Abstract
Retinoblastoma (RB), the most common malignant retinal tumor among children under 3 years old, is lethal if left untreated. Early diagnosis, together with timely and effective treatment, is important to improve retinoblastoma-related outcomes. Circular RNAs (circRNAs), a new class of non-coding RNAs with the capacity to regulate cellular activities, have great potential in retinoblastoma diagnosis and treatment. Recent studies have identified circular RNAs that regulate multiple cellular processes involved in retinoblastoma, including cell viability, proliferation, apoptosis, autophagy, migration, and invasion. Six circular RNAs (circ-FAM158A, circ-DHDDS, circ-E2F3, circ-TRHDE, circ-E2F5, and circ-RNF20) promote disease progression and metastasis in retinoblastoma and function as oncogenic factors. Other circular RNAs, such as circ-TET1, circ-SHPRH, circ-MKLN1, and circ-CUL2, play tumor suppressive roles in retinoblastoma. At present, the studies on the regulatory mechanism of circular RNAs in retinoblastoma are not very clear. The purpose of this review is to summarize recent studies on the functional roles and molecular mechanisms of circular RNAs in retinoblastoma and highlight novel strategies for retinoblastoma diagnosis, prognosis, and treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Retinoblastoma
Retinoblastoma (RB), the most common malignant retinal tumor among children under 3 years of age, is lethal if left untreated. Patients with retinoblastoma typically present with leukocoria or strabismus signs in the early stages of the disease, with the development of proptosis, buphthalmos, or hypopyon in some cases in later stages (Ortiz and Dunkel 2016; Global Retinoblastoma Study et al. 2020). According to statistics, retinoblastoma accounts for 2 − 4% of all tumors in children under 5 years old, and around 8000 children worldwide are diagnosed with retinoblastoma each year (Dimaras et al. 2015). In recent years, with the continuous development of treatment strategies, retinoblastoma is considered a curable intraocular malignancy in high-income countries (Ancona-Lezama et al. 2020). Thus, increasing attention is focused on how to preserve vision and improve the quality of life of retinoblastoma patients.
In contrast to high-income countries, in low- and middle-income countries, where more than 80% of retinoblastoma cases occur, the prognosis of retinoblastoma patients is poor, and metastasis rates are high (18.9%) (Global Retinoblastoma Study et al. 2020). There are delays in the diagnosis and treatment of retinoblastoma in low- and middle-income countries (Ancona-Lezama et al. 2020), with the median age at the time of diagnosis in low- and middle-income countries being 14.1 months as compared with 30.5 months in high-income countries (Global Retinoblastoma Study et al. 2020). In addition, only 23% of retinoblastoma patients in low- and middle-income countries are detected through screening as compared with 72% of patients in high-income countries (Kivela and Hadjistilianou 2017). Due to the delays in the diagnosis and treatment of retinoblastoma in low- and middle-income countries, the global retinoblastoma patient survival rate is less than 30% [4]. Specifically, reported retinoblastoma survival rates in Africa, Asia, and Latin America are approximately 30%, 60%, and 80% as compared with 95 − 97% in Europe and North America (Shields et al. 2015).
At present, the treatment for retinoblastoma is based on the International Classification of Retinoblastoma Staging, which includes chemotherapy, radiotherapy, and enucleation (Shields and Shields 2010; Fabian et al. 2018; Ancona-Lezama et al. 2020). Combination therapy can significantly improve the overall survival rate of patients with retinoblastoma (Sun et al. 2020). However, the timing of the treatment is vital. In the absence of prompt treatment, retinoblastoma can develop and spread by direct growth through the optic nerve into the central nervous system or through the blood to the lungs, bone, and other distant systemic organs (Correa-Acosta et al. 2018). Delayed treatment of retinoblastoma can result in metastasis (Teixo et al. 2015). Secondary malignancies are one of the leading factors responsible for high mortality in retinoblastoma patients (Teixo et al. 2015). In addition to the risk of mortality, retinoblastoma is associated with a psychological burden for families and high economic costs. Thus, in order to improve the therapeutic outcome in retinoblastoma patients, the disease needs to be diagnosed at an early stage, followed by effective treatment.
Circular RNAs
Circular RNAs (circRNAs) are a new class of non-coding single-stranded RNAs without free 3′ poly (A) tails or 5′ caps (Ren et al. 2020). Unlike linear RNAs, circRNAs have a covalently closed-loop structure. As early as the 1970s, scientists knew of the existence of circRNAs in cells (Hsu and Coca-Prados 1979). However, they did not become the focus of widespread attention until decades later. With the development of next-generation sequencing and computational analysis, circRNAs, as important transcriptional regulators, began to be correctly recognized (Memczak et al. 2013). Subsequently, scientists revealed the biogenesis and biological characteristics of circRNAs. circRNAs are commonly generated by back splicing of exons from precursor micro-RNA (Xu et al. 2017; Kristensen et al. 2019; Chen 2020). They are classified into three types based on the sequences from which they originate, including exons, introns, and exon–intron circRNAs (Henry and Hayes 2012). They are abundant in eukaryotes, conserved in evolution, highly stable and tissue specific, and play crucial roles in many tissues (Xu et al. 2017; Kristensen et al. 2019; Chen 2020).
Due to the increasing number of studies on circRNAs, we now have a clearer understanding of their mechanisms and functions. circRNAs are known to regulate the expression of multiple genes in cells and participate in the regulation of cellular activities by acting as micro-RNA sponges and sequestering RNA-binding proteins (Tang et al. 2017; Kristensen et al. 2019; Chen 2020). They also regulate transcription and alternative splicing, binding proteins, forming pseudogenes, and expressing peptides via a variety of mechanisms (Tang et al. 2017; Kristensen et al. 2019; Chen 2020). circRNAs are involved in the biological process of cell proliferation and apoptosis, and play a key role in neurogenesis and the development of various organs (Rahmati et al. 2021). According to numerous studies, abnormal expression of circular RNAs plays an important role in the pathological process of a variety of diseases, such as diabetes (Li et al. 2017), immune and inflammatory diseases (Chen et al. 2019c; Lin et al. 2020), tumors, and cardiovascular diseases (Altesha et al. 2019; Goodall and Wickramasinghe 2021; Wen et al. 2021; Zhang et al. 2021a, c; Sayad et al. 2022a). Especially, circRNAs play an important role in the development of various cancers, such as lung cancer (Hu et al. 2018), stomach cancer (Shan et al. 2019), liver cancer (Song et al. 2018), renal cell carcinoma (Sayad et al. 2022b), and prostate cancer (Taheri et al. 2021). The expression levels of circRNAs are correlated with the occurrence and development of the tumor and act as oncogenes or tumor suppressors (Rahmati et al. 2021; Sayad et al. 2022b). Therefore, circRNAs are considered to have great potential in tumor diagnosis and treatment (Li et al. 2015; Lei et al. 2019; De Palma et al. 2022). This review focused on recent studies on the functional roles and molecular mechanisms of circRNAs in retinoblastoma (Figs. 1 and 2, and Tables 1 and 2) with the aim of highlighting novel strategies for retinoblastoma diagnosis, prognosis, and treatment.
circRNAs contribute to several “hallmarks of cancer” in RB
The “hallmarks of cancer” were proposed as a set of functional capabilities acquired by human cells, including sustaining proliferative signaling, evading growth suppressor, resisting cell death, activating invasion and metastasis, inducing or accessing vasculature, etc., that are crucial for their ability to form malignant tumors (Hanahan 2022). The abnormal expressions of circRNAs in RB cells were reported to promote the malignant behavior of cancer cells and influence the occurrence and development of RB (Wang et al. 2020; Huang et al. 2021; Jiang et al. 2021a, b; Zhang et al. 2022b). For the oncogenic circRNAs in RB, including circ-FAM158A, circ-DHDDS, circ-E2F3, circ-TRHDE, circ-E2F5, and circ-RNF20, their increased expressions could promote cell viability, migration, invasion, and autophagy, and inhibit cell cycle arrest and apoptosis of RB cells (Fig. 1 and Table 1) (Chen et al. 2020; Liu et al. 2020; Wang et al. 2020; Zhang et al. 2020, 2022b; Zhao et al. 2020; Huang et al. 2021; Jiang et al. 2021a, b; Yu et al. 2021; Zheng et al. 2021; Zuo et al. 2021; An et al. 2022; Han et al. 2022; Jiang et al. 2022; Liang et al. 2022). On the other hand, the decreased expressions of the tumor-suppressor circRNAs, such as circ-TET1, circ-SHPRH, circ-MKLN1, and circ-CUL2, also could promote cell proliferation, migration, invasion, and inhibit cell cycle arrest and apoptosis of RB cells (Fig. 2 and Table 1) (Xing et al. 2018; Fu et al. 2021; Xu et al. 2021; Zhang et al. 2022a). Xenograft animal experiments have also demonstrated that some circRNAs can affect tumor growth and metastasis, affecting tumor size and volume, such as circ-TET1, circ-DHDDS, and circ-FAM158A (Liu et al. 2020; Fu et al. 2021; Yu et al. 2021). In summary, circRNAs in RB cells and tissues contribute to several “hallmarks of cancer” and can influence the malignant phenotype and influence the initiation and development of RB.
