bags were removed immediately after flowering finished. The number of mature fruits was counted in each inflorescence in late July, and fruit set was measured in terms of the ratio of the number of mature fruits to the number of flower buds.
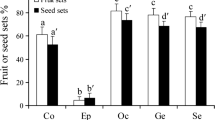
Fruit set obtained from the pollination experiment was analyzed using a linear logistic mixed model (Crawley 2005). The model included the fixed effects of the treatments and control and the random effects of inflorescences. The fixed-effect term in a linear predictor of the model was a categorical variable of four levels of the treatments and control. The random effect term was a random variable following the normal distribution that represented the individuality of each inflorescence. In order to examine differences in fruit set among the treatments and control, the best model was explored over all models under 15 possible groupings of the four levels using Akaike’s information criteria (AIC) (Burnham and Anderson 2002), defined as AIC = −2log(L) + 2K, where log(L) is the maximized log likelihood and K is the number of parameters in the model. Fruit set was identical in the levels that were grouped but different in the levels that were not grouped. Because the model is a member of the generalized linear mixed model (GLMM), the parameters were estimated using the glmmML package in R (R Development Core Team 2005). The package seeks the maximum likelihood estimates of the parameters for the fixed effects and the standard deviation of the random effects.
Study plot and tree survey
A plot (240 × 260 m, 6.24 ha) was established in a cool-temperate deciduous broad-leaved forest in the Tomakomai Experimental Forest, Hokkaido University (42º43′N, 141º34′E, 90 m altitude). At the center of the plot, there is a crane system with a 25-m tall tower, a 41-m long jib covering a circular area of 0.53 ha, and a gondra hung from the jib that enables access to the forest canopy. In the plot, all P. ssiori trees more than 100 mm in diameter at breast height (dbh) were mapped because they potentially produced flowers in the forest. Their dbh (mm) was measured in May 2003.
Pollinator observation
Flower visitors were observed in a 5 × 5-m area of the crown surface of P. ssiori trees using the crane system between 7:00 and 14:00 on 16 June and between 8:00 and 13:00 on 17 June in 2003. The number of insect visits to inflorescences was counted, and their taxonomic group was recorded. In order to describe pollination distance by main pollinators, the distance between their consecutive visits to inflorescences was observed. When a pollinator visited an inflorescence observed, its flight to the next visit to another inflorescence was traced, and the distance of the flight was recorded in terms of eight distance classes, 0–1, 1–2, 2–4, 4–6, 6–8, 8–10, 10–15, and >15 m. Whether the flight was within a tree or between trees was also recorded.
Sampling and microsatellite genotyping
Shoots with fresh leaves or buds were collected from all the trees mapped in the plot in 2003. Nine maternal trees accessible from the crane system were selected at the center of the plot. In June 2003, 12–15 inflorescences were sampled from each of the nine maternal trees, and the number of flowers was counted in each of the 127 inflorescences sampled. In August 2003, these inflorescences were collected to count the number of mature fruits, and fruit set was measured for each inflorescence. In P. ssiori, a flower with two ovules becomes a mature fruit with a single seed. A total of 300 seeds were sampled from the mature fruits collected, namely, 40 seeds from each of six maternal trees and 20 seeds from each of three maternal trees.
Total DNA was extracted from fresh leaves or buds of the trees mapped using the FastDNA Kit (Q-BIOgene, Irvine, CA, USA) and from embryos of the seeds sampled using PrepMan Ultra (Applied Biosystems, Foster City, CA, USA).
The genotypes of the trees and seeds were determined in eight microsatellite loci developed in P. persica: UDP96-008, UDP97-403, UDP98-408, UDP98-410, and UDP98-412 (Testolin et al. 2000), and BPPCT001, BPPCT002, and BPPCT014 (Dirlewanger et al. 2002). Polymerase chain reactions (PCR) were carried out in 15 μL of a mixture containing 2.5 mM MgCl2, 0.5 units of AmpliTaq Gold DNA polymerase in a GeneAmp PCR Gold buffer (Applied Biosystems), 0.4 μM of each primer, 0.2 mM of each dNTP, and approximately 10 ng of template DNA, using a PTC-200 thermal cycler (Bio-Rad, Hercules, CA, USA) programmed for 9 min at 95°C followed by 30 cycles of 30 s at 95°C, 30 s at an optimized annealing temperature, and 45 s at 72°C. The length of the PCR products was measured using an ABI PRISM 3100-Avant genetic analyzer and Genescan analysis software (Applied Biosystems).
Genet identification and paternity assignment
A genet was defined as a group of tree(s) with identical genotypes in the eight microsatellite loci. Genetic parameters for the genet population in the plot were estimated using F-STAT 2.9.1 (Goudet 1995).
