Abstract
Maintenance of growth is important for sustaining yield under stress conditions. Hence, identification of genes involved in cell division and growth under abiotic stress is utmost important. Ras-related nuclear protein (Ran) is a small GTPase required for nucleocytoplasmic transport, mitotic progression, and nuclear envelope assembly in plants. In the present study, two Ran GTPase genes TaRAN1 and TaRAN2 were identified though genome-wide analysis in wheat (T. aestivum). Comparative analysis of Ran GTPases from wheat, barley, rice, maize, sorghum, and Arabidopsis revealed similar gene structure within phylogenetic clades and highly conserved protein structure. Expression analysis from expVIP platform showed ubiquitous expression of TaRAN genes across tissues and developmental stages. Under biotic and abiotic stresses, TaRAN1 expression was largely unaltered, while TaRAN2 showed stress specific response. In qRT-PCR analysis, TaRAN1 showed significantly higher expression as compared to TaRAN2 in shoot and root at seedling, vegetative, and reproductive stages. During progressive drought stress, TaRAN1 and TaRAN2 expression increase during early stress and restored to control level expression at higher stress levels in shoot. The steady-state level of transcripts was maintained to that of control in roots under drought stress. Under cold stress, expression of both the TaRAN genes decreased significantly at 3 h and became similar to control at 6 h in shoots, while salt stress significantly reduced the expression of TaRAN genes in shoots. The analysis suggests differential regulation of TaRAN genes under developmental stages and abiotic stresses. Delineating the molecular functions of Ran GTPases will help unravel the mechanism of stress induced growth inhibition in wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ran (Ras-related nuclear protein) belongs to RAS super family of small monomeric GTPases of 20 to 40 kDa. These are small molecular switches that are activated by GTP and inactivated by hydrolysis of GTP to GDP. Small G proteins constitute a large super family present in humans, and from yeast to plants. Based on structure and function, GTPase super family has been classified into five families: Ras, Rab, Rho, Arf, and Ran (Kahn et al. 1992). Structurally, all the members of RAS super family are characterized by the presence of conserved G domain responsible for nucleotide binding (GTP or GDP) and GTP hydrolysis. The switch between GTP bound to GDP bound state of G protein during GTP hydrolysis is regulated by two classes of proteins namely guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). GEFs catalyze the formation of the GTP-bound active form, while GAPs are required for activating the intrinsically weak GTP hydrolytic activity of the small GTP-binding proteins, thereby converting these into inactive GDP bound GTPase. The variable residues outside the conserved G domain as well as the conformational changes acquired by GTP and GDP bound form are recognized by different effector proteins which contribute to diverse signaling, cellular, and physiological processes (Takai et al. 2001).
The small GTPase super family from Arabidopsis and rice has 93 and 111 members respectively. Analyses reveal that these proteins belong to four families’ viz. Rab, Arf, Rho, and Ran. The true Ras family is absent in Arabidopsis and rice. However, they have ROPs (Rho-related GTPase from plants) which is unique subfamily of Rho-family GTPases in plants (Vernoud et al. 2003; Jiang and Ramachandran 2006). ROP GTPases have multifunctional roles in cellular pathways controlling plant growth, differentiation, and development and play an important role in signaling pathways related to hormone and biotic-abiotic stress response (Molendijk et al. 2004, Nagawa et al. 2012). Rab and Arf GTPases are regulators of vesicle trafficking. These play an important role in cell polarity and polarized growth and thus contribute to root and pollen growth, auxin transport, cell wall synthesis, etc. (Nibau et al. 2006; Lycett 2008). Vesicle trafficking is a common function for ROP, Rab, and Arf (Molendijk et al. 2004), while Ran is exclusively involved in nucleocytoplasmic transport, microtubular mitotic assembly, cell cycle control, and formation of nuclear envelope (NE) in plants (Moore 1998; Wang et al. 2006; Zhao et al. 2006; Ma et al. 2007). Ran is also unique as it is the only GTPase family protein with nuclear localization and shuttles between nucleus and cytoplasm (Moore 1998; Xu et al. 2016). Rab GTPases are the largest family with 57 and 47 members in Arabidopsis and rice respectively, followed by Arf and ROP, while Ran is the smallest family with 4 members (Vernoud et al. 2003; Jiang and Ramachandran 2006). Furthermore, Rab, Arf, and ROP protein are further classified into subclasses and exhibit sequence and functional divergence, while Ran proteins show high conservation (Jiang and Ramachandran 2006). Overexpression studies with wheat and rice RAN1 revealed that these GTPases are involved in mitotic process in shoot and root apical meristems and these are regulated by auxin (Wang et al. 2006; Xu and Cai 2014). Plants overexpressing OsRAN1, OsRAN2, and AtRAN1 were found to be low temperature tolerant (Chen et al. 2011; Xu and Cai 2014; Xu et al. 2016). However, overexpression of OsRAN2 in Arabidopsis and rice conferred hypersensitivity to salt and osmotic stress (Zang et al. 2010). Thus, Ran proteins regulate plant growth and development through their cellular processes and may contribute differentially to abiotic stress response in plants. Abiotic stresses affect growth and development of plants and hence severely affect the yield of the crop plants. Maintenance of growth is an important aspect for sustaining yield under stress condition. Therefore, identification of genes involved in cell division and growth and analysis of their response under abiotic stress becomes utmost important.