Oncogenic circRNAs in RB
circ-FAM158A
circ-FAM158A (has_circ_0000527) is generated from the endoplasmic reticulum membrane protein complex subunit 9 (EMC9), which is also known as FAM158A (Yi et al. 2019). Many studies have demonstrated significantly higher expression of circ-FAM158A in retinoblastoma tissues and cell lines than that in normal tissues and cells (Chen et al. 2020; Zhang et al. 2020; Yu et al. 2021; Zheng et al. 2021; Zuo et al. 2021; Liang et al. 2022). These studies also showed that circ-FAM158A promotes proliferation, invasion, and migration of retinoblastoma cells and functions as an oncogenic factor in retinoblastoma development via a molecular mechanism involving competing endogenous RNA (Fig. 1) (Chen et al. 2020; Zhang et al. 2020; Yu et al. 2021; Zheng et al. 2021; Zuo et al. 2021; Liang et al. 2022).
As demonstrated by gain-and-loss function experiments and bioinformatics analysis, circ-FAM158A affects the development of retinoblastoma cells via the miR-646/BCL2 axis (Chen et al. 2020). According to the results of in vitro experiments, overexpression of circ-FAM158A promotes the viability, migration, and invasion of retinoblastoma cells, while miR-646 has the opposite effects, and circ-FAM158A upregulates the expression of a target gene, BCL2, which encodes a critical antiapoptotic protein, via sponging miR-646, thereby promoting the development of retinoblastoma (Chen et al. 2020). Studies show that there is a negative interaction between circ-FAM158A and miR-646 (Chen et al. 2020; Zhang et al. 2020). Another study showed that circ-FAM158A regulates the expression levels of the trans-membrane protein LRP6 by adsorbing miR-646 and promoting retinoblastoma progression (Zhang et al. 2020). LRP6 functions as a co-receptor in the Wnt/β-catenin signaling cascade, which is essential in embryonic development and tumor genesis (Tamai et al. 2000; He et al. 2004).
As demonstrated using a xenograft murine model, silencing of circ-FAM158A inhibits tumor growth in vivo (Yu et al. 2021). In this study, circ-FAM158A promoted the development of retinoblastoma by decoying miR-98-5p and elevating the expression of the X-linked inhibitor of apoptosis protein (XIAP) (Yu et al. 2021), a member of the inhibitor of apoptosis family of proteins, mainly caspase inhibitors (Schimmer et al. 2006). Similarly, knockdown of circ-FAM158A inhibits tumor growth via the miR-138–5p/SLC7A5 axis (Zheng et al. 2021). SLC7A5, as a target of miR-138–5p, promotes retinoblastoma cell progression (Zheng et al. 2021). HDAC9 is a class II histone deacetylase (HDAC) inhibitor (Zhou et al. 2001). The HDAC9 gene functions as an oncogene in some cancers (Zhou et al. 2001; Yang et al. 2015; Rastogi et al. 2016). miR-27a-3p is a target of circ-FAM158A, and miR-27a-3p inhibition reverses the role of circ-FAM158A downregulation (Zuo et al. 2021). According to previous research, circ-FAM158A modulates the development of retinoblastoma via the miR-27a-3p/HDAC9 pathway by decreasing the expression of miR-27a-3p and significantly increasing that of HDAC9 (Zuo et al. 2021). In addition, knockdown of circ-FAM158A suppresses the P13K/AKT signaling pathway, which is crucial for the growth and survival of cancer cells (Zuo et al. 2021).
SMAD2 is an intracellular mediator of the transforming growth factor beta (TGFbeta) signaling pathway (Liu et al. 2004), which is involved in the progression of many cancers, such as glioblastoma (Chen et al. 2019b), gastric carcinoma (Shinto et al. 2010), and colorectal cancer (Zhang et al. 2021b). Another study has shown that circ-FAM158A drives retinoblastoma progression via the miR-1236-3p/SMAD2 pathway (Liang et al. 2022). The expression of miR-123-3p was decreased in RB tissues and cells (Liang et al. 2022). circ-FAM158A sponges miR-1236-3p, which directly targets SMAD2 (Liang et al. 2022). miR-1236-3p inhibition can reverse the effect of circ-FAM158A knockdown on RB cell malignant behavior, while SMAD2 overexpression can abolish the inhibitory effect of miR-1236-3p overexpression on RB cell progression (Liang et al. 2022). In addition, high expression of circ-FAM158A was associated with advanced tumor-node-metastasis (TNM) stage, optic nerve invasion, and choroidal invasion (Liang et al. 2022).
In conclusion, circ-FAM158A is upregulated in retinoblastoma patients, and it plays a carcinogenic role in retinoblastoma. Thus, it represents a potential new therapeutic target and diagnostic biomarker for retinoblastoma.
circ-DHDDS
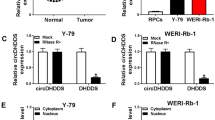
circ-DHDDS (hsa_circ_0000034) is located on chromosome 1 (26,772,806–26,774,151) and is derived from the dehydrodolichol diphosphate synthase (DHDDS) gene (Wang et al. 2020; Jiang et al. 2021a). As shown in several studies, the expression level of circ-DHDDS is significantly upregulated in retinoblastoma tissues and cells (Table 1) (Liu et al. 2020; Wang et al. 2020; Jiang et al. 2021a). The expression level of circ-DHDDS is related to the disease stage and clinical characteristics of retinoblastoma patients, with high expression strongly correlated with the degree of optic nerve invasion and International Intraocular Retinoblastoma Classification (IIRC) stage (Sun et al. 2020). As shown in a xenograft murine model, knockdown of circ-DHDDS inhibits proliferation, migration, invasion, and autophagy, as well as promoting apoptosis of retinoblastoma cells (Liu et al. 2020; Wang et al. 2020; Jiang et al. 2021a). circ-DHDDS is also associated with cell cycle arrest, with inhibition of circ-DHDDS inducing the arrest of more retinoblastoma cells in the G0/G1 phase of the cell cycle (Wang et al. 2020). Therefore, circ-DHDDS may serve as a potential diagnostic/prognostic biomarker and therapeutic target for retinoblastoma.
As an oncogene, circ-DHDDS directly sponges miR-361-3p to promote the development of retinoblastoma (Fig. 1) (Liu et al. 2020; Sun et al. 2020; Wang et al. 2020; Jiang et al. 2021a). As demonstrated in several studies, miR-361-3p functions as a tumor suppressor, and its expression is downregulated in many cancers (Chen et al. 2019a; Xu et al. 2020). Furthermore, circ-DHDDS accelerates the progression of retinoblastoma via the miR‑361‑3p/ADAM19 axis (Jiang et al. 2021a). circ-DHDDS acts as a competing endogenous RNA for miR‑361‑3p and regulates the expression of ADAM19, a trans-membrane soluble protein and specific target of miR‑361‑3p, which regulates the cell phenotype through cell adhesion and proteolysis (Jiang et al. 2021a). The autolytic processing activity of cysteine domain in ADAM19 is attributed to proteolytic activity (Tanabe et al. 2010; Hoyne et al. 2016).