Paternal genets of the 300 seeds genotyped were assigned with the simple exclusion method using CERVUS 3.0 (Kalinowski et al. 2007). The maximum likelihood paternity was also assigned with regard to genotyping error and unsampled fathers. The proportion of seeds for which paternal genets were not assigned within the plot with the simple exclusion method was regarded as the rate that fathers were not sampled. Because the rate of genotyping error often ranged from 0.13% to 0.74% in microsatellites (Hoffman and Amos 2005), a 0.5% genotyping error was assumed. The maximum likelihood of paternity was obtained on the basis of strict and relaxed Δ thresholds, which represented 95 and 80% confidence levels, respectively, by means of 10,000 randomization tests. The paternity of each seed was assigned to the genet with the highest Δ scores that significantly exceeded the Δ thresholds. The paternity determined with the simple exclusion method was verified by the maximum likelihood paternity and used in the following analysis of male reproductive success.
Modeling of male reproductive success
A statistical model was constructed to evaluate the relative ability of individual trees to sire a seed using the neighborhood model (Burczyk et al. 2002) and a modified version of it (Goto et al. 2006; Kitamoto et al. 2006). Although the neighborhood models often include mating with unsampled trees outside the plot, our model did not include it because the properties of these trees were not observed. Our model estimated the male fertility of individual trees from the conditional probability that a paternal genet of each seed is present in the plot. According to the definition of conditional probability, it can be proved that an effect of the male fertility of trees located outside the plot can be ignored if this effect is identical for every tree in the plot. The neighborhood models generally use the Mendelian segregation probability of seed genotypes to calculate the likelihood without assigning the paternity of the seeds, but our model did not use it because their paternity had been assigned already in the previous step of our analysis.
The paternity of seed i of a maternal tree is assigned to genet j, which contains tree k, i.e., tree k is an element of the set of all trees of genet j (k ∈ N j ) (Fig. 1). It is assumed that pollen dispersed from tree k of genet j fertilized seed i with probability proportional to a non-negative variable f ijk representing the male fertility of individual trees. The probability σ ij that genet j sires seed i is derived as the total fertility over all trees in genet j normalized by the total fertility over all trees of all genets in the plot:
where N is the set of all genets in the plot. Four factors are assumed to determine male fertility f ijk , as follows:
where {β 0, …, β 3} are parameters to be estimated (Fig. 1); tree size T k is the dbh (mm) of tree k; distance D ik is the horizontal distance (m) between the maternal tree of seed i and tree k; genetic identity I ij is 1 if the maternal tree of seed i belongs to genet j and 0 otherwise; and clonal growth G j is 1 if genet j contains two or more trees and 0 if it consists of a single tree.
Variables in a modified neighborhood model of male reproductive success. Tree k belongs to genet j. Tree k′ and a maternal tree of seed i belong to genet j′. Tree sizes T k and T k′ are the dbh (diameter at breast height) of trees k and k′, respectively. The distances from the maternal tree of seed i to trees k and k′ are D ik and D ik′ , respectively. Clonal growth G j of genet j is 0 because genet j consists of a single tree, and G j′ of genet j’is 1 because genet j′ contains two or more trees. Siring probability σ ij or σ ij′ indicates the probability that genet j or j′, respectively, sired seed i. Genetic identity I ij between the maternal tree of seed i and genet j is 0, because the maternal tree does not belong to genet j, and I ij′ is 1, because the maternal tree belongs to genet j′
Parameters {β 0, …, β 3} represent the effects of the four factors on the male reproductive success of individual trees. Parameter β 0 is the effect of tree size, which reflects the floral display in the crown scale and geitonogamy within a tree. Parameter β 1 indicates the effect of distance on pollen dispersal. Parameter β 2 is the effect of genetic identity on seed production, which depends on self-incompatibility, selective abortion, and inbreeding depression during embryo development. If P. ssiori shows complete avoidance of selfing, β 2 is negatively infinite, and if it is more self-compatible, β 2 increases. Parameter β 3 is the effect of clonal growth, which represents the floral display in the genet scale and geitonogamy between trees within a genet. If the male reproductive success of individual trees is larger in genets with two or more clonal trees than in genets with a single tree, β 3 is positive; otherwise, it is negative.
The likelihood equation was defined as a product of siring probability σ ij over the set of all seeds whose paternity was determined (i ∈ O):
The maximum likelihood estimates of parameters {β 0, …, β 3} were obtained by maximizing the likelihood defined above. The likelihood maximization was performed using the Nelder–Mead algorithm (Press 1992) implemented in Perl language. The AIC was evaluated for the full-model with the four parameters and for sub-models with 14 possible subsets of the parameters (Burnham and Anderson 2002). Finally, the model with the smallest AIC value was selected as the best model to predict the siring probability of genets and the male fertility of trees.