Wheat is the second most important food crop. Sustaining yield under abiotic stress in wheat is necessary for global food security. The availability of the fully annotated genome of wheat has paved the way for dissecting this complex hexaploid genome and identification and characterization of genes for crop improvement (IWGSC 2018). In this study, Ran GTPase family genes were identified in wheat through in silico genome-wide analysis of wheat genome. A phylogenetic and comparative analysis of wheat Ran GTPases in terms of gene structure and domain with that of Arabidopsis and cereals such as rice, barley, maize, and sorghum was also performed. Expression analysis of TaRAN genes during growth and development and to environmental stresses was carried out using publicly available datasets. The transcriptional regulation of wheat Ran genes was further analyzed under progressive moisture deficit stress, cold and salt stress by qRT-PCR. The results contribute to knowledge on Ran GTPases from wheat and other cereals and lay basis for detailed functional analysis of RAN genes in wheat.
Materials and methods
Identification of RAN family genes and phylogenetic analysis
For identification of RAN family genes in wheat, protein sequences of Arabidopsis RAN genes namely AtRAN1, AtRAN2, AtRAN3, and AtRAN4 were downloaded from TAIR database (https://www.arabidopsis.org) and used as query against wheat (T. aestivum, cv. Chinese spring) reference genome sequence in the Ensembl Plants database (http://plants.ensembl.org/index.html; IWGSC 2018). A profile HMM search was also performed using conserved domains of Arabidopsis Ran family against wheat genome database to identify the RAN homeologs (Finn et al. 2015). RAN genes were identified in Rice (Oryza sativa subsp. Japonica cv. Nipponbare) using MSU Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). RAN family genes from maize cv. B73, sorghum cv. BTx623, and barley cv. Morex were identified using Ensembl Plants database (http://plants.ensembl.org/index.html; B73_RefGen_v4; Sorghum_bicolor_NCBIv3, Hordeum vulgare_ IBSC_v2). These sequences were analyzed for Ran specific domain using SMART tool (Letunic et al. 2015). One protein sequence each from wheat and rice was corrected with FGENESH+ using already published sequence of OsRan1. Isoelectric point (pI) of RAN proteins was computed using ExPASy tool (http://web.expasy.org/compute_pi/). Sub-cellular localization prediction was performed using CELLOv2.5 (Yu et al. 2006). Multiple sequence alignment of Ran protein sequences was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Phylogenetic tree was constructed by MEGA version X (Kumar et al. 2018) using neighbor-joining tree method with pair wise deletion and Poisson correction with a bootstrap analysis of 1000 replicates. Exon/Intron organization of RAN family genes were generated using Gene Structure Display Server (Hu et al. 2015).
Cis-acting regulatory element
The promoter sequence comprising 2000 bp upstream to the translation start site (ATG) were downloaded from the wheat genome database (http://plants.ensembl.org/index.html, IWGSC 2018). The cis-acting regulatory motifs in the promoter regions were predicted using PLACE database (https://www.dna.affrc.go.jp/PLACE/?action=newplace).
Digital expression analysis
Tissue, developmental, biotic, and abiotic stress-specific expression patterns of the identified TaRAN genes were analyzed using RNAseq datasets in expVIP platform (Borrill et al. 2016).
Plant material and abiotic stress treatments
Wheat (T. aestivum) cv. HD2329 was used for all the experiments. For drought treatment, seeds were germinated on the moist filter paper. After 48 h of germination, uniformly germinated seeds were planted in the 6-inch pots filled with soil. Plants were grown for 21 days during which the plants were watered optimally. After 21 days, drought stress was imposed by with-holding water. The entire experiment was conducted during natural wheat growing conditions in net-house. The root and shoot samples were collected from stressed and non-stressed (control) plants at 5th, 9th, and 11th day of drought stress imposition. The level of drought stress was estimated by measuring the leaf relative water content (RWC) (Barrs and Weatherley 1962) and soil moisture content (SMC) (Gravimetric method). The plants watered optimally (RWC > 90%; SMC ≥ 20%) served as control. For cold and salt stress treatments, the wheat seedlings were grown in hydroponics supplemented with Hoagland solution at 22 °C under 16 h/8 h light/dark conditions. The 7-day-old seedlings were subjected to salt stress by adding salt (final concentration of NaCl 150 mM) to Hoagland solution. For cold stress, seedlings were kept at 4 °C ± 1 °C. The root and shoot samples were collected at 3 h and 6 h post treatments. Seedlings grown in Hoagland solution at 22 °C served as control.
For developmental stage study, root and shoot tissues of uniformly germinated 7-day-old seedlings (seedling stage) and 25-day-old plants (Vegetative stage) were collected. At full spike emergence stage (reproductive stage), samples for spike, flag leaf, penultimate leaf, and roots were collected, frozen in liquid nitrogen, and stored at -80 °C till further use.