WNT3A and STX17 are two other downstream targets of miR‑361‑3p in retinoblastoma (Liu et al. 2020; Wang et al. 2020). As a molecular sponge of miR-361-3p, circ-DHDDS regulates the expression of WNT3A, thereby stimulating the development of retinoblastoma (Wang et al. 2020). As a member of the Wnt family, WNT3A plays a key role in regulating cell self-renewal, proliferation, differentiation, and motility (He et al. 2015). The Wnt/β-catenin pathway is involved in cell proliferation, tissue homeostasis, and tissue regeneration (Chen et al. 2019a). Abnormal activation of this pathway plays a role in the development of cancer (Jung and Park 2020). The final step of autophagy is membrane fusion between autophagosomes and lysosomes (Itakura and Mizushima 2013). STX17, localized to the outer membrane of intact autophagosomes, has a unique hairpin-type structure and is mediated by two transmembrane domains, each of which contains a glycine zipper motif (Itakura and Mizushima 2013). The unique structure of STX17 enables it to specifically localize to intact autophagosomes, which is important for autophagosome-lysosome fusion (Itakura and Mizushima 2013; Huang et al. 2018). As demonstrated in experimental research, circ-DHDDS upregulates the expression of STX17 to induce cell proliferation, migration, invasion, autophagy, and tumor growth, and to suppress apoptosis of human retinoblastoma by inhibiting miR-361-3p (Liu et al. 2020).
circ-E2F3
circ-E2F3 (hsa_circ_0075804) is located on chromosome 6 (length: 720 bp) and is derived from E2F transfection factor 3 (E2F3) gene (Huang et al. 2021). In retinoblastoma tissues and cells, the expression levels of circ-E2F3 are markedly high, and it functions as an oncogenic factor in retinoblastoma development (Fig. 1, Table 1) (Huang et al. 2021; Han et al. 2022; Zhang et al. 2022b). As demonstrated in loss of function experiments, knockdown of circ-E2F3 inhibits proliferation, migration, and invasion and induces apoptosis of retinoblastoma cells (Huang et al. 2021; Han et al. 2022; Zhang et al. 2022b). In addition, as shown in xenograft studies, circ-E2F3 knockdown has an inhibitory effect on tumors in vivo (Huang et al. 2021; Han et al. 2022; Zhang et al. 2022b).
As a sponge for miR-204-5p, circ-E2F3 regulates the expression of rho-associated protein kinase 1 (ROCK1) (Huang et al. 2021). ROCK1 is a member of the ROCK family, which is the immediate downstream targets of RhoA, ubiquitously expressed serine-threonine protein kinases, and plays an important role in tumor cell migration and invasion (Noma et al. 2006; Hu et al. 2019). Many studies have reported that ROCK1 expression is high in tumors and that it promotes the proliferation and migration of tumors (Cai et al. 2014; Wu et al. 2014; Wan et al. 2019). The circ-E2F3/miR-138-5p/PEG10 axis promotes malignant behavior in retinoblastoma cells (Zhang et al. 2022b). circ-E2F3 functions as a molecular sponge for miR-138-5p, and miR-138-5p silencing largely impairs the effects of circ-E2F3 knockdown in RB cells (Zhang et al. 2022b). PEG10 is a retrotransposon-derived gene located on human chromosome 7q21 and is highly conserved among mammalian species (Lux et al. 2010; Chikamori et al. 2019). It promotes the proliferation, migration, and invasion of tumors (Kai et al. 2008; Peng et al. 2017). In addition, circ-E2F3 enhances the stability of E2F3 mRNA by binding to heterogeneous ribosomal protein K (HNRNPK), leading to the development of retinoblastoma (Zhao et al. 2020). Knockdown of circ-E2F3 results in an increase in the expression of apoptotic proteins, which promotes apoptosis and inhibits cell proliferation (Zhao et al. 2020). HNRNPK functions both as an RNA/DNA-binding protein and as a p53 co-regulator (Chen et al. 2021). As such, it participates in a variety of cellular processes and initiates apoptosis under conditions of DNA damage (Chen et al. 2021).
LIM and SH3 protein 1 (LASP1) is an actin-binding protein and plays important roles in cancer progression (He et al. 2013), such as prostate cancer (Dejima et al. 2017), nasopharyngeal carcinoma (Jiang et al. 2018), and glioblastoma (Zhong et al. 2018). circ-E2F3 regulates the expression of LASP1 by sponging miR-1287-5p and affects the biological process of retinoblastoma (Han et al. 2022). In RB tissues and cells, the expression levels of circ-E2F3 and LASP1 were upregulated, and the expression level of miR-1287-5p was downregulated (Han et al. 2022). Knockdown of circ-E2F3 can inhibit the growth, invasion, and survival of RB cells and hinder the development of tumors in vivo, while the inhibition of miR-1287-5p largely reverses the functional effects of circ-E2F3 knockdown (Han et al. 2022). LASP1 overexpression can also reverse the inhibition of RB cell proliferation, survival, and invasion by upregulating the expression of miR-1287-5p (Han et al. 2022).
circ-TRHDE
circ-TRHDE (hsa_circ_0099198) is located on chromosome 12 (73,012,670–73,015,531) and is derived from the thyrotropin-releasing hormone-degrading enzyme (TRHDE) (Jiang et al. 2021b). According to the literature, circ-TRHDE acts as an oncogenic factor in retinoblastoma and facilitates the progression of retinoblastoma via the miR-1287/LRP6 axis (Fig. 1, Table 1) (Jiang et al. 2021b). In RB tissues and cells, the expression levels of circ-TRHDE and LRP6 are increased, whereas those of miR-1287 are decreased (Jiang et al. 2021b). Depletion of circ-TRHDE induces retinoblastoma cell cycle arrest and apoptosis and inhibits the proliferation, migration, and invasion of retinoblastoma cells (Jiang et al. 2021b). As a sponge for miR-1287, circ-TRHDE negatively regulates the function of miR-1287 and upregulates the expression of LRP6 (Jiang et al. 2021b). According to a number of studies, miR-1287 plays an inhibitory role in many cancers, such as non-small cell lung cancer, hepatocellular carcinoma, and triple-negative breast cancer (Li et al. 2018; Lu et al. 2018; Schwarzenbacher et al. 2019). LRP6 functions as a co-receptor in the Wnt/β-catenin signaling cascade, which is essential in embryonic development and tumor genesis (Tamai et al. 2000; He et al. 2004). The discovery of the circ-TRHDE/miR-1287/LPR6 axis may provide a new therapeutic target for retinoblastoma therapy.
circ-E2F5
E2F transcription factor 5 (E2F5) is a member of the E2F family of transcription factors, which are involved in regulation of various cellular processes, including cellular proliferation, apoptosis, differentiation, and DNA damage response (Inagaki et al. 2020). circ_0084811, also known as circ-E2F5, regulates cell proliferation and apoptosis in RB through the miR-18a-5p/miR-18b-5p/E2F5 axis (Jiang et al. 2022). The expression of circ-E2F5 was significantly increased in RB cells (Jiang et al. 2022). circ-E2F5 upregulates E2F5 expression via sponging miR-18a-5p, miR-18b-5p or directly (Jiang et al. 2022). Overexpression of circ-E2F5 can promote cell proliferation and inhibit cell apoptosis, while knockdown of circ-E2F5 can inhibit cell proliferation and promote cell apoptosis (Jiang et al. 2022). miR-18a-5p inhibitor or miR-18b-5p inhibitor can partially restore the effect caused by hsa_circ_E2F5 silencing, and E2F5 overexpression completely reversed this effect (Jiang et al. 2022).
circ-RNF20
circ-RNF20, also known as hsa_circ_0087784, aggravates the malignancy of retinoblastoma via miR-132-3p/PAX6 axis (An et al. 2022). In RB tissues and cells, the expression levels of cir-cRNF20 and PAX6 were increased, and the expression levels of miR-132-3p were significantly decreased (An et al. 2022). circ-RNF20 directly targets miR-132-3p, and PAX6 is a target gene of miR-132-3p (An et al. 2022). PAX6 is a transcription factor essential for normal development of the eyes and nervous system (Pinson et al. 2006). Deficiency of circ-RNF20 inhibits cell proliferation, migration, and invasion, and induces apoptosis in vitro, and miR-132-3p inhibition or PAX6 overexpression reverses circ-RNF20-mediated effects on RB cell malignant behavior (An et al. 2022). By constructing a xenogeneic tumor model, it was found that circ-RNF20 silencing could inhibit the formation of tumors in vivo. In addition, circ-RNF20 was found to be associated with advanced TNM stage and poor overall survival. The expression levels of circ-RNF20 were higher in patients with TNM stage III tumors compared with patients with TNM stage I/II tumors; RB patients with higher circ-RNF20 expression had a lower overall survival than those with lower circ-RNF20 expression (An et al. 2022). circ-RNF20 can promote the growth, migration, and invasion of RB cells and inhibit the apoptosis (An et al. 2022).