Modeling of female reproductive success
Another statistical model was constructed to evaluate fruit set in individual inflorescences. It was assumed that the reception of pollen from trees adjacent to the inflorescences, which is called pollination index hereafter, affected fruit set. The pollination index was calculated from the size of the adjacent trees and the distance to them, which were likely to affect the production and dispersal of pollen, respectively. Because the pollination process had been evaluated in the previous analysis of male reproductive success, the estimates of parameters obtained from the best model for male reproductive success were used to calculate the pollination index. The pollination index was divided into three components: the within-tree geitonogamy index (pollination within a maternal tree), the among-tree geitonogamy index (pollination from other clonal trees), and the xenogamy index (pollination from non-clonal trees in the plot). The weighted sum of these pollination indices was assumed to be a linear predictor of fruit set π i in inflorescence i with logit link function as follows:
where {γ 0, …, γ 3} are parameters to be estimated; P i is the within-tree geitonogamy index; Q i is the among-tree geitonogamy index; and R i is the xenogamy index for inflorescence i. These pollination indices are defined as:
where genetic identities I ij and I ik are 1 if the maternal tree of inflorescence i belongs to genet j and if inflorescence i belongs to tree k, and 0 otherwise; the pollination indices for inflorescence i are derived from tree size T k (mm) of tree k of genet j (k ∈ N j ) and distance D ik (m) between the maternal tree of inflorescence i and tree k and the estimated parameters {β 0, β 1}.
Parameters {γ 1, …, γ 3} represent the effects of the three pollination indices on female reproductive success in individual inflorescences. Parameter γ 1 is the effect of the maternal tree size, which reflects not only geitonogamy within a maternal tree but also floral display in the crown scale. Parameter γ 2 is the effect of clonal growth, which represents geitonogamy between clonal trees within a genet and floral display of clonal trees around the maternal tree. Parameter γ 3 is the effect of the reception of pollen from non-clonal trees in the plot, which represents the local mate availability for outcross pollination.
The model included the fixed effects of an intercept γ 0 and the three factors with parameters {γ 1, …, γ 3} and the random effects of individual inflorescences (Crawley 2005). The functional form of the model is equivalent to that of the GLMM of the binomial family with a logit link function. Thus, the parameters for the fixed effects and the standard deviation of the random effects were estimated using the glmmML package. The AIC was evaluated for the full-model with the four parameters and for sub-models with seven possible subsets of the parameters (Burnham and Anderson 2002). Finally, a model that minimized the AIC value was selected as the best model to predict fruit set in individual inflorescences.
Results
Mating system
In the pollination experiment, the model with different fruit sets for all the treatments and the control had the smallest AIC value (Table 1). Fruit set in the bag treatment was the smallest (0.06), followed by that in the self treatment (0.15). Fruit set in the outcross treatment was the highest (0.57). Fruit set in the control (0.31) was intermediate between the self and outcross treatments. Because models with the random effects had much smaller AIC values than those without them, the individuality of each inflorescence resulted in over-dispersion of the fruit set observed.
Pollinator behavior
Assemblages of flower visitors were similar between the two observation dates (Table 2). Bumble bees (Bombus, Apidae) were the most abundant (82%, n = 753), followed by dipteran insects (13%), in particular, syrphid flies (Syrphidae). Most bumble bees imbibed nectar and collected pollen from several flowers while moving along inflorescences.
The patterns of bumble bee flights between consecutive visits to inflorescences were similar between the two observation dates (Fig. 2). Most flights were within a tree (89%, n = 607). Flights were the most frequent in the shortest distance class (0–1 m, 54%), and the frequency of flights gradually decreased as the distance increased. The frequency of flights more than 15 m long was 9%.
Clonal and genetic structure
There were 212 trees with more than 100 mm dbh in the 6.24-ha plot (Fig. 3). The maximum and mean dbhs of the trees were 517 and 175 mm, respectively. According to microsatellite genotypes in the eight loci, the 212 trees belonged to 59 genets. The probability that multilocus genotypes were identical by chance was extremely small (1.1 × 10−7). Therefore, the possibility that different genets had the same multilocus genotype could be ignored. Among the 59 genets, 42 consisted of a single tree, whereas 17 contained two or more trees, in which the maximum and mean numbers of clonal trees were 65 and 10, respectively (Fig. 3). The genets with clonal trees had mutually exclusive patches, and the largest one occupied 0.4 of a hectare. Nine maternal trees from which inflorescences were collected belonged to two genets that consisted of 14 and 28 clonal trees.