Expression analysis by qRT-PCR
RNA isolation was carried out using TRIzol reagent (Invitrogen) according to manufacturer’s instructions. RNA was treated with ezDNase™ Enzyme (Invitrogen) to remove residual DNA. First strand synthesis was carried out using 2 μg of RNA using first strand cDNA synthesis kit (Invitrogen) according to kit protocol. Quantitative RT-PCR analysis was carried out by using the Realplex4 system (Eppendorf) using KAPA-SYBR® FASTqPCR mix (KAPA Biosystems). The 10 μl master mix contained 1x KAPA SYBR®FAST qPCR Master Mix, 1 μl (1:10 diluted) of cDNA, and 0.4 μM each of forward and reverse primers (online resource 1; Table S1). A two-step qRT-PCR protocol with initial denaturation of 95 °C for 3 min, followed by 40 cycles of denaturation and annealing/extension at 95 °C for 3 sec and 60 °C for 45 sec, was carried out. Expression data were normalized using endogenous control gene, ADPR (ADP-ribosylation factor) expression (Paolacci et al. 2009). Relative fold change was calculated using 2-ΔΔCt method (Livak and Schmittgen 2001). Each data point represents average of three biological and three technical replicates. Graphs were prepared in GraphPad prism 5.0. Statistical significance between mean values was calculated using student’s T test.
Results and discussion
Identification of Ran GTPases and comparative analysis
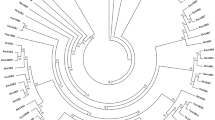
The genome-wide search for RAN genes led to identification of 5 homologs in wheat genome. Domain analysis revealed that five of the Ran GTPases were characterized by RAS (PF00071) and Ran domain (SM00176). For phylogenetic analysis, Ran GTPase family genes were also identified in rice, barley, maize, and sorghum genomes. The analysis revealed that rice and barley genome encode 2 and 3 RAN genes respectively, while the number of RAN genes encoded by sorghum and maize genomes was 5 each. All these Ran family members were also confirmed by SMART domain analysis. One gene each from sorghum (SORBI_3010G182500) and maize (Zm00001d011328) were truncated and lacked three G motifs, respectively, and hence were not included in further analysis. In rice, previously 4 RAN genes have been reported (Jiang and Ramachandran 2006). However, our genome-wide analysis and sequence analysis revealed that of the two genes reported on chromosome 1 and chromosome 4, AP002844 matched with the two RAN genes, LOC_Os5g49890 and LOC_Os06g39875, identified in rice genome, while AL606650 (BAC clone) sequence did not code for any RAN gene. Phylogenetic analysis of wheat Ran GTPases with that of Arabidopsis, barley, rice, maize, and sorghum revealed three major clades (Fig. 1). Clade I was further divided in to three sub-clades. Sub-clade I included LOC_Os5g49890 (OsRan2), HORVu1Hr1G090760 (HvRan1), and three wheat homeologs on chromosome 1 of genome A, B and D, thereby represented a homeolog-triad. These wheat homeologs on chromosome 1 showed 100% identity with already characterized Ran GTPase named as TaRAN1 (Wang et al. 2006). Hence, for consistency, these clade I members were named as TaRAN1. The sub genomes were represented by suffix A, B, or D viz. TaRan1A, TaRan1B, and TaRan1D. Sub-clade II and sub-clade III had one gene from sorghum and two genes from maize in each, while SORBI_3003G217800 was ungrouped member in clade I. Clade II included two sub-clades where sub-clade I had HORVu1Hr1G84420 and two wheat homeologs on chromosome 7B and 7D (homeolog-diad) and thereby named as TaRan2 (TaRan2B and TaRan2D). Second sub-clade grouped one gene each from sorghum and rice, while AtRan4 (At5G55080) remained as ungrouped member. Clade III was constituted by AtRan1, AtRan2, AtRAN3, and HORVu1Hr1G30130 (Fig. 1). In spite of having large and polyploid genome, the number of RAN genes in wheat is lower than that of Arabidopsis, barley, maize, and sorghum though it has similar number to that of rice. However, existence of homeologs in wheat might compensate for fewer genes. Furthermore, two homeologs of TaRAN1 and one homeolog of TaRAN2 have been predicted to code for one alternate splice (AS) forms each which might add to the functional diversity of the Ran GTPases in wheat. Likewise, AS forms have been predicted for some of the RAN genes in sorghum, rice, barley, and maize, whereas Arabidopsis RAN genes do not have any alternate splice forms (Table 1). The pI (iso-electric point) of majority of proteins was within 6.38 to 6.65, except SbRan4 (pI 7.03), OsRan1 (7.61), AtRan4 (pI of 8.16), and HvRan3 (pI 8. 48) (Table 1). Sub-cellular localization prediction revealed that all the Ran GTPases from Arabidopsis and five of the cereals were distributed between cytoplasm and nucleus (Table 1). This is in accordance with cyclic movement of Ran GTPase in active and inactive form between nucleus and cytoplasm respectively (Moore 1998; Vernoud et al. 2003).
Gene structure and motif analysis
Exon-intron structure is an important feature to study gene evolution and function. For gene structure analysis, primary or full length transcripts representing each gene were considered. The genes in clade I had 7 exons each (Fig. 2), while genes in clade II had 6 (sub-clade I) and 7 exons (sub-clade II). This shows conservation in primary gene structure except HORVu1Hr1G30130 which had no introns. Genes in clade I mostly showed variation in length of 1st intron, while in clade II, first introns were shorter and 2nd intron length varied among genes. AtRAN1 and AtRAN2 genes had 4 exons each, while AtRAN3 and AtRAN4 had 7 and 5 exons respectively.