Tumor-suppressor circRNAs in RB
circ-TET1
circ-TET1 (has_circ_0093996) is derived from the Tet methylcytosine dioxygenase 1 (TET1) gene. As a member of TET protein family, TET1 affects the DNA methylation/demethylation cycle and is an important tumor suppressor (Tian et al. 2017), which is downregulated in various cancers (Duan et al. 2017; Tian et al. 2017). In retinoblastoma tissues and cells, the expression levels of circ-TET1 are decreased, whereas those of miR-492 and miR-494-3p are increased (Fu et al. 2021). circ-TET1 inhibits the proliferation, migration, and invasion of retinoblastoma cells and accelerates the apoptosis and cell cycle arrest of retinoblastoma cells (Fig. 2, Table 2) (Fu et al. 2021). As demonstrated in a xenograft mouse model, overexpression of circ-TET1 inhibits tumor growth in vivo (Fu et al. 2021). Upregulation of circ-TET1 inhibits the Wnt/β-catenin pathway by absorbing miR-492 and miR-494-3p (Fu et al. 2021). Abnormal activation of the Wnt/β-catenin signaling pathway is related to the malignant behavior of tumors (Rosenbluh et al. 2014). This regulatory axis may provide a new and promising therapeutic strategy for retinoblastoma therapy (Fu et al. 2021).
circ-SHPRH
circ-SHPRH (hsa_-circ_0001649) is a transcription product of the Snf2 histone linker Phd ring helicase (SHPRH) gene (Xing et al. 2018). As a reported tumor suppressor, circ-SHPRH inhibits proliferation, migration, and invasion and induces cell apoptosis (Ji et al. 2018; Xing et al. 2018; Xu et al. 2018; Zhang et al. 2018). In a previous retinoblastoma study, decreased expression of circ-SHPRH was significantly associated with tumor size, an advanced IIRC stage, and an aggressive phenotype of retinoblastoma (Table 2) (Xing et al. 2018). The decrease in the expression of circ-SHPRH was independent of age, sex, and optic nerve involvement, suggesting that a reduction in the expression of circ-SHPRH in tissues might be an independent prognostic factor after surgery in retinoblastoma patients (Xing et al. 2018). In the same study, patients with low expression had poorer 5-year overall survival after surgery, which suggests that low expression of circ-SHPRH could predict a poor prognosis in retinoblastoma patients (Xing et al. 2018). This study also noted that downregulation of circ-SHPRH promoted the growth of retinoblastoma cells, partly by regulating the AKT/mTOR signaling pathway (Fig. 2) and that the expression of circ-SHPRH was negatively correlated with the expression of p-AKT and p-mTOR (Xing et al. 2018). The P13K-AKT-mTOR signaling network controls many characteristics of cancer cells such as the cell cycle, survival, metabolism, motility, and genomic instability (Fruman and Rommel 2014). Taken together, the literature suggests that upregulation of circ-SHPRH may be a potentially promising strategy for retinoblastoma treatment.
circ-MKLN1
circ-MKLN1 is also termed as hsa_circ_0082415 (Lyu et al. 2019). Aberrant expression of circ-MKLN1 and PDCD4, with reduced expression of both, as well as significant elevation of miR-425-5p expression, has been observed in retinoblastoma tissues and cells (Lyu et al. 2019; Xu et al. 2021). Overexpression of circ-MKLN1 represses the proliferation, migration, and invasion of retinoblastoma cells in vitro and tumor growth (i.e., tumor volume and weight) in vivo (Xu et al. 2021). In the development of retinoblastoma, circ-MKLN1 functions as a sponge for miR-425-5p (Fig. 2) (Xu et al. 2021). Overexpression of miR-425-5p reverses the effects of circ-MKLN1 on the proliferation, invasion, and migration of retinoblastoma cells (Xu et al. 2021). Likewise, knockdown of PDCD4, which is as the target gene of miR-425-5p, reversed circ-MKLN1 overexpression-mediated inhibitory effect on cell proliferation of retinoblastoma (Xu et al. 2021). PDCD4 maps to chromosome band 10q24 in the human genome (Soejima et al. 1999), and it encodes a 469 amino acid protein with two basic domains at the N-terminus and C-terminus and two conserved α-helical MA-3 domains (Lankat-Buttgereit and Goke 2009). PDCD4 is a proinflammatory protein that promotes activation of the transcription factor NF-κB and inhibits IL-10 (Sheedy et al. 2010). Based on recent research, PDCD4 is considered a tumor suppressor gene and a potential target for anticancer therapy (Lankat-Buttgereit and Goke 2009). In many cancers, such as skin cancer (Matsuhashi et al. 2007), human gliomas (Afonja et al. 2004), and tongue tumors (Carinci et al. 2005), the expression of PDCD4 is downregulated. Acting as a tumor suppressor, at transcriptional and translational levels, PDCD4 is involved in tumor progression, the cell cycle, and differentiation, regulating a variety of proteins in cells, and these proteins regulate different signal transduction pathways (Lankat-Buttgereit and Goke 2009).
circ-CUL2
circ-CUL2 is also known as hsa_circ_0000234, which is derived from back-splicing of the CUL2 mRNA (from exon 2 to exon 4) (Peng et al. 2020). E2F transcription factor 2 (E2F2) is a prototypical transcription factor implicated in transcriptional regulation, cell cycle, and tumorigenesis (Du et al. 2022). circ-CUL2 inhibits the proliferation, invasion, and migration of retinoblastoma cells by regulating the miR-214-5p/E2F2 axis (Zhang et al. 2022a). In retinoblastoma tumor tissues and cells, circ-CUL2 and E2F2 were underexpressed, whereas miR-214-5p was overexpressed (Zhang et al. 2022a). circ-CUL2 negatively regulates the expression of miR-214-5p, and miR-214-5p negatively regulates the expression of E2F2 (Zhang et al. 2022a). miR-214-5p promotes proliferation, invasion, and migration of retinoblastoma cells (Zhang et al. 2022a). Overexpression of miR-214-5p or inhibition of E2F2 partially reversed the inhibitory effects of circ-CUL2 on retinoblastoma cell invasion and migration (Zhang et al. 2022a). The discovery of this expression axis may provide a new strategy for the treatment of retinoblastoma patients.
The potential clinical applications of circRNAs in RB
Diagnostic potentials of circRNAs in RB
Retinoblastoma, the most common malignant retinal tumor among children under 3 years old, is lethal if left untreated (Global Retinoblastoma Study et al. 2020). The clinical diagnosis of RB is based on fundus changes and imaging findings, such as B-ultrasound, optical coherence tomography, and brain and orbital magnetic resonance imaging (Manrique et al. 2021). In addition, studies have found that children with a family history of RB and a known RB1 mutation in one or both parents have a significantly higher risk of RB, so prenatal genetic testing and close follow-up are important for early diagnosis and treatment (AlAli et al. 2018; Skalet et al. 2018). Early diagnosis, together with timely and effective treatment, is important to improve retinoblastoma-related outcomes, so the discovery of new diagnostic biomarkers has important implications for improving the treatment of RB. circRNA has a closed circular structure that is not affected by RNA exonuclease, is more stable in expression (Xu et al. 2017), and has great potential in cancer diagnosis (Najafi 2022a, b). These significantly different expressed oncogenic circRNAs (circ-FAM158A, circ-DHDDS, circ-E2F3, circ-TRHDE, circ-E2F5, and circ-RNF20) and tumor-suppressor circRNAs (circ-TET1, circ-SHPRH, circ-MKLN1, and circ-CUL2) in retinoblastoma tissues compared with normal tissues (Tables 1 and 2),have great potentials as biomarkers for RB diagnostic, and further research in large human samples is needed.