Spatial distribution of Prunus ssiori trees in the study plot. Circles show trees, and their sizes indicate the dbh (diameter at breast height). Filled circles at the center of the plot show nine maternal trees. Inflorescences of these maternal trees were collected to measure fruit set and to assign paternal genets of their seeds. Contours enclosing two or more trees indicate the ranges of genets with the clonal trees. Trees that are not enclosed by the contours have no clonal trees (i.e., genets with a single tree)
In the 59 genets, 91 alleles were observed in the eight loci, which ranged from 2 to 18 alleles over the loci. Nei’s gene diversity H E in individual loci was 0.717 on average and ranged from 0.184 to 0.891. Wright’s fixation index F IS in individual loci was −0.025 on average and ranged from −0.105 to 0.034. The multilocus estimate of F IS was not significantly different from zero (10,000 randomization test, P = 0.088). There was no significant deviation from Hardy–Weinberg equilibrium in any loci according to F IS (P > 0.083). The estimated frequency of null alleles was negative in all the loci, ranging from −0.065 to −0.009, indicating that null alleles could be ignored.
Paternity assignment
The paternity exclusion probability was 0.995 over the eight loci according to the genets in the plot. In the simple exclusion method, the paternity of 300 seeds sampled from the nine maternal trees was assigned to a single genet in the plot for 107 (36%) seeds (Fig. 4) and to multiple genets in the plot for 26 (9%) seeds, and the paternity of 167 (55%) seeds was excluded from all genets in the plot. The maximum likelihood paternity of 76 and 159 seeds was assigned to genets in the plot on the basis of strict (4.17) and relaxed (0.81) Δ thresholds, respectively. In these paternity assignments, there was no self-fertilization. The simple exclusion method showed a consistent result with the maximum likelihood assignments. The paternity of 76 seeds using the strict Δ threshold was identical to the subset of the paternity of 107 seeds assigned to a single genet using the simple exclusion, which was also identical to the subset of the paternity of 159 seeds using the relaxed Δ threshold. In the following analysis, the paternity of 107 seeds in the simple exclusion method was used (Fig. 4).
Number of seeds sired by genets in the study plot. Paternal genets of the seeds were assigned in the simple exclusion method. Figures enclosed with contours indicate the number of seeds sired by genets with two or more trees, and figures without enclosing contours indicate those sired by genets with a single tree. Gray circles show nine maternal trees, and their sizes indicate the dbh (diameter at breast height) of the maternal trees. Their seeds were collected for paternity assignment
Male reproductive success
Among the five best models that predicted the siring probability of genets and the male fertility of trees, the signs of four parameters were consistent when the parameters were selected (Table 3). According to the four parameters in the best model, the tree size had a positive effect on the male fertility of individual trees (Table 3, tree size β 0), which increased in proportion to their dbh (mm) to the power 0.85 (Fig. 5). The distance from maternal trees had a negative effect (Table 3, distance β 1). For example, the male fertility of trees at a 100 m distance from a maternal tree was 0.69 times as high as that of trees at a 50 m distance from it. Male reproductive success decreased substantially (by a factor of approximately 10−8) when the maternal trees belonged to the same genet, i.e., in the case of geitonogamy within a genet (Table 3, identity β 2). The male fertility of trees was lower in genets with two or more trees than in those with a single tree (Table 3, clonal growth β 3), and the former was 0.44 times as high as the latter (Fig. 5).
Effects of the mean dbh (diameter at breast height) of trees that belong to a genet on the number of seeds sired by the genet per tree. The number of seeds sired by individual genets was divided by the number of trees of the genets to evaluate the male fertility of individual trees. The numbers of seeds of nine maternal trees were summed. Circles show paternal genets with a single tree, and crosses show paternal genets with two or more trees. Curves indicate values expected in a typical genet, which were estimated from the parameters of the best model (Table 3). The typical genet j does not contain the maternal trees of seed i (I ij = 0) and the mean distances between the maternal trees and tree k of genet j (D ik = 77.2–89.3 m). Solid and broken curves indicate genet j with a single tree (G j = 0) and ten trees (the mean size of genets with clonal trees; G j = 1), respectively. The expected number of seeds sired by genet j was obtained from a product of siring probability σ ij and the number of seeds whose paternity was assigned in each maternal tree (1–26)
Female reproductive success
Fruit set in individual inflorescences was 0.071 on average and ranged from 0.000 to 0.317 among the 127 inflorescences sampled from the nine maternal trees. According to the parameters (β 0 = 0.85, β 1 = −0.0074) estimated from the best model for male reproductive success, a model with three parameters, i.e., an intercept and two geitonogamy indices, was the best at predicting fruit set in individual inflorescences (Table 4). The individuality of each inflorescence resulted in variation in the intercept, the standard deviation of which was 0.99–1.02.
In the best five models, reception of pollen within maternal trees and from its clonal trees had negative effects on fruit set when the parameters were selected (Table 4, within-tree and among-tree geitonogamy indices γ 1 and γ 2, respectively). The within-tree geitonogamy index ranged from 0.10 to 0.20. In the best model, fruit set in an inflorescence with the smallest index was 2.53 times as high as that with the largest index (Fig. 6). The among-tree geitonogamy index ranged from 1.04 to 2.35. The index varied mainly between two maternal trees in one genet and seven in the other genet (Fig. 7) because the number of clonal trees in these two genets was different (14 and 29) (Fig. 3). In the best model, fruit set in an inflorescence with the smallest index was 1.85 times as high as that with the largest index (Fig. 7). The xenogamy index, the reception of pollen from trees of other genets, which ranged from 7.30 to 8.70, varied less than the former geitonogamy indices. Probably because of its small variation, its effect on fruit set was excluded from the best model, and the sign of its parameter was inconsistent among the second, third, and fourth best models (Table 4, xenogamy index γ 3).