The protein sequences were further analyzed for presence of conserved motifs in the G domain. The G domain is conserved across RAS superfamily. It consists of five motifs (G boxes) namely G1(GxxxxGK[S/T] (where x is any amino acid), G2 (XTX), G3 (DXXG), G4 (N/T)(K/Q)xD, and G5 ((T/G/C)(C/S)AK) (Yang 2002; Toma-Fukai and Shimizu 2019). Among these, G1 motif, also called as the P-loop, binds to β-phosphates and Mg2+ ion. The conserved threonine (T) residue of G2 contacts γ-phosphate and Mg2+ ion. G3 motif is involved in hydrolysis. The G4 and G5 recognize guanine base and confer nucleotide specificity (Toma-Fukai and Shimizu 2019). The protein sequence alignment of the primary transcripts from Arabidopsis, wheat, rice, barley, maize, and sorghum revealed conserved G boxes (Fig. 3, online resource 1; Fig. S1). All the proteins except AtRan4 (AT5G55080) contained an acidic tail at C terminal which is unique to Ran GTPases and highly conserved across species. Absence of acidic tails in AtRan4 has been reported earlier, and it was suggested that AtRan4 might have different function (Vernoud et al. 2003). Interestingly, sequence analysis of splice forms of wheat showed presence of at least one alternate splice form of TaRAN2B (TaRan2B.1;TraesCS7B02G254000.1) which had complete G domain (G1 to G5 motifs) but lacked acidic tail (online resource 1, Fig. S2a). Therefore, the analysis was extended to other cereals, and it was found that there was at least one splice form in rice (OsRan2.2; LOC_Os05g49890.2) and sorghum (SbRan2.1; SORBI_3003G238000.1) which also had conserved G domain motifs but lacked acidic tail (online resource 1, Fig. S2b,c). There was no splice form in maize and barley with complete G domain sans acidic tail (online resources 1, Fig. S2d & e). Acidic tail is required for interaction of Ran with Ran-binding proteins (RanBP1) (Haizel et al. 1997). RanBPs are localized in cytoplasm and stimulates the GTPase activity of Ran and promote the formation of Ran-GDP (Clarke and Zhang 2008). Deletion of C-terminal domain causes cytoplasmic retention of Ran and inhibits the nuclear import of proteins (Lounsbury et al. 1996). Furthermore, effector binding domain (KKYEPTIGVEV) which is required for interaction with Ran GTPase activating protein (RanGAP) was found conserved in all the full length proteins except in AtRan4 (Fig. 3, online resource 1, Fig. S2). AtRan4 had partially conserved effector binding domain (Fig. 3; Vernoud et al. 2003). Both RanGAP and RanBP stimulate the conversion of RanGTP to RanGDP. RanBP acts as co-activator and on binding to Ran increases its affinity for RanGAP by 10-fold (Moore 1998). This suggests that absence of acidic tail and hence RanBP binding/stimulation might slow down the conversion RanGTP to RanGDP. Therefore, it seems plausible that these proteins without acidic tail from wheat, rice, and sorghum might be having role in regulation of nucleotide state (RanGTP/RanGDP) which still remains to be analyzed.
Multiple sequence alignment of Ran GTPases from wheat, rice, and Arabidopsis showing conserved domain and motifs. Red boxes represent five G domain motifs. Highly conserved residues are highlighted in yellow, conserved residues in cyan; specificity determining position (SDPs) residues are shaded in grey. Blue box represents effector binding (RanGAP) motif and green box represents acidic tail
In addition to having conserved residues for GTPase activity, the G domain residues have sequence specific residues named as specificity determining positions (SDPs) (Rojas et al. 2012). These SDPs are important as these differentiate between RAS subfamilies. Furthermore, GTP/GDP-based conformational changes in switch I and switch II of G domain along with the SDPs outside the conserved G motifs govern the effector or protein-protein interaction and biological function of the RAS subfamilies. The SDPs (residues in gray shade Fig. 3) were found to be highly conserved among the full length Ran GTPases from wheat, barley, rice, sorghum, maize, and Arabidopsis except AtRan4 which showed V30L (G1) and K130P (G4) change respectively (Fig. 3, online resource 1, Fig. S2).
