Abstract
Main conclusion
The SiMBR genes in foxtail millet were identified and studied. Heterologous expression of SiMBR2 in Arabidopsis can improve plant tolerance to drought stress by decreasing the level of reactive oxygen species.
Abstract
Foxtail millet (Setaria italica L.), a C4 crop recognized for its exceptional resistance to drought stress, presents an opportunity to improve the genetic resilience of other crops by examining its unique stress response genes and understanding the underlying molecular mechanisms of drought tolerance. In our previous study, we identified several genes linked to drought stress by transcriptome analysis, including SiMBR2 (Seita.7G226600), a member of the MED25 BINDING RING-H2 PROTEIN (MBR) gene family, which is related to protein ubiquitination. Here, we have identified ten SiMBR genes in foxtail millet and conducted analyses of their structural characteristics, chromosomal locations, cis-acting regulatory elements within their promoters, and predicted transcription patterns specific to various tissues or developmental stages using bioinformatic approaches. Further investigation of the stress response of SiMBR2 revealed that its transcription is induced by treatments with salicylic acid and gibberellic acid, as well as by salt and osmotic stresses, while exposure to high or low temperatures led to a decrease in its transcription levels. Heterologous expression of SiMBR2 in Arabidopsis thaliana enhanced the plant's tolerance to water deficit by reducing the accumulation of reactive oxygen species under drought stress. In summary, this study provides support for exploring the molecular mechanisms associated with drought resistance of SiMBR genes in foxtail millet and contributing to genetic improvement and molecular breeding in other crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Foxtail millet (Setaria italica L.) is a crop that originated in China and has been cultivated for over 8000 years (Diao et al. 2014). It serves as an important food and forage in the arid and semi-arid regions of Asia (He et al. 2015). Foxtail millet has become an important model C4 crop for studying stress resistance due to its strong drought tolerance, small gene pool (515 Mb), self-pollination and short life cycle (He et al. 2015; Peng and Zhang 2020; Fan et al. 2021; Li et al. 2022; Liu et al. 2023; Wen et al. 2024).
The ubiquitin–proteasome system (UPS) is a main way to break down proteins, which is important for changing the activity of proteins that control gene activity and is part of many signalling pathways (Zhou et al. 2024). In plants, the UPS facilitates plant response to internal and external stimuli by rapidly regulating its proteome and plays a crucial role in regulating plant growth, development, and physiology (Chen and Hellmann 2013; Mandal et al. 2018; Xu and Xue 2019). The UPS is characterized by the 26S proteasome and a triad of enzymes responsible for substrate protein ubiquitination: E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) enzymes. E3 ligases mainly choose which proteins to break down, moving ubiquitin from the E2-UPS complex to the chosen proteins (Mandal et al. 2018; Liu et al. 2021). The E3 ligases involve four main types, namely HECT (homologous to the E6-AP carboxyl terminus), RING (really interesting new gene), U-box, and cullin-RING ligases (CRL) (Chen and Hellmann 2013). RING proteins have a region with many cysteine amino acids, about 40–60 in length. In Arabidopsis, the genome encodes over 460 RING proteins, categorized into seven subtypes based on structural motifs: RING-H2, RING-HC, RING-v, RING-C2, RING-D, RING-S/T, and RING-G (Stone et al. 2005; Sun et al. 2019; Han et al. 2022).
Among Arabidopsis RING proteins, Iñigo et al. (2012) reported a RING-H2 group protein that binds to MEDIATOR25 (MED25)/PHYTOCHROME AND FLOWERING TIME1 (PFT1). This led to their naming as MED25-BINDING RING-H2 PROTEIN (MBR). MBR proteins contain the RING-H2 domain close to its C-terminus. Based on protein structure and sequence similarity, four members belong to this small group in Arabidopsis (Iñigo et al. 2012; Zhang et al. 2017b). MBR1 and MBR2 interact with MED25 and promote the degradation of MED25, which delays the flowering time of plants (Iñigo et al. 2012). Furthermore, these MBRs interact with the transcriptional repressor TCP INTERACTOR-CONTAINING EAR MOTIF PROTEIN1 (TIE1), contributing to its degradation and promoting leaf development (Zhang et al. 2017b).