Effects of within-tree geitonogamy index on fruit set in individual inflorescences of nine maternal trees. Within-tree geitonogamy P i for inflorescence i was obtained from the size of maternal trees with inflorescence i. A curve indicates the values expected from the estimated parameters of the best model (Table 4) in typical inflorescence i with the mean of the among-tree geitonogamy index (Q i = 1.96)
Effects of the among-tree geitonogamy index on fruit set in individual inflorescences of nine maternal trees. The among-tree geitonogamy Q i for inflorescence i was obtained from the size of clonal trees and the distance to them from maternal trees with inflorescence i. A curve indicates the values expected from the estimated parameters of the best model (Table 4) in typical inflorescence i with the mean of the within-tree geitonogamy index (P i = 0.15)
Discussion
Our pollination experiment in an arboretum demonstrated that fruit set in the bag and self treatments was positive but lower than that in the outcross treatment (Table 1). In another study (Niwa 1999), the bag treatment resulted in a positive fruit set (0.06). These findings indicate the potential of autogamy and partial self-compatibility in P. ssiori. The genus Prunus is well known to have gametophytic self-incompatibility with the multi-allelic S locus, but there are self-compatible phenotypes with non-functional S alleles (Ushijima et al. 2004). Such self-compatible factors may be responsible for the positive fruit set in the bag and self treatments (Porcher and Lande 2005). Although contamination by outcross pollen during the treatments could have caused the positive fruit set, the contamination was unlikely to result in fruit set as high as the ratios observed in the pollination experiment (0.06 and 0.15). Our paternity assignment in the plot, however, showed no selfed seeds. This result suggests selective fertilization with outcross pollen, selective abortion of selfed embryos, and inbreeding depression during embryo development when both self and outcross pollen are available (Lloyd 1992; Burd 1998; Hufford and Hamrick 2003).
The main diurnal pollinators of P. ssiori seemed to be bumble bees in the plot, because of their abundance in the daytime and behavior on its flowers (Table 2). Their flights between consecutive visits to inflorescences were frequent in short distances within a tree, and the frequency of flights decreased with increasing distance (Fig. 2), which is common in bumble bees (Nuortila et al. 2002). Thus, bumble bees frequently transfer pollen within a tree, and the frequency of pollen dispersal decreases as the distance increases. Such pollen dispersal predicted from the flight pattern is consistent with the negative effect of distance on male fertility estimated from our model for male reproductive success (Table 3). On the other hand, pollen dispersal over long distances did occur in P. ssiori because bumble bees potentially travel over several kilometers (Cresswell et al. 2000) and carry pollen over to flowers during a foraging trip (Cresswell et al. 1995). Consistently, more than half of the seeds collected from maternal trees at the center of the plot with 240 and 260 m sides were likely to have paternal genets outside the plot. This result indicates that most pollination distances were longer than 100 m. In P. mahaleb, which is also pollinated by bumble bees, in an isolated habitat where almost all pollination events could be observed, the average pollination distance was 125 m (García et al. 2005). This evidence is consistent with the result in P. ssiori.
The somatic mutations during clonal growth suggested to occur in another P. ssiori population (Nagamitsu et al. 2004) were not observed in the plot. Wright’s fixation index F IS of genets in the plot was not significantly different from zero. These findings suggest that all genets in the plot originated from seedlings and had been randomly mating with each other. Some genets had many clonal trees, but most genets consisted of a single tree, which is similar to the results of our previous study (Nagamitsu et al. 2004). Because clonal propagation in P. ssiori was observed only in trees more than 70 years old (Ogawa 1997), the presence of genets with a single tree suggests that seedling recruitment has occurred recently.
The spatial distribution of clonal trees in the plot showed mutually exclusive patches (Fig. 3), as observed in our previous study (Nagamitsu et al. 2004). These findings confirm the phalanx form of clonal growth in P. ssiori. Phalanx clonal growth is likely to facilitate geitonogamy within a genet (Charpentier 2001) whereas the patchy aggregations of clonal trees seem to provide floral display over the scale of tree crowns.