Cis- regulatory elements in TaRAN promoters
For in silico prediction of conserved cis-regulatory elements (CREs) in promoters of the TaRAN genes, 2 kb region upstream of translation start site of both TaRAN1and TaRAN2 and their homeologs was carried out. In addition to core elements such as TATA box and CAAT box, the promoters revealed presence of many diverse cis-regulatory elements. These cis-elements were grouped into seven groups viz. tissue and development specific expression, nutrient response, phytohormone regulated, biotic and abiotic stress response, enhancer/silencing elements, and miscellaneous elements (Fig. 4, online resource 2, Table S2). Tissue-specific elements such as seed/endosperm/embryo-specific, mesophyll, and pollen-specific cis-elements were overrepresented in all the promoters followed by root-specific and guard cell-specific elements (Fig. 4a). Elements related to cell cycle (E2F binding & MYB binding site for CYCB1) and proliferation were also present (1 to 4 times) in all the TaRAN promoters. Element for circadian expression was unique to TaRAN1 (B and D). Owing to their role in cell division and mitosis, presence of CREs specific for cell cycle and meristem specific transacting factors in RAN promoters is quite evident. Furthermore, there was a high presentation of sugar responsive elements in all the TaRAN promoters. Sugars are source of carbon and energy and also play an important role in signaling and especially cell cycle regulations (Wang and Ruan 2013). Therefore, presence of at least nine diverse sugar response elements suggests an important role of sugar signaling or sugar mediated regulation of TaRAN genes (Fig. 4b, online resource 2, Table S2). Interestingly, transgenic rice overexpressing OsRAN1 were found to have high sugar levels and high mitotic index as compared to wild-type rice and also showed cold tolerance (Xu and Cai 2014). Among phytohormone regulated elements, GA response and ARRIAT1 (cytokinin response) followed by CGCGBOXAT (ethylene, ABA and light response) element were highly represented. Auxin and salicylic acid regulated ASF1MOTIFCAMV elements was present in all the promoters (Fig. 4c, online resource 2, Table S2). Among abiotic stress response elements, low temperature responsive elements and dehydration responsive elements followed by etiolation inducible elements showed high frequency. Biotic response elements, i.e., elicitor, wound, and diseases’ response elements, were present in moderate number except GT1GMSCAM4 element (pathogen and NaCl response) which was present 8 times in TaRAN1B promoter (Fig. 4d & e, online resource 2, Table S2).
Cis-regulatory elements in promoter region of TaRAN genes. The in silico prediction of cis-elements was carried out using PLACE database. The number of cis-elements is represented by numerals as well as shades in the boxes with white as 0 and darkest shade represents highest number. a Tissue and development specific elements, b nutrient and metabolite specific elements, c phytohormone regulated elements, d,e abiotic and biotic stress responsive elements, and f expression modulators elements
Other CREs which were conspicuously present in these promoters were expression modulator elements such as enhancer elements (EE-1, EE-2, SV40 enhancer); silencing elements, i.e. S1F and silencing element binding factor (SEBF); and high expression and constitutive expression elements (Fig. 4f, online resource 2, Table S2). There were several other motifs which were grouped under miscellaneous which included binding site for AGL15 (AGAMOUS-Like 15), GBF4 (G-box binding factor 4), VOZ- and maize P binding site, etc. (online resource 2, Table S2). Thus, presence of such diverse elements in promoter region suggests the ubiquitous expression and function of TaRAN genes under normal growth and development as well as biotic and abiotic stress response of wheat.
Digital expression analysis
To analyze the tissue, developmental and stress-specific expression pattern of the TaRAN genes RNAseq data available in the expVIP database was explored. The expression was analyzed in different tissues at seedling, vegetative, and reproductive stages. All the three homeologs of TaRAN1were ubiquitously and highly expressed during all the developmental stages (Fig. 5a). The highest expression of TaRAN1A with fold change of ≥ 9 was observed in shoot and root apical meristems during both seedling and vegetative stage. The high expression of RAN1 in meristematic tissues corroborates with its role in mitotic processes in plants and animals (Ren et al. 1993; Quimby and Dasso 2003; Wang et al. 2006). Transgenic Arabidopsis overexpressing TaRAN1 showed a high number of new organ primordia around the shoot apical point which is primarily driven by higher cell division. The transgenic plants overexpressing OsRAN1 and TaRAN1 also showed higher number of cells in root meristem as compared to wild type (Wang et al. 2006; Xu and Cai 2014).
Expression analysis of TaRAN genes in a different tissues and developmental stages, and under b abiotic and c biotic stresses in wheat. Expression analysis was performed using wheat RNAseq datasets from expVIP database. Numbers in the boxes show expression of TaRAN genes based on log transformed TPM (transcripts per million) value. The color scale of heatmap shows the level of gene expression, red color; high level expression bright yellow; low level gene expression
Among reproductive tissues, highest expression of TaRAN1A was reported in microspores (fold change 8.9) followed by embryo, seed coat, and pistil. Similar expression pattern was observed for TaRAN1B and TaRAN1C except in microspores where instead of full length transcripts of TaRAN1B, splice form TaRAN1B.2 was expressed (Fig. 5a). The splice forms TaRAN1B.2 and TaRAN1D.1 were also expressed in most of the tissues albeit at very low and variable levels (online resource 1, Fig. S3a).
Full length transcripts of TaRAN2 homeologs, TaRAN2B and TaRAN2D, were also ubiquitously expressed in all the tissues and developmental stages except in embryo (Fig. 5a). Higher expression (5.5- to 7.1-fold) was recorded in flag leaf, endosperm, seed coat, and microspore. The splice form TaRAN2B.1 expressed in almost all the tissues except in embryo and microspores. The expression of both TaRAN1 and TaRAN2 was observed across all the tissues and developmental stages. However, expression levels of TaRAN1 homeologs were significantly higher compared to TaRAN2. Higher level expression of OsRAN1 has been reported in rice panicles (Xu and Cai 2014). Silencing of OsRAN2 in rice by RNAi led to pleiotropic developmental abnormalities including abnormal inflorescence, fewer and less viable pollen grains, and male sterile plants (Zang et al. 2010). This suggests that Ran GTPases may have an important role in development of reproductive organs in plants.