At present, the SiMBR gene family in foxtail millet has received limited research attention. In our earlier study, we compared the transcriptomes among three foxtail millet cultivars with varying drought resistance and found 46 genes linked to drought resistance (Guo et al. 2022). One of these genes, Seita.7G226600 (SiMBR2), is part of the MBR gene family related to the protein ubiquitination system in foxtail millet. Under drought stress, its transcription level decreases. To better understand the role of SiMBR2 in abiotic stress responses, we retrieved and characterized the SiMBR gene family from the foxtail millet database, identifying ten members. We examined their phylogenetic relationships, motifs, chromosome locations, gene structures, promoter regions, and predicted expression patterns across different tissues. Additionally, we studied how SiMBR2 responds at the transcriptional level to various abiotic stresses and hormone treatments. In Arabidopsis, over-expression of SiMBR2 improved plant resistance to drought stress by improving its capacity to neutralize reactive oxygen species (ROS). Our findings support further research into the molecular mechanisms behind SiMBR gene-related drought resistance in foxtail millet and could help improve genetics and breeding methods for other crops.
Materials and methods
Identification of genes from the SiMBR family in foxtail millet
The complete foxtail millet genome was obtained from the Phytozome database (https://phytozome-next.jgi.doe.gov/, accessed on 30 July 2023; Goodstein et al. 2012). The Arabidopsis MBR gene sequences (Iñigo et al. 2012) were submitted to the Pfam database (http://pfam.sanger.ac.uk/, accessed on 30 July 2023) to obtain Pfam ID PF13639 of the MBR Ring finger domain and download the Hidden Markov Model (HMM) profile. Utilizing the TBtools software (version 2.056; Chen et al. 2020), the Simple HMM Search command was employed to scan the complete protein sequences of foxtail millet, with a selection threshold of candidate genes set at an E-value of 1e-4. After removing all redundant sequences, the obtained candidate genes were submitted to the SMART (http://smart.embl-heidelberg.de/, accessed on 30 July 2023) and the National Center for Biotechnology Information Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 30 July 2023) for further confirmation of MBR gene family members. The MBR genes in other plants (Brachypodium distachyon, Hordeum vulgare, Sorghum bicolor, Oryza sativa, and Zea mays) were also preliminarily identified using the methods mentioned above.
The SiMBR proteins were further analyzed using ExPASy (https://web.expasy.org/protparam/, accessed on 1 August 2023) to determine their theoretical isoelectric point (pI), molecular weight (MW), and amino acid composition based on protein sequence data (Gasteiger et al. 2003). WoLF PSORT (https://wolfpsort.hgc.jp, accessed on 1 August 2023) was applied to predict the subcellular localization of these proteins (Horton et al. 2007).
Phylogenetic analysis and chromosome location of genes in the SiMBR family
Candidate SiMBR genes were extracted from the Phytozome database, which provided chromosomal positioning of the genes. The physical map was subsequently generated using TBtools software (Chen et al. 2020). Phylogenetic trees were constructed using neighbor-joining (NJ) methods and generated after 1000 bootstrap replications using MEGA 7.0 software (Kumar et al. 2016).
Motif composition and gene structure analysis of SiMBR family genes
The Multiple EM for Motif Elicitation (MEME) online tool (https://meme-suite.org/meme/tools/meme, accessed on 30 July 2023) was utilized to detect motifs within the candidate SiMBR proteins (Bailey et al. 2009). A MEME search was conducted with the following specifications: the number of repetitions is any and a maximum of 20 motifs, and the motif composition was drawn using TBtools software (Chen et al. 2020). Candidate SiMBR gene annotations were extracted from the Phytozome database, from which the genes in exon–intron sequence information were obtained. The exon/intron structure of each SiMBR gene was analyzed using the online Gene Structure Display Server 2.0 (https://gsds.gao-lab.org/, accessed on 30 July 2023; Hu et al. 2015).
Cis-acting element analyses and tissue expression patterns of SiMBR family genes
Then PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 31 July 2023; Lescot et al. 2002) was used to analyze the 2000 bp upstream cis-acting elements in the SiMBR genes. Finally, the results were visualized using TBtools software (Chen et al. 2020). The transcriptomic data for roots, stems, leaves, germinated grains and grains during the filling period S1-S5 (corresponding to the early, early, middle, late, and late filling stages, respectively) were downloaded from the MDSi database (https://foxtail-millet.biocloud.net/home, accessed on 1 August 2023; Li et al. 2023). Utilizing Transcripts Per Kilobase per Million mapped reads data, a heat map representing the expression levels of SiMBR genes across various tissues was generated with TBtools software (Chen et al. 2020).