Our models fitted to the paternity of seeds showed that the male fertility of individual trees was lower in genets with clonal trees than in genets with a single tree (Table 3; Fig. 5). This result indicates a siring advantage of trees in genets without clonal growth. This evidence suggests that pollen discounting due to geitonogamous pollination between clonal trees reduces male reproductive success, which has been demonstrated in a few clonal plants (Routley et al. 2004; Trapnell and Hamrick 2006). In addition to the negative effect of genet size (clonal growth), there was a positive effect of tree size (ramet growth) on male fertility (Table 3; Fig. 5). These contrasting effects of plant growth between genets and ramets were not examined in the previous studies.
Floral aggregations in clonal patches may make pollinators frequently forage within the patches, waste pollen on self-pollination, and rarely bring pollen out of the patches. Such pollinator behavior is consistent with the foraging behavior of main pollinators, i.e., bumble bees. They can adjust turning rates and flight distances according to the floral density so that they quickly leave patches with few flowers and stay long in patches with many flowers (Cresswell 1997). Large patches of clonal trees seem to provide enough rewards to fill the crops of foragers. Thus, bumble bees may forage on a single genet during each foraging trip. Consistently, radar-tracked bumble bees often exhibited strong site fidelity and frequently traveled to a single destination for each trip (Osborne et al. 1999).
In contrast to geitonogamy, floral display is expected to attract pollinators and positively affect reproductive success (Charpentier 2001). The positive effect of tree size on the male fertility of individual trees suggests that floral display in the scale of tree crowns affects male reproductive success. Bumble bees can respond to floral display in various spatial scales, such as inflorescences, tree crowns, and aggregations of flowering trees (Kato and Hiura 1999). Although it is difficult to explain the reason that the scale of tree crowns is effective in attracting bumble bees, the emergence of the crowns of large trees beyond the forest canopy may be responsible for the attraction. Such tall crowns of flowering trees are conspicuous, and bumble bees may use them as landmarks to help their foraging flights over long distances (Plowright and Galen 1985). In a lily species, higher flowers in larger racemes attracted more pollinators than lower flowers (Ishii 2004). Thus, more pollen was removed from the larger racemes, leading to an increase in male reproductive success as the size of floral display increased (Ishii 2004). In contrast to the floral display of individual tree crowns, the floral aggregations of clonal trees in P. ssiori may not be effective at attracting its pollinators.
Fruit set in individual inflorescences decreased as both the within-tree and among-tree geitonogamy indices increased in our models (Table 4; Figs. 6, 7). This result indicates the negative effects of both tree enlargement and clonal growth on female reproductive success, which has been shown in several clonal plants (Eriksson and Bremer 1993; Eckert 2000; Wolf et al. 2000; Wang et al. 2005; Clark-Tapia et al. 2006). Unfortunately, the full range of genet size variation in the plot was not investigated because the maternal trees were sampled from only two genets with clonal trees. The among-tree geitonogamy index differed more between these two genets than within each of them. In this case, this index was confounded by factors that varied between the two genets, such as differences in the genetic properties of self-incompatibility, resource availability for fruit production, and maternal conditions affecting fruit development. These confounding factors could affect fruit set instead of the among-tree geitonogamy index. Therefore, the effect of tree enlargement on fruit set was clear although the effect of clonal growth on maternal success was not evident.
Limitation of outcross pollen seems to be responsible for reduced fruit set in large maternal trees. The availability of outcross pollen can be indicated by the xenogamy index in our models. However, the effect of the xenogamy index on fruit set was not clear (Table 4) because the maternal trees that clumped at the center of the plot had similar availability of outcross pollen. On the other hand, our pollination experiment showed pollen limitation in the arboretum because of the lower fruit set in the control than in the outcross treatment (Table 1). The natural fruit set in the plot (0.07) was lower than that in the arboretum (0.31), where trees of different genets were densely planted. These results suggest that pollen limitation occurs in the plot. In addition to pollen limitation, interference by self pollen received from geitonogamous pollination is also likely to reduce the female fertility of large maternal trees (Barrett 2002). Clogged stigma surface, inhibited growth of pollen tubes, and disturbed fertilization by self pollen are plausible mechanisms for the interference (Waser and Price 1991).
In conclusion, a negative effect of geitonogamy among clonal trees on at least male reproductive success of individual trees was demonstrated for P. ssiori. The positive effect of floral display on male fertility was found only on the scale of individual trees. Thus, clonal growth may reduce reproductive efficiency per plant module, although reproductive output per genet can increase because of multiplication of the plant modules. In habitats where clonal multiplication is favorable and seedling recruitment is limited spatially and temporally, lifetime reproductive success can increase by means of clonal expansion and genet longevity despite the reduction in the marginal gain of reproductive success. It remains a task for future studies to evaluate the adaptive significance of clonal growth in P. ssiori.