The expression of TaRAN transcripts was analyzed under different abiotic stresses such as drought, heat, drought and heat combined, PEG, and phosphorus starvation. The relative change in transcripts of TaRAN1 remained largely unaffected under all the abiotic stresses as evident from the expression levels at 1 h and 6 h of abiotic stresses (Fig. 5b). TaRAN2 homeologs were differentially regulated by abiotic stresses. Under drought stress, TaRAN2 transcripts were moderately upregulated at 6 h compared to 1 h, and under cold stress, also TaRAN2 transcript levels were higher in vegetative shoots after 2 weeks and in microspores (reproductive tissue). However, there was no significant change in TaRAN2 transcripts under heat, combined drought and heat, and osmotic stress (Fig. 5b, online resource 1, Fig. S3b).
There was no significant effect of duration of disease (powdery mildew, stripe rust) and pathogen (Fusarium pseudograminearum, Magnaporthe oryzae) on the expression of expression of TaRAN1 homeologs. In contrast, TaRAN2 showed increase in transcript levels from 2.1- (24 h) to 3.8-fold (72 h) under powdery mildew disease. For stripe rust pathogen (CYR 31), TaRAN2 expression decreased after 72 h but with stripe rust pathogen 87/66; TaRAN2B and TaRAN2D expression increased from 2.2- and 3-fold on the 1st day to 4- and 4.9-fold on the 11th day respectively (Fig. 5c, online resource 1, Fig. S3c). The differential response of TaRAN1 and TaRAN2 to different stresses and diseases suggests that these genes may have distinct role under these conditions.
Expression analysis of TaRAN genes during developmental stages and abiotic stress conditions by qRT-PCR
The expression of TaRAN genes was analyzed in shoot and root at three developmental stages, viz., seedling, vegetative, and reproductive stages. TaRAN1 expression levels were significantly higher as compared to TaRAN2 which showed very low level expression in both shoot and root at all these stages (Fig. 6a,b). At the seedling stage, there was a similar level of TaRAN1 expression in shoot (8 fold) and root (9.75 fold), while at the vegetative stage, expression of TaRAN1was significantly higher in shoots (24-fold) compared to roots (3.5-fold). This suggests that TaRAN1 is differentially regulated in shoot and root at vegetative stage. Overexpression of TaRAN1 and AtRAN1 in Arabidopsis has been shown to increase rosette leaves and vegetative growth of plants (Wang et al. 2006; Xu et al. 2016). Rice transgenic plants overexpressing TaRAN1 and OsRAN1 showed higher tiller number and delayed flowering as compared to wild-type plants (Wang et al. 2006; Xu and Cai 2014). It was suggested that RAN genes help in new organ primordial formation which subsequently leads to higher rosette and tiller number. Interestingly, the roots of these RAN overexpressing transgenics were shorter and lateral root number was also reduced. Thus RAN may have different roles in root and shoot development. At reproductive stage, expression levels were analyzed in leaf (penultimate to flag leaf), root, flag leaf, and spike. Among these, flag leaf and spike showed highest expression (11.58-fold and 9.64-fold) of TaRAN1 followed by root (5.8-fold) and shoot (3.8-fold) (Fig. 6c). The expression of TaRAN2 remained significantly low in all these tissues (Fig. 6c). Analysis of expVIP data also showed higher expression of TaRAN1 genes in reproductive organs especially pistil and microspores (Fig. 5a). Similar expression pattern was reported for OsRAN1 which showed highest expression in spike followed by sheath root and leaf (Xu and Cai 2014). AtRAN1 expressions were also found to be relatively higher in siliques and cauline leaf (Xu et al. 2016). The down- or upregulation of RAN gene expression has been associated with male sterility (Wu et al. 2007; Zang et al. 2010). This suggests that RAN genes play vital role in male reproductive development in plants.