Therapeutic potentials of circRNAs in RB
Current treatments for RB include enucleation of the eye, chemotherapy, and focused therapy (Manrique et al. 2021). Survival and salvage rates for RB are still low worldwide due to the health care gap between countries (Manrique et al. 2021). At present, the treatment of RB is also focused on improving the affordability and availability of treatment, reducing related side effects and complications, and prolonging the effective period of treatment (Manrique et al. 2021). With the development of the research on circRNA, the role of circRNAs in RB has been taken more and more attention. Many studies have shown that the oncogenic circRNAs, including circ-FAM158A, circ-DHDDS, circ-E2F3, circ-TRHDE, circ-E2F5, and circ-RNF20 (Fig. 1 and Table 1), and the tumor-suppressor circRNAs, including circ-TET1, circ-SHPRH, circ-MKLN1, and circ-CUL2 (Fig. 2 and Table 2), participate in the occurrence and development of RB through affecting the proliferation, apoptosis, migration, and invasion of RB cells. Studies in xenograft mouse models have also demonstrated that the knockdown of circ-FAM158A (Yu et al. 2021), circ-E2F3 (Huang et al. 2021; Han et al. 2022; Zhang et al. 2022b), circ-RNF20 (An et al. 2022), and overexpression of circ-TET1 (Fu et al. 2021) and circ-MKLN1 (Xu et al. 2021) inhibit tumor formation and growth in vivo. Therefore, the discovery of circRNA or its pathway of action may provide a new therapy for RB.
Prognostic potentials of circRNAs in RB
The overall prognosis of RB patients is associated with tumor size, tumor location, presence of subretinal fluid, and histopathology characteristics (AlAli et al. 2018). circRNA was reported to be correlated with the clinicopathological features of RB, such as tumor size, TNM stages, and lymph node metastasis (Tables 1 and 2). circ-FAM158A was related to advanced TNM stages, optic nerve invasion, and choroidal invasion in RB patients (Liang et al. 2022). High expression of circ-DHDDS strongly correlated with IIRC stage and optic nerve invasion degree of RB (Sun et al. 2020). An increased expression level of circ-RNF20 was found to be associated with advanced tumor, lymph node, and metastatic stage, and indicates a poor overall survival of RB patients (An et al. 2022). A decreased expression level of circ-SHPRH was associated with tumor size, advanced IIRC stage, and aggressive phenotype of RB, and indicates a poor prognosis (Xing et al. 2018). These circRNAs all represent as potential biomarkers for prognosis of RB.
Perspective
RB is the most common intraocular cancer of childhood, which is caused by mutation of the RB1 gene (Dimaras et al. 2012). It has a high mortality risk due to its origin, complex clinical presentation, and metastasis potential. Therefore, early detection, diagnosis, and treatment are critical to improve the cure rate and reduce mortality. circRNAs regulate the expression of multiple genes and participate in the regulation of cellular activities by sponging microRNAs, acting as sequestering agents of RNA blinding proteins (RBPs), regulating transcription and alternative splicing, binding proteins, forming pseudogenes, and expressing peptides (Tang et al. 2017; Kristensen et al. 2019; Chen 2020). Due to their stability, tissue-specific roles, and crucial roles in many cancers, circRNAs are considered to have great potential in tumor diagnosis and treatment. In recent years, with the advancement of high-throughput sequencing technology, we now have a better grasp of the roles of circRNAs in RB. Several studies reported that specific circRNAs may affect the proliferation, metastasis, and apoptosis of RB cells by functioning as a sponge for microRNAs and participate in the development of RB (Figs. 1 and 2) (Chen et al. 2020; Wang et al. 2020; Huang et al. 2021; Jiang et al. 2021a; Yu et al. 2021). This review outlined recent progress in understanding of the roles of circRNAs in RB with the aim of highlighting novel strategies for the diagnosis, prognosis, and treatment of RB (Tables 1 and 2).
As noted above, studies suggested that circRNAs may act as potential diagnostic and therapeutic targets, as well as key prognostic markers in clinical practice (Chen et al. 2020; Zhang et al. 2020; Fu et al. 2021; Huang et al. 2021; Jiang et al. 2021b; Yu et al. 2021; Zuo et al. 2021). As a novel biomarker, circRNAs are helpful and promising for the diagnosis, treatment, and prognosis of cancer. However, there are still many challenges in the application of circRNAs. First of all, the current understanding of circRNAs is incomplete, with many unknown circRNAs likely waiting to be discovered. Second, the majority of recent studies that focused on circRNAs in RB have used tumor tissues and normal tissues or tumor cells and normal cells as samples (Wang et al. 2020; Huang et al. 2021; Zheng et al. 2021; Zhang et al. 2022b). In future studies, samples such as blood, urine, and tear samples will be available for selection. Finally, the ultimate goal of the various experiments we are currently conducting is to apply circRNAs to the clinical diagnosis and treatment of RB. Therefore, in this regard, the safety and efficacy of circRNAs are very important, which require us to conduct a large number of clinical studies and experiments to verify and evaluate. For the time being, the sample size of study about circRNA in RB is not large enough and there is a lack of certain clinical experimental studies, and many studies are in the exploratory stage, so circRNAs are not yet fully applicable to clinical practice. However, we firmly believe that as our understanding of the pathogenesis of RB improves, circRNAs will be widely used in the diagnosis, treatment, and prognosis of RB, greatly reducing the mortality rate and improving the cure rate, alleviating the pain of patients, and bringing happiness to their lives.
Data Availability
All data generated during this study are included in this published article.
Abbreviations
- RB :
-
Retinoblastoma
- circRNA :
-
Circular RNA
- EMC9 :
-
Endoplasmic reticulum membrane protein complex subunit 9
- XIAP :
-
X-linked inhibitor of apoptosis protein
- HDAC :
-
Histone deacetylase
- TGFbeta :
-
Transforming growth factor beta
- TNM :
-
Tumor-node metastasis
- E2F3 :
-
E2F transfection factor 3
- ROCK1 :
-
Rho-associated protein kinase 1
- HNRNPK :
-
Heterogeneous ribosomal protein K
- LASP1 :
-
LIM and SH3 protein 1
- TRHDE :
-
Thyrotropin-releasing hormone-degrading enzyme
- E2F5 :
-
E2F transcription factor 5
- TET1 :
-
Tet methylcytosine dioxygenase 1
- SHPRH :
-
Snf2 histone linker Phd ring helicase
- E2F2 :
-
E2F transcription factor 2
- RBPs :
-
RNA blinding proteins
References
Afonja O, Juste D, et al (2004) Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene 23(49):8135–8145. https://doi.org/10.1038/sj.onc.1207983
AlAli A, Kletke S, et al (2018) Retinoblastoma for pediatric ophthalmologists. Asia Pac J Ophthalmol (Phila) 7(3):160–168. https://doi.org/10.22608/apo.201870
Altesha MA, Ni T et al (2019) Circular RNA in cardiovascular disease. J Cell Physiol 234(5):5588–5600. https://doi.org/10.1002/jcp.27384
An D, Yang J et al (2022) circRNF20 aggravates the malignancy of retinoblastoma depending on the regulation of miR-132-3p/PAX6 axis. Open Med (wars) 17(1):955–968. https://doi.org/10.1515/med-2022-0483