Plant growth conditions and treatment
In the current study, Setaria italica “Jingu21” was utilized. Seeds were sourced from Prof. Lizhen Zhang's laboratory at the School of Life Science, Shanxi University, China. The seeds were cultivated in plastic pots filled with a 1:1 mixture of vermiculite and nutrient soil under the following conditions: a temperature range of 23–26 °C, light intensity of 50,000 Lux, relative humidity between 30 and 50%, and a photoperiod 16 h of light/8 h of darkness. At the two-leaf stage, four days post-germination, uniform seedlings were selected and transferred to individual plastic pots, with three plants per pot. For the temperature stress experiments, after a 14-day growth period, “Jingu 21” seedlings were subjected to heat stress (40 °C day /32 °C night) and cold stress (4 °C), for 0 and 24 h in a controlled temperature incubator. To induce long-term drought stress, watering was discontinued for an additional two weeks following the initial 14-day growth period. For other abiotic stresses and phytohormone treatment, the seedlings of “Jingu 21” grown for ten days were transferred to 1/2 MS liquid medium for four days and then treated with salt (200 mmol/l NaCl), PEG6000 (15%), NaHCO3 (150 mmol/l), ABA (100 µmol/l), 6-BA (75 µmol/l), IAA (10 µmol/l), α-naphthaleneacetic acid (NAA; 10 nmol/l), gibberellic acid (GA3; 1 mmol/l), methyljasmonat (MJ; 100 µmol/l), and salicylic acid (SA; 10 mmol/l) for 0 and 24 h. Each stress treatment had three replicates. Leaf samples were collected and immediately frozen in liquid nitrogen, then stored at –80 °C to facilitate RNA extraction later.
RNA extraction and qRT-PCR
The transcription levels of SiMBR2 and antioxidant enzyme genes (AtCAT1, AtPOD1 and AtCSD1) in plants were detected by qRT-PCR. Total RNA Extraction and qRT-PCR were performed as described before (Zhang et al. 2022). Total RNA was isolated from leaf tissues using the TransZol™ UP Plus RNA Kit (TransGen Biotech, Beijing, China). The extracted RNA was reverse-transcribed into cDNA using the EasyScript® One-step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech). The qRT-PCR reaction mixture had a total volume of 10 µl, comprising 5 µl of TransStart® Tip Green qPCR SuperMix (TransGen Biotech), 1 µl of diluted cDNA template, 0.8 µl of each primer (10 µmol/l), and 3.2 µl of RNase free ddH2O. The qRT-PCR thermal cycling conditions were set as 94℃ for 30 s, followed by 45 cycles of 94℃ for 15 s and 58℃ for 30 s. All primers (as listed in Suppl. Table S1) were synthesized by Shanghai Sangon Biotech. For foxtail millet, SiAct2 (Seita.8G043100) and RNA POL II (Seita.2G142700) served as internal controls (Kumar et al. 2013; Zhang et al. 2022). For Arabidopsis, AtACT2 (AT3G18780) and AtTIP41-like (TAP42 239 INTERACTING PROTEIN OF 41 kDa, AT4G34270) were used as internal reference (Czechowski et al. 2005; Škiljaica et al. 2022). Each experiment was performed with three technical replicates and three biological replicates.
Clone of SiMBR2
The open reading frames of SiMBR2 (Seita.7G226600) were amplified from “Jingu21” using gene-specific primers designed with attB1/B2 Gateway attachment sites. The BP reactions in pDONR207 (Invitrogen) produced Entry clones, confirmed by sequencing provided by Sangon Biotech (Shanghai, China). The destination clones were created using LR Clonase II (Invitrogen) through an LR reaction. For phenotypic analysis in Arabidopsis, the construct was transferred into the pUBN-GFP-Dest vector via LR reaction, as described before (Grefen et al. 2010).
Generation of transgenic Arabidopsis plants and physiology analysis
Stable Columbia-0 Arabidopsis lines expressing SiMBR2 were obtained through floral dipping (Clough and Bent 1998; Zhang et al. 2015) with Agrobacterium GV3101 carrying pUBN-GFP-SiMBR2, and transformants were selected on a medium containing 50 mg/l Basta (phosphinothricin). The transgene expression in homozygous T3 lines was confirmed by immunoblot analysis, and three independently transformed lines were selected for further experimental use.