References
Barrett SCH (2002) Sexual interference of the floral kind. Heredity 88:154–159. doi:10.1038/sj.hdy.6800020
Burczyk J, Adams WT, Moran GF, Griffin AR (2002) Complex patterns of mating revealed in a Eucalyptus regnans seed orchard using allozyme markers and the neighbourhood model. Mol Ecol 11:2379–2391. doi:10.1046/j.1365-294X.2002.01603.x
Burd M (1998) “Excess” flower production and selective fruit abortion: a model of potential benefits. Ecology 79:2123–2132
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information - theoretic approach. Springer, New York
Charpentier A (2001) Consequences of clonal growth for plant mating. Evol Ecol 15:521–530. doi:10.1023/A:1016057503722
Clark-Tapia R, Alfonso-Corrado C, Mandujano MC, Molina-Freaner F (2006) Reproductive consequences of clonal growth in Stenocereus eruca, a rare clonal cactus of the Sonoran desert. Evol Ecol 20:131–142. doi:10.1007/s10682-005-5379-x
Crawley MJ (2005) Statistics: an Introduction using R. Wiley, West Sussex
Cresswell JE (1997) Spatial heterogeneity, pollinator behavior and pollinator-mediated gene flow: bumblebee movements in variously aggregated rows of oil-seed rape. Oikos 78:546–556. doi:10.2307/3545616
Cresswell JE, Bassom AP, Bell SA, Collins SJ, Kelly TB (1995) Predicted pollen dispersal by honey-bees and three species of bumble-bees foraging on oil-seed rape: a comparison of three models. Funct Ecol 6:829–841. doi:10.2307/2389980
Cresswell JE, Osborne JL, Goulson D (2000) An economic model of the limits to foraging range in central place foragers with numerical solutions for bumblebees. Ecol Entomol 25:249–255. doi:10.1046/j.1365-2311.2000.00264.x
Dirlewanger E, Cosson P, Tavaud M, Aranzana MJ, Poizat C, Zanetto A et al (2002) Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet 105:127–138. doi:10.1007/s00122-002-0867-7
Eckert CG (2000) Contributions of autogamy and geitonogamy to self-fertilization in a mass-flowering, clonal plant. Ecology 81:532–542
Eriksson O, Bremer B (1993) Genet dynamics of the clonal plant Rubus saxatillis. J Ecol 81:533–542. doi:10.2307/2261531
García C, Arroyo JM, Godoy JA, Jordano P (2005) Mating patterns, pollen dispersal, and the ecological maternal neighborhood in a Prunus mahaleb L. population. Mol Ecol 14:1821–1830. doi:10.1111/j.1365-294X.2005.02542.x
Goto S, Shimatani K, Yoshimaru H, Takahashi Y (2006) Fat-tailed gene flow in the dioecious canopy tree species Fraxinus mandshurica var. japonica revealed by microsatellites. Mol Ecol 15:2985–2996
Goudet J (1995) FSTAT (ver. 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Handel SN (1985) The intrusion of clonal growth patterns on plant breeding systems. Am Nat 125:367–384. doi:10.1086/284348
Harder LD, Barrett CH (1995) Mating cost of large floral display in hermaphrodite plants. Nature 373:512–515. doi:10.1038/373512a0
Hoffman JI, Amos W (2005) Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol Ecol 14:599–612. doi:10.1111/j.1365-294X.2004.02419.x
Hufford KM, Hamrick JL (2003) Viability selection at three early life stages of the tropical tree, Platypodium elegans (Fabaceae, Papilionoideae). Evol Int J Org Evol 57:518–526
Ishii HS (2004) Increase of male reproductive components with size in an animal-pollinated hermaphrodite, Narthecium asiaticum (Liliaceae). Funct Ecol 18:130–137. doi:10.1111/j.1365-2435.2004.00826.x
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106. doi:10.1111/j.1365-294X.2007.03089.x
Kato E, Hiura T (1999) Fruit set in Styrax obassia (Styracaceae): the effect of light availability, display size, and local floral density. Am J Bot 86:495–501. doi:10.2307/2656810
Kitamoto N, Ueno S, Takenaka A, Tsumura Y, Washitani I, Ohsawa R (2006) Effect of flowering phenology on pollen flow distance and the consequence for spatial genetic structure within a population of Primula sieboldii (Primulaceae). Am J Bot 93:226–233. doi:10.3732/ajb.93.2.226
Klinkhamer PGL, de Jong TJ, de Bruyn GJ (1989) Plant size and pollinator visitation in Cynoglossum officinale. Oikos 54:201–204. doi:10.2307/3565267
Lloyd DG (1992) Self- and cross-fertilization in plants. II. the selection of self-fertilization. Int J Plant Sci 153:370–380. doi:10.1086/297041
Lovett Doust L (1981) Population dynamics and local specialization in a clonal perennial (Ranunclus repens). I. the dynamics of ramets in contrasting habitat. J Ecol 69:743–755. doi:10.2307/2259633