Ancona-Lezama D, Dalvin LA et al (2020) Modern treatment of retinoblastoma: a 2020 review. Indian J Ophthalmol 68(11):2356–2365. https://doi.org/10.4103/ijo.IJO_721_20
Cai, H., L. Lin, et al. (2014). Combined microRNA-340 and ROCK1 mRNA profiling predicts tumor progression and prognosis in pediatric osteosarcoma. Int J Mol Sci 15(1):560–573. https://doi.org/10.3390/ijms15010560
Carinci F, Lo Muzio L et al (2005) Potential markers of tongue tumor progression selected by cDNA microarray. Int J Immunopathol Pharmacol 18(3):513–524. https://doi.org/10.1177/039463200501800311
Chen C-C, Yang J-H et al (2021) Arginine methylation of hnRNPK inhibits the DDX3-hnRNPK interaction to play an anti-apoptosis role in osteosarcoma cells. Int J Mol Sci 22(18):9764. https://doi.org/10.3390/ijms22189764
Chen L, Nan A et al (2019a) Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer 18:13. https://doi.org/10.1186/s12943-019-0943-0
Chen LL (2020) The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 21(8):475–490. https://doi.org/10.1038/s41580-020-0243-y
Chen NN, Chao DL et al (2020) Circular RNA has_circ_0000527 participates in proliferation, invasion and migration of retinoblastoma cells via miR-646/BCL-2 axis. Cell Biochem Funct 38(8):1036–1046. https://doi.org/10.1002/cbf.3535
Chen W, Huang B et al (2019b) MiR-145 inhibits EGF-induced epithelial-to-mesenchymal transition via targeting Smad2 in human glioblastoma. Onco Targets Ther 12:3099–3107. https://doi.org/10.2147/ott.S202129
Chen X, Yang T et al (2019c) Circular RNAs in immune responses and immune diseases. Theranostics 9(2):588–607. https://doi.org/10.7150/thno.29678
Chikamori H, Ishida Y et al (2019) Distinctive expression pattern of Peg10 in the mouse brain. Eur J Anat 23(5):361–368
Correa-Acosta A, Gonzalez-Alviar ME et al (2018) Retinoblastoma and optic nerve enhancement in a brain magnetic resonance scan: is it always a metastasis? Arch Sociedad Esp Oftalmol 93(5):251–254. https://doi.org/10.1016/j.oftal.2017.10.010
De Palma FDE, Salvatore F et al (2022) Circular RNAs as potential biomarkers in breast cancer. Biomedicines 10(3):725. https://doi.org/10.3390/biomedicines10030725
Dejima T, Imada K et al (2017) Suppression of LIM and SH3 domain protein 1 (LASP1) negatively regulated by androgen receptor delays castration resistant prostate cancer progression. Prostate 77(3):309–320. https://doi.org/10.1002/pros.23269
Dimaras H, Corson TW et al (2015) Retinoblastoma. Nat Rev Dis Primers 1:15021. https://doi.org/10.1038/nrdp.2015.21
Dimaras H, Kimani K et al (2012) Retinoblastoma. Lancet 379(9824):1436–1446. https://doi.org/10.1016/s0140-6736(11)61137-9
Du K, Sun S et al (2022) E2F2 promotes lung adenocarcinoma progression through B-Myb- and FOXM1-facilitated core transcription regulatory circuitry. Int J Biol Sci 18(10):4151–4170. https://doi.org/10.7150/ijbs.72386
Duan H, Yan Z et al (2017) TETI inhibits EMT of ovarian cancer cells through activating Wnt/beta-catenin signaling inhibitors DKK1 and SFRP2. Gynecol Oncol 147(2):408–417. https://doi.org/10.1016/j.ygyno.2017.08.010
Fabian ID, Onadim Z et al (2018) The management of retinoblastoma. Oncogene 37(12):1551–1560. https://doi.org/10.1038/s41388-017-0050-x
Fruman DA, Rommel C (2014) PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 13(2):140–156. https://doi.org/10.1038/nrd4204
Fu C, Wang S et al (2021) CircTET1 inhibits retinoblastoma progression via targeting miR-492 and miR-494-3p through Wnt/beta-catenin signaling pathway. Curr Eye Res 46(7):978–987. https://doi.org/10.1080/02713683.2020.1843685
Global Retinoblastoma Study G, Fabian ID, et al (2020) Global retinoblastoma presentation and analysis by national income level. JAMA Oncol 6(5):685-695. https://doi.org/10.1001/jamaoncol.2019.6716
Goodall GJ, Wickramasinghe VO (2021) RNA in cancer. Nat Rev Cancer 21(1):22–36. https://doi.org/10.1038/s41568-020-00306-0
Han Q, Ma L et al (2022) Circ_0075804 regulates the expression of LASP1 by targeting miR-1287-5p and thus affects the biological process of retinoblastoma. Curr Eye Res 47(7):1077–1086. https://doi.org/10.1080/02713683.2022.2053164
Hanahan D (2022) Hallmarks of cancer: new dimensions. Cancer Discov 12(1):31–46. https://doi.org/10.1158/2159-8290.CD-21-1059
He B, Yin B et al (2013) Overexpression of LASP1 is associated with proliferation, migration and invasion in esophageal squamous cell carcinoma. Oncol Rep 29(3):1115–1123. https://doi.org/10.3892/or.2012.2199
He S, Lu Y et al (2015) Wnt3a: functions and implications in cancer. Chin J Cancer 34:50. https://doi.org/10.1186/s40880-015-0052-4
He X, Semenov M et al (2004) LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development 131(8):1663–1677. https://doi.org/10.1242/dev.01117
Henry NL, Hayes DF (2012) Cancer biomarkers. Mol Oncol 6(2):140–146. https://doi.org/10.1016/j.molonc.2012.01.010
Hoyne G, Rudnicka C et al (2016) Genetic and cellular studies highlight that A disintegrin and Metalloproteinase 19 is a protective biomarker in human prostate cancer. BMC Cancer 16:151. https://doi.org/10.1186/s12885-016-2178-4
Hsu MT, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280(5720):339–340. https://doi.org/10.1038/280339a0
Hu C, Zhou H et al (2019) ROCK1 promotes migration and invasion of non-small-cell lung cancer cells through the PTEN/PI3K/FAK pathway. Int J Oncol 55(4):833–844. https://doi.org/10.3892/ijo.2019.4864
Hu W, Bi Z-Y et al (2018) Emerging landscape of circular RNAs in lung cancer. Cancer Lett 427:18–27. https://doi.org/10.1016/j.canlet.2018.04.006
Huang J, Yang Y et al (2018) MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J Cell Biochem 119(5):3853–3863. https://doi.org/10.1002/jcb.26464
Huang Y, Xue B et al (2021) Circ-E2F3 acts as a ceRNA for miR-204-5p to promote proliferation, metastasis and apoptosis inhibition in retinoblastoma by regulating ROCK1 expression. Exp Mol Pathol 120:104637. https://doi.org/10.1016/j.yexmp.2021.104637
Inagaki Y, Wu D et al (2020) Knockdown ofE2F5induces cell death via the TP53-dependent pathway in breast cancer cells carrying wild-typeTP53. Oncol Rep 44(5):2241–2252. https://doi.org/10.3892/or.2020.7761
Itakura E, Mizushima N (2013) Syntaxin 17: the autophagosomal SNARE. Autophagy 9(6):917–919. https://doi.org/10.4161/auto.24109
Ji W, Qiu C et al (2018) Hsa_circ_0001649: a circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun 497(1):122–126. https://doi.org/10.1016/j.bbrc.2018.02.036
Jiang G, Qu M et al (2022) hsa_circ_0084811 regulates cell proliferation and apoptosis in retinoblastoma through miR-18a-5p/miR-18b-5p/E2F5 axis. Biomed Res Int 2022:6918396. https://doi.org/10.1155/2022/6918396
Jiang N, Zong D et al (2018) Expression and prognostic value of LASP1 in nasopharyngeal carcinoma. Bulletin of Chinese Cancer 27(12):949–955
Jiang Y, Xiao F et al (2021a) Circular RNA has_circ_0000034 accelerates retinoblastoma advancement through the miR-361-3p/ADAM19 axis. Mol Cell Biochem 476(1):69–80. https://doi.org/10.1007/s11010-020-03886-5
Jiang Y, Xiao F et al (2021b) Hsa_circ_0099198 facilitates the progression of retinoblastoma by regulating miR-1287/LRP6 axis. Exp Eye Res 206:108529. https://doi.org/10.1016/j.exer.2021.108529