Expression profile of TaRAN1 and TaRAN2 at different developmental stages. Expression of TaRAN1 and TaRAN2 in shoot and root tissues at a seedling (7-day-old seedlings); b vegetative (25-day-old plants), and c reproductive stage (full spike emergence) in shoot, root, flag leaf, and spike of wheat (Triticum aestivum). Error bars represent the SE of three biological replicates. Asterisks indicates significant difference between expression of TaRAN1 and TaRAN2 (*p < 0.05, **p < 0.001, ***p < 0.0001) by student’s T test
The response of TaRAN genes was analyzed under progressive drought stress. For this, 21-day-old wheat seedlings were subjected to moisture deficit stress, and shoot and root samples were collected at 5, 9, and 11 days of stress imposition. The relative water content (RWC) of stressed plants at these time points was 94.2%, 72.6%, and 54.0%, and soil moisture content (SMC) was 15.15%, 7.54%, and 6.82% respectively (online resource 1 Fig. S4). The RWC and SMC of the non-stressed (control) plants were in the range of 91.6–94.3% and 19.18–22.12% respectively (online resource 1, Fig. S4). There was a 1.5-fold (p ˂ 0.05) increase in expression of TaRAN1 and TaRAN2 in shoot tissues after 5 days of drought stress. TaRAN1 expression decreased significantly after the 9th day, but after 11 days, expression was similar to that of control (Fig. 7a). TaRAN2 expression remained similar to that of control after 9 and 11 days of drought stress (Fig. 7a). There was no significant change in expression levels of TaRAN1 and TaRAN2 in roots under drought stress except that TaRAN2 showed upregulation (p ˂ 0.05) on 9 days after drought stress (Fig. 7b). Similar expression profile was recorded for both the genes under drought stress in expVIP expression analysis (Fig. 5b). Thus, under higher moisture deficit, the steady-state expression levels of TaRAN genes were largely maintained to that of control plants in both shoot and root. TaRAN1 and TaRAN2 showed a significant (p ˂ 0.05) decrease in shoots after 3 h of cold stress (Fig. 7c). TaRAN1 expression became similar to control after 6 h, while TaRAN2 increased up to 1.3-fold (p ˂ 0.05) in shoots. In roots, both TaRAN1 and TaRAN2 were significantly downregulated after 3 h and 6 h of cold stress (Fig. 7d). It was shown that OsRAN1 and OsRAN2 were most responsive to cold stress (Chen et al. 2011; Xu and Cai 2014). AtRAN1 showed an increase in expression after 12 h of cold acclimation (Xu et al. 2016).
Expression of TaRAN1 and TaRAN2 in shoot and root tissues under abiotic stresses. a,b Drought stress, 21-day-old plants were subjected to moisture deficit stress by with-holding water. Optimally watered (non-stressed) plants served as control. Relative expression was analyzed at 5th day, 9th day, and 11th day of drought stress. c,d Cold stress (4 °C) and e,f salt stress (150 mm); 7-day-old seedlings were subjected to cold and salt stress and relative expression was analyzed at 3 h and 6 h post stress imposition. Seedlings grown in Hoagland solution at 22 °C served as control. Error bars represent the SE of three biological replicates. Asterisks indicates significant difference between expression of TaRAN1 and TaRAN2 (*p < 0.05, **p < 0.001) by student’s T test
Salt stress significantly reduced the expression of TaRAN1 and TaRAN2 in shoots (Fig. 7e). In roots after initial decrease at 3 h, both the TaRAN gene showed expression similar to that of control (Fig. 7f). AtRAN1 showed a 4-fold increase under salt stress, whereas OsRAN1and OsRAN2 expression remained unaffected or downregulated by salt stress (Zang et al. 2010; Xu and Cai 2014). The response of RAN genes to salt stress appears to be common in rice and wheat. The effect of drought stress on Arabidopsis and rice RAN genes has not been analyzed. However, osmotic stress (PEG) has been shown to reduce the expression of OsRAN1 and OsRAN2 genes (Zang et al. 2010; Chen et al. 2011; Xu and Cai 2014). AtRAN and OsRAN also respond differently to exogenous ABA treatment. AtRAN1 and AtRAN3 showed increase in expression, while OsRAN2 showed a decrease in expression on treatment with ABA (Zang et al. 2010; Xu et al. 2016). This suggests that in spite of being highly conserved in structure and function, Ran GTPases may play differential role in stress response in different species.
In conclusion, genome-wide analysis of Ran GTPases in wheat genome led to identification of two RAN genes. The comparative analysis of wheat Ran GTPases with that of barley, rice, maize and sorghum reveal that these are highly conserved proteins. However, their expression under abiotic stresses may be species specific. In wheat, after initial upregulation and downregulation in drought and cold stress, respectively, expression of both the RAN genes is restored to control levels, while in salt stress, expression of these genes is significantly downregulated in shoot. The analysis suggests differential regulation of TaRAN genes under developmental stages and abiotic stresses. As abiotic stresses inhibit cell division and growth, delineating molecular functions of Ran GTPases will help unravel the mechanism of stress induced growth inhibition in wheat.