Nagamitsu T, Ogawa M, Ishida K, Tanoushi H (2004) Clonal diversity, genetic structure, and mode of recruitment in a Prunus ssiori population established after volcanic eruptions. Plant Ecol 174:1–10. doi:10.1023/B:VEGE.0000046054.87587.8b
Niwa S (1999) Relationships between inflorescence sizes and fruit-set ratios in four trees growing in Hokkaido. Bull Higashi Taisetsu Mus Nat Hist 21:25–30 (in Japanese with English summary)
Nuortila C, Tuomi J, Laine K (2002) Inter-plant distance affects reproductive success in two clonal dwarf shrubs, Vaccinium myrtillus and Vaccinium vitis-idaea (Ericaceae). Can J Bot 80:875–884. doi:10.1139/b02-079
Ogawa M (1997) Life history of Prunus ssiori and its characteristics. Ecohabitat JISE Res 4:87–92 (in Japanese with English summary)
Ogawa M, Aiba Y, Watanabe N (1999) Morphological and anatomical characteristics of sprouting root of Prunus ssiori. J Jpn For Soc 81:36–41 (in Japanese with English summary)
Osborne JL, Clark SJ, Morris RJ, Williams IH, Riley JR, Smith AD et al (1999) A landscape-scale study of bumble bee foraging range and constancy, using harmonic radar. J Appl Ecol 36:519–533. doi:10.1046/j.1365-2664.1999.00428.x
Plowright RC, Galen C (1985) Landmarks or obstacles: the effects of spatial heterogeneity on bumble bee foraging behavior. Oikos 44:459–464. doi:10.2307/3565787
Porcher E, Lande R (2005) Loss of gametophytic self-incompatibility with evolution of inbreeding depression. Evol Int J Org Evol 59:46–60
Press WH (1992) Numerical recipes in C: the art of scientific computing. Cambridge University Press, Cambridge
R Development Core Team (2005) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org
Reusch TBH (2001) Fitness-consequences of geitonogamous selfing in a clonal marine angiosperm (Zostera marina). J Evol Biol 14:129–138. doi:10.1046/j.1420-9101.2001.00257.x
Routley M, Kron P, Husband B (2004) The consequences of clone size for paternal and maternal success in domestic apple (Malus × domestica). Am J Bot 91:1326–1332. doi:10.3732/ajb.91.9.1326
Smouse PE, Sork VL (2004) Measuring pollen flow in forest trees: an exposition of alternative approaches. For Ecol Manage 197:21–38
Testolin R, Marrazzo T, Cipriani G, Quarta R, Verde I, Dettori MT et al (2000) Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome 43:512–520. doi:10.1139/gen-43-3-512
Thompson JD (2001) How do visitation patterns vary among pollinators in relation to floral display and floral design in a generalist pollination system? Oecologia 126:386–394. doi:10.1007/s004420000531
Trapnell DW, Hamrick JL (2006) Floral display and mating patterns within populations of the neotropical epiphytic orchid, Laelia rubescens (Orchidaceae). Am J Bot 93:1010–1018. doi:10.3732/ajb.93.7.1010
Ushijima K, Yamane H, Watari A, Kakehi E, Ikeda K, Hauck NR et al (2004) The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J 39:573–586. doi:10.1111/j.1365-313X.2004.02154.x
Wang Y, Wang Q-F, Guo Y-H, Barrett SCH (2005) Reproductive consequences of interactions between clonal growth and sexual reproduction in Nymphoides peltata: a distylous aquatic plant. New Phytol 165:329–336. doi:10.1111/j.1469-8137.2004.01234.x
Waser N, Price M (1991) Reproductive costs of self-pollination in Ipomopsis aggregata (Polemoniaceae): are ovules usurped? Am J Bot 78:1036–1043. doi:10.2307/2444892
Wilson ASG, van der Kamp BJ, Ritland C (2005) Opportunities for geitonogamy in the clonal herb Maianthemum dilatatum. Can J Bot 83:1082–1087. doi:10.1139/b05-096
Wolf A, Harrison S, Hamrick J (2000) Influence of habitat patchiness on genetic diversity and spatial structure of a serpentine endemic plant. Conserv Biol 14:454–463. doi:10.1046/j.1523-1739.2000.98499.x
Acknowledgments
The authors would like to thank the staff of the Tomakomai Experimental Forest, Field Science Center, Hokkaido University for the use of the crane system and the plot around it. We also thank Mayuko Hotta, Takashi Kohyama, Gaku Kudo, and Takeshi Osawa for supporting our field research and Takayuki Kawahara and Yukiko Sakamoto for assisting with our microsatellite analysis. Comments with regard to our manuscript by Susumu Goto resulted in significant improvement of our manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mori, Y., Nagamitsu, T. & Kubo, T. Clonal growth and its effects on male and female reproductive success in Prunus ssiori (Rosaceae). Popul Ecol 51, 175–186 (2009). https://doi.org/10.1007/s10144-008-0099-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10144-008-0099-z