Jung Y-S, Park J-I (2020) Wnt signaling in cancer: therapeutic targeting of Wnt signaling beyond beta-catenin and the destruction complex. Exp Mol Med 52(2):183–191. https://doi.org/10.1038/s12276-020-0380-6
Kai W, Yunfeng P et al (2008) Expression of imprinted gene PEGIO in human gastric adenocarcinoma tissues and significance. Journal of Jilin Univeristy Medicine Edition 34(2):309–312
Kivela TT, Hadjistilianou T (2017) Neonatal retinoblastoma. Asia Pac J Oncol Nurs 4(3):197–204. https://doi.org/10.4103/apjon.apjon_18_17
Kristensen LS, Andersen MS et al (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20(11):675–691. https://doi.org/10.1038/s41576-019-0158-7
Lankat-Buttgereit B, Goke R (2009) The tumour suppressor Pdcd4: recent advances in the elucidation of function and regulation. Biol Cell 101(6):309–317. https://doi.org/10.1042/bc20080191
Lei B, Tian Z et al (2019) Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci 16(2):292–301. https://doi.org/10.7150/ijms.28047
Li X, Zhao Z et al (2017) Hsa-circRNA11783-2 in peripheral blood is correlated with coronary artery disease and type 2 diabetes mellitus. Diab Vasc Dis Res 14(6):510–515. https://doi.org/10.1177/1479164117722714
Li Y, Hu J et al (2018) Upregulated circular RNA circ_0016760 indicates unfavorable prognosis in NSCLC and promotes cell progression through miR-1287/GAGE1 axis. Biochem Biophys Res Commun 503(3):2089–2094. https://doi.org/10.1016/j.bbrc.2018.07.164
Li Y, Zheng Q et al (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25(8):981–984. https://doi.org/10.1038/cr.2015.82
Liang T, Fan M et al (2022) Circ_0000527 drives retinoblastoma progression by regulating miR-1236-3p/SMAD2 pathway. Curr Eye Res 47(4):624–633. https://doi.org/10.1080/02713683.2021.2007535
Lin L, Zhou G, et al. (2020) Which long noncoding RNAs and circular RNAs contribute to inflammatory bowel disease? Cell Death Dis 11(6). https://doi.org/10.1038/s41419-020-2657-z
Liu H, Yuan HF, et al. (2020) Circular RNA circ_0000034 upregulates STX17 level to promote human retinoblastoma development via inhibiting miR-361–3p. Eur Rev Med Pharmacol Sci 24(23):12080–12092. https://doi.org/10.26355/eurrev_202012_23997
Liu Y, Festing MH et al (2004) Generation of novel conditional and hypornorphic alleles of the Smad2 gene. Genesis 40(2):118–123. https://doi.org/10.1002/gene.20072
Lu J, Tang L et al (2018) Mir-1287 suppresses the proliferation, invasion, and migration in hepatocellular carcinoma by targeting PIK3R3. J Cell Biochem 119(11):9229–9238. https://doi.org/10.1002/jcb.27190
Lux H, Flammann H et al (2010) Genetic and molecular analyses of PEG10 reveal new aspects of genomic organization, transcription and translation. PLoS One 5(1):e8686. https://doi.org/10.1371/journal.pone.0008686
Lyu J, Wang Y et al (2019) Reduction of circular RNA expression associated with human retinoblastoma. Exp Eye Res 184:278–285. https://doi.org/10.1016/j.exer.2019.03.017
Manrique M, Akinbolue D et al (2021) Update on the treatment of retinoblastoma. NeoReviews 22(7):e423–e437. https://doi.org/10.1542/neo.22-7-e423
Matsuhashi S, Narisawa Y et al (2007) Expression patterns of programmed cell death 4 protein in normal human skin and some representative skin lesions. Exp Dermatol 16(3):179–184. https://doi.org/10.1111/j.1600-0625.2006.00531.x
Memczak S, Jens M et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495(7441):333–338. https://doi.org/10.1038/nature11928
Najafi S (2022a) Circular RNAs as emerging players in cervical cancer tumorigenesis; a review to roles and biomarker potentials. Int J Biol Macromol 206:939–953. https://doi.org/10.1016/j.ijbiomac.2022.03.103
Najafi S (2022b) The emerging roles and potential applications of circular RNAs in ovarian cancer: a comprehensive review. J Cancer Res Clin Oncol. https://doi.org/10.1007/s00432-022-04328-z
Noma K, Oyama N et al (2006) Physiological role of ROCKs in the cardiovascular system. Am J Physiol Cell Physiol 290(3):C661–C668. https://doi.org/10.1152/ajpcell.00459.2005
Ortiz MV, Dunkel IJ (2016) Retinoblastoma. J Child Neurol 31(2):227–236. https://doi.org/10.1177/0883073815587943
Peng L, Sang HM, et al (2020) circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142–3p/ROCK2. Mol Cancer 19(1). https://doi.org/10.1186/s12943-020-01270-x
Peng Y-P, Zhu Y et al (2017) PEG10 overexpression induced by E2F–1 promotes cell proliferation, migration, and invasion in pancreatic cancer. J Exp Clin Cancer Res 36:30. https://doi.org/10.1186/s13046-017-0500-x
Pinson J, Simpson TI, et al (2006) Positive autoregulation of the transcription factor Pax6 in response to increased levels of either of its major isoforms, Pax6 or Pax6(5a), in cultured cells. Bmc Dev Biol 6. https://doi.org/10.1186/1471-213x-6-25
Rahmati Y, Asemani Y et al (2021) CiRS-7/CDR1as; an oncogenic circular RNA as a potential cancer biomarker. Pathol Res Pract 227:153639. https://doi.org/10.1016/j.prp.2021.153639
Rastogi B, Raut SK et al (2016) Overexpression of HDAC9 promotes oral squamous cell carcinoma growth, regulates cell cycle progression, and inhibits apoptosis. Mol Cell Biochem 415(1–2):183–196. https://doi.org/10.1007/s11010-016-2690-5
Ren S, Lin P et al (2020) Circular RNAs: promising molecular biomarkers of human aging-related diseases via functioning as an miRNA sponge. Mol Ther Methods Clin Dev 18:215–229. https://doi.org/10.1016/j.omtm.2020.05.027
Rosenbluh J, Wang X et al (2014) Genomic insights into WNT/13-catenin signaling. Trends Pharmacol Sci 35(2):103–109. https://doi.org/10.1016/j.tips.2013.11.007
Sayad A, Najafi S et al (2022a) The role of circular RNAs in pancreatic cancer: new players in tumorigenesis and potential biomarkers. Pathol Res Pract 232:153833. https://doi.org/10.1016/j.prp.2022.153833
Sayad A, Najafi S et al (2022b) Circular RNAs in renal cell carcinoma: functions in tumorigenesis and diagnostic and prognostic potentials. Pathol Res Pract 229:153720. https://doi.org/10.1016/j.prp.2021.153720
Schimmer AD, Dalili S et al (2006) Targeting XIAP for the treatment of malignancy. Cell Death Differ 13(2):179–188. https://doi.org/10.1038/sj.cdd.4401826
Schwarzenbacher D, Klec C et al (2019) MiR-1287-5p inhibits triple negative breast cancer growth by interaction with phosphoinositide 3-kinase CB, thereby sensitizing cells for PI3Kinase inhibitors. Breast Cancer Res 21:20. https://doi.org/10.1186/s13058-019-1104-5
Shan C, Zhang Y, et al (2019) Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer 18(1). https://doi.org/10.1186/s12943-019-1069-0
Sheedy FJ, Palsson-McDermott E et al (2010) Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol 11(2):141-U159. https://doi.org/10.1038/ni.1828
Shields CL, Say EAT et al (2015) Rescue intra-arterial chemotherapy following retinoblastoma recurrence after initial intra-arterial chemotherapy. J Francais D Ophtalmol 38(6):542–549. https://doi.org/10.1016/j.jfo.2015.03.004
Shields CL, Shields JA (2010) Retinoblastoma management: advances in enucleation, intravenous chemoreduction, and intra-arterial chemotherapy. Curr Opin Ophthalmol 21(3):203–212. https://doi.org/10.1097/ICU.0b013e328338676a
Shinto O, Yashiro M, et al (2010) Phosphorylated Smad2 in advanced stage gastric carcinoma. Bmc Cancer 10. https://doi.org/10.1186/1471-2407-10-652