References
Barrs HD, Weatherley PE (1962) A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci 15(3):413–428. https://doi.org/10.1071/BI9620413
Borrill P, Ramirez-Gonzalez R, Uauy C (2016) expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol 170:2172–2186. https://doi.org/10.1104/pp.15.01667
Chen N, Xu Y, Wang X, Du C, Du J, Yuan M, Xu Z, Chong K (2011) OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ 34:52–64. https://doi.org/10.1111/j.1365-3040.2010.02225.x
Clarke PR, Zhang C (2008) Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 9:464–477. https://doi.org/10.1038/nrm2410
Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR (2015) HMMER web server: 2015 update. Nucleic Acids Res 43:W30–W38. https://doi.org/10.1093/nar/gkv397
Haizel T, Merkle T, Pay A, Fejes E, Nagy F (1997) Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J 11:93–103. https://doi.org/10.1046/j.1365-313x.1997.11010093.x
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Jiang SY, Ramachandran S (2006) Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol Genomics 24:235–251. https://doi.org/10.1152/physiolgenomics.00210.2005
Kahn RA, Der CJ, Bokoch GM (1992) The ras superfamily of GTP-binding proteins: guidelines on nomenclature. FASEB J 6:2512–2513. https://doi.org/10.1096/fasebj.6.8.1592203
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:D257–D260. https://doi.org/10.1093/nar/gku949
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lounsbury KM, Richards SA, Carey KL, Macara IG (1996) Mutations within the Ran/TC4 GTPase. Effects on regulatory factor interactions and subcellular localization. J Biol Chem 271:32834–32841. https://doi.org/10.1074/jbc.271.51.32834
Lycett G (2008) The role of Rab GTPases in cell wall metabolism. J Exp Bot 59:4061–4074. https://doi.org/10.1093/jxb/ern255
Ma L, Hong Z, Zhang Z (2007) Perinuclear and nuclear envelope localizations of Arabidopsis Ran proteins. Plant Cell Rep 26:1373–1382. https://doi.org/10.1007/s00299-007-0367-y
Molendijk AJ, Ruperti B, Palme K (2004) Small GTPases in vesicle trafficking. Curr Opin Plant Biol 7:694–700. https://doi.org/10.1016/j.pbi.2004.09.014
Moore MS (1998) Ran and nuclear transport. J Biol Chem 273:22857–22860. https://doi.org/10.1074/jbc.273.36.22857
Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z (2012) ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol 10:e1001299. https://doi.org/10.1371/journal.pbio.1001299
Nibau C, Wu HM, Cheung AY (2006) RAC/ROP GTPases: 'hubs' for signal integration and diversification in plants. Trends Plant Sci 11:309–315. https://doi.org/10.1016/j.tplants.2006.04.003
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11. https://doi.org/10.1186/1471-2199-10-11
Quimby BB, Dasso M (2003) The small GTPase Ran: interpreting the signs. Curr Opin Cell Biol 15:338–344. https://doi.org/10.1016/s0955-0674(03)00046-2
Ren M, Drivas G, D'Eustachio P, Rush MG (1993) Ran/TC4: a small nuclear GTP-binding protein that regulates DNA synthesis. J Cell Biol 120:313–323. https://doi.org/10.1083/jcb.120.2.313
Rojas AM, Fuentes G, Rausell A, Valencia A (2012) The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol 196:189–201. https://doi.org/10.1083/jcb.201103008
Takai Y, Sasaki T, Matozaki T (2001) Small GTP-binding proteins. Physiol Rev 81:153–208. https://doi.org/10.1152/physrev.2001.81.1.153
Toma-Fukai S, Shimizu T (2019) Structural insights into the regulation mechanism of small GTPases by GEFs. Molecules 24(18):3308. https://doi.org/10.3390/molecules24183308
Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131:1191–1208. https://doi.org/10.1104/pp.013052
Wang L, Ruan YL (2013) Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci 4:163. https://doi.org/10.3389/fpls.2013.00163
Wang X, Xu Y, Han Y, Bao S, Du J, Yuan M, Xu Z, Chong K (2006) Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol 140:91–101. https://doi.org/10.1104/pp.105.071670
Wu JY, Shen JR, Mao XZ, Liu KD, Wei LP, Liu PW, Yang GS (2007) Isolation and analysis of differentially expressed genes in dominant genic male sterility (DGMS) Brassica napus L. using substractive PCR and cDNA microarray. Plant Sci 172:204–211
Xu P, Cai W (2014) RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J Exp Bot 65:3277–3287. https://doi.org/10.1093/jxb/eru178
Xu P, Zang A, Chen H, Cai W (2016) The small G protein AtRAN1 regulates vegetative growth and stress tolerance in Arabidopsis thaliana. PLoS One 11:e0154787. https://doi.org/10.1371/journal.pone.0154787
Yang Z (2002) Small GTPases: versatile signaling switches in plants. Plant Cell 14(Suppl):S375–S388. https://doi.org/10.1105/tpc.001065
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64:643–651. https://doi.org/10.1002/prot.21018
Zang AP, Xu XJ, Neill S, Cai WM (2010) Overexpression of OsRAN2 in rice and Arabidopsis renders trans-genic plants hypersensitive to salinity and osmotic stress. J Exp Bot 61:777–789. https://doi.org/10.1093/jxb/erp341
Zhao Q, Leung S, Corbett AH, Meier I (2006) Identification and characterization of the Arabidopsis orthologs of nuclear transport factor 2, the nuclear import factor of Ran. Plant Physiol 140:869–878. https://doi.org/10.1104/pp.105.075499
The International Wheat Genome Sequencing Consortium (IWGSC), Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, Poland J, Ronen G. et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361(6403):eaar7191. https://doi.org/10.1126/science.aar7191
Acknowledgments
SC gratefully acknowledges ICAR for JRF fellowship.
Funding
This study was funded by ICAR-National Institute for Plant Biotechnology, New Delhi.
Author information
Authors and Affiliations
Contributions
Experimentation by SC; data analysis and manuscript editing by M; in silico analysis by SKM and MD; conceptualization by MD; manuscript writing JCP and MD. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Choudhury, S., Mansi, Muthusamy, S.K. et al. Genome-wide identification of Ran GTPase family genes from wheat (T. aestivum) and their expression profile during developmental stages and abiotic stress conditions. Funct Integr Genomics 21, 239–250 (2021). https://doi.org/10.1007/s10142-021-00773-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-021-00773-0