Skalet AH, Gombos DS et al (2018) Screening children at risk for retinoblastoma: consensus report from the American Association of Ophthalmic Oncologists and Pathologists. Ophthalmology 125(3):453–458. https://doi.org/10.1016/j.ophtha.2017.09.001
Soejima H, Miyoshi O et al (1999) Assignment of the programmed cell death 4 gene (PDCD4) to human chromosome band 10q24 by in situ hybridization. Cytogenet Cell Genet 87(1–2):113–114. https://doi.org/10.1159/000015408
Song M, Xia L et al (2018) Circular RNA in liver: health and diseases. Circular Rnas: Biogenesis and Functions. J Xiao 1087:245–257
Sun Z, Zhang A et al (2020) Circular RNA hsa_circ_0000034 promotes the progression of retinoblastoma via sponging microRNA-361-3p. Bioengineered 11(1):949–957. https://doi.org/10.1080/21655979.2020.1814670
Taheri M, Najafi S et al (2021) The role and clinical potentials of circular RNAs in prostate cancer. Front Oncol 11:781414. https://doi.org/10.3389/fonc.2021.781414
Tamai K, Semenov M et al (2000) LDL-receptor-related proteins in Wnt signal transduction. Nature 407(6803):530–535. https://doi.org/10.1038/35035117
Tanabe C, Hotoda N et al (2010) ADAM19 autolysis is activated by LPS and promotes non-classical secretion of cysteine-rich protein 2. Biochem Biophys Res Commun 396(4):927–932. https://doi.org/10.1016/j.bbrc.2010.05.025
Tang CM, Zhang M et al (2017) CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep 7:40342. https://doi.org/10.1038/srep40342
Teixo R, Laranjo M et al (2015) Retinoblastoma: might photodynamic therapy be an option? Cancer Metastasis Rev 34(4):563–573. https://doi.org/10.1007/s10555-014-9544-y
Tian Y, Pan F et al (2017) Association of TET1 expression with colorectal cancer progression. Scand J Gastroenterol 52(3):312–320. https://doi.org/10.1080/00365521.2016.1253767
Wan X, Xiang J et al (2019) Long noncoding RNA POU3F3 promotes cancer cell proliferation in prostate carcinoma by upregulating rho-associated protein kinase 1. J Cell Biochem 120(5):8195–8200. https://doi.org/10.1002/jcb.28101
Wang H, Li M et al (2020) CircDHDDS/miR-361-3p/WNT3A axis promotes the development of retinoblastoma by regulating proliferation, cell cycle, migration, and invasion of retinoblastoma cells. Neurochem Res 45(11):2691–2702. https://doi.org/10.1007/s11064-020-03112-0
Wen ZJ, Xin H et al (2021) Emerging roles of circRNAs in the pathological process of myocardial infarction. Mol Ther Nucleic Acids 26:828–848. https://doi.org/10.1016/j.omtn.2021.10.002
Wu Y-J, Tang Y et al (2014) Expression and significance of Rac1, Pak1 and Rock1 in gastric carcinoma. Asia Pac J Clin Oncol 10(2):E33–E39. https://doi.org/10.1111/ajco.12052
Xing L, Zhang L et al (2018) Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomed Pharmacother 105:326–333. https://doi.org/10.1016/j.biopha.2018.05.141
Xu J, Yang B et al (2020) LncRNA BBOX1-AS1 upregulates HOXC6 expression through miR-361-3p and HuR to drive cervical cancer progression. Cell Prolif 53(7):e12823. https://doi.org/10.1111/cpr.12823
Xu L, Long H et al (2021) CircMKLN1 suppresses the progression of human retinoblastoma by modulation of miR-425-5p/PDCD4 axis. Curr Eye Res 46(11):1751–1761. https://doi.org/10.1080/02713683.2021.1927110
Xu T, Wu J et al (2017) Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics 18(Suppl 6):680. https://doi.org/10.1186/s12864-017-4029-3
Xu Y, Yao Y et al (2018) Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun 496(2):455–461. https://doi.org/10.1016/j.bbrc.2018.01.077
Yang R, Wu Y, et al (2015) HDAC9 promotes glioblastoma growth via TAZ-mediated EGFR pathway activation. Oncotarget 6(10):7644–7656. https://doi.org/10.18632/oncotarget.3223
Yi YX, Liu YY et al (2019) Reconstruction and analysis of circRNA-miRNA-mRNA network in the pathology of cervical cancer. Oncol Rep 41(4):2209–2225. https://doi.org/10.3892/or.2019.7028
Yu B, Zhao J et al (2021) Circ_0000527 promotes retinoblastoma progression through modulating miR-98-5p/XIAP pathway. Curr Eye Res 46(9):1414–1423. https://doi.org/10.1080/02713683.2021.1891255
Zhang C, Ding R et al (2021a) Circular RNA in tumor metastasis. Mol Ther Nucleic Acids 23:1243–1257. https://doi.org/10.1016/j.omtn.2021.01.032
Zhang H, Qiu X et al (2022a) CircCUL2 suppresses retinoblastoma cells by regulating miR-214-5p/E2F2 axis. Anticancer Drugs 33(1):e218–e227. https://doi.org/10.1097/cad.0000000000001190
Zhang L, Wu J et al (2020) Circ_0000527 promotes the progression of retinoblastoma by regulating miR-646/LRP6 axis. Cancer Cell Int 20:301. https://doi.org/10.1186/s12935-020-01396-4
Zhang W, Wu G et al (2021b) circ_SMAD2 regulate colorectal cancer cells proliferation through targeting miR-1258/RPN2 signaling pathway. J Cancer 12(6):1678–1686. https://doi.org/10.7150/jca.50888
Zhang X, Qiu S et al (2018) Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomark 22(1):135–142. https://doi.org/10.3233/cbm-171109
Zhang Y, Dou X et al (2022b) Circ_0075804 promotes the malignant behaviors of retinoblastoma cells by binding to miR-138-5p to induce PEG10 expression. Int Ophthalmol 42(2):509–523. https://doi.org/10.1007/s10792-021-02067-7
Zhang Y, Jia DD et al (2021c) The emerging function and clinical significance of circRNAs in thyroid cancer and autoimmune thyroid diseases. Int J Biol Sci 17(7):1731–1741. https://doi.org/10.7150/ijbs.55381
Zhao W, Wang S et al (2020) Circular RNA (circ-0075804) promotes the proliferation of retinoblastoma via combining heterogeneous nuclear ribonucleoprotein K (HNRNPK) to improve the stability of E2F transcription factor 3 E2F3. J Cell Biochem 121(7):3516–3525. https://doi.org/10.1002/jcb.29631
Zheng T, Chen W et al (2021) Circular RNA circ-FAM158A promotes retinoblastoma progression by regulating miR-138-5p/SLC7A5 axis. Exp Eye Res 211:108650. https://doi.org/10.1016/j.exer.2021.108650
Zhong C, Chen Y, et al (2018) LIM and SH3 protein 1 regulates cell growth and chemosensitivity of human glioblastoma via the PI3K/AKT pathway. Bmc Cancer 18. https://doi.org/10.1186/s12885-018-4649-2
Zhou XB, Marks PA et al (2001) Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci USA 98(19):10572–10577. https://doi.org/10.1073/pnas.191375098
Zuo X, Fu C, et al (2021) Hsa_circ_0000527 Downregulation suppresses the development of retinoblastoma by modulating the miR-27a-3p/HDAC9 pathway. Curr Eye Res 1-12. https://doi.org/10.1080/02713683.2021.1925697
Acknowledgements
Reviewers are thanked for their suggestions.
Funding
This research was funded by National Natural Science Foundation of China [81500763]; Shandong Provincial Natural Science Foundation, China [ZR2020MC059]; China Postdoctoral Science Foundation [2019M652311]; special support for post-doc creative funding in Shandong province; and Applied Research Program for Post-Doctoral in Qingdao.
Author information
Authors and Affiliations
Contributions
X.D.H. designed the research; F.L. wrote the manuscript; K.Y.Y., P.H.G., and T.J.Z. organized the material; X.D.H. and F.L. revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, F., Yin, YK., Zhang, JT. et al. Role of circular RNAs in retinoblastoma. Funct Integr Genomics 23, 13 (2023). https://doi.org/10.1007/s10142-022-00942-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-022-00942-9