Arabidopsis seeds (wild-type and stable transgenic lines) were grown in soil under a 12/12 h (day/night), 23/20 °C (day/night) cycle with 8000 Lux light intensity. Homozygous T3 lines were verified for transgene expression, and three independently transformed lines were chosen for the experiments. For drought stress, watering was stopped for two weeks after four weeks of growth. The water content in the soil was measured by a soil moisture analyzer (TZA-1 K-G, Zhejiang Tuopu Yunnong Technology Co., Ltd). The MDA content (MDA-2-Y), the H2O2 content (H2O2-2-Y), the glutathione (GSH) content (GSH-2-W), the CAT activity (CAT-2-Y), and the POD activity (POD-2-Y) of the leaf sample were measured using kits purchased from Suzhou Keming Biotechnology co. Ltd (China). To verify protein expression, the immunoblot analysis of plant tissues was performed as described before (Zhang et al. 2017a) using a commercial anti-GFP antibody (Abcam).
Statistical analysis
Statistical analysis of independent experiments is reported as means ± SE as appropriate, with significance determined by Student’s t-test or ANOVA.
Results
Identification of SiMBR genes in foxtail millet
Based on data of Iñigo et al. (2012), this study got the Arabidopsis MBR gene sequences and used them to receive an HMM profile from the Pfam database. Subsequently, this profile was used to find MBR genes in different plants. We found 10 genes in Setaria italica, as shown in Table 1. More genes were found in Oryza sativa (rice), Brachypodium distachyon, Zea mays (maize), Sorghum bicolor (sorghum), and Hordeum vulgare (barley), as detailed in Suppl. Table S2. The SiMBR proteins in foxtail millet ranged from 347 to 707 amino acids long and had molecular weights between 37.68 and 76.9 kDa. Most of these proteins were predicted to be in the nucleus, except for SiMBR6 (Seita.3G158100), which is in both the nucleus and peroxisomes (Table 1). Like the proteins in Arabidopsis, the SiMBR proteins have a RING domain, either C3H2C3-type or RING-H2 type, that holds two zinc ions with eight metal-binding residues (as illustrated in Fig. S1).
Phylogeny analysis, chromosome location, gene structure, and motif analyses of SiMBR genes in foxtail millet
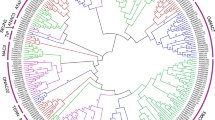
Phylogenetic analysis, using MBR gene protein sequences from various species, showed that MBR proteins are divided into five groups, as shown in Fig. 1. The data also suggested that the MBR gene family is conserved across these species, with a similar distribution of genes within each group. The SiMBR genes in foxtail millet are closely related to those in maize and sorghum, showing a closer evolutionary connection with these two plants than with the others. This confirms the conservation of the MBR gene family and highlights the importance of studying the evolution of plant MBR genes to understand their functions in plant biology.
The SiMBR genes in foxtail millet are spread across six of its nine chromosomes (Fig. 2). Chromosomes 1, 4, and 6 each have a single gene, while chromosomes 3 and 5 have two genes each, and chromosome 7 harboured three genes. This distribution indicates that the SiMBR genes are broadly dispersed throughout various chromosomes in foxtail millet. The analysis of the SiMBR protein motifs found 15 common motifs, illustrated in Fig. 3 (left panel). Each motif shares similar protein sequences (Fig. S2). The motif 1 contains the C3H2C3-type or RING-H2-type Zn binding site. Depending on motifs, these proteins can also be classified into five groups based on their similarities. Similarly, genes in the same group had similar numbers of introns and exons, further supporting the classification from the phylogenetic tree shown in Fig. 3. These results give more information on the structure and evolution of SiMBR genes in foxtail millet.
Protein motifs and gene structures of SiMBR from foxtail millet. Left In the SiMBR2 proteins, 15 motifs were identified by the MEME tool, represented by different colors (1–15) and depicted by TBtools. Right The exon–intron structure of these SiMBR2 genes was predicted by GSDS 2.0. The green boxes represent the coding sequence (CDS), the black lines represent introns, and the yellow boxes represent up/downstream untranslated regions (UTR)
Cis-acting regulatory elements analysis of SiMBR genes in foxtail millet
To understand how SiMBR genes are controlled, we looked at the 5' upstream region of each gene for cis-acting elements. We found 35 different cis-elements related to resistance mechanisms, as summarized in Fig. 4 and Suppl. Table S3. Among these, cis-elements related to plant growth, such as the O2-site, CAT-box, and A-box, were identified in 3, 4, and 8 SiMBR genes, respectively. Furthermore, all SiMBR genes contained abscisic acid response elements (ABREs). Methyl jasmonate-responsive elements, the CGTCA-motif and TGACG-motif, were in 7 SiMBR genes. The auxin response-related TGA-element and the salicylic acid response-related TCA-element were in 4 and 5 SiMBR genes, respectively. Many cis-acting elements related to plant stress responses were also present, such as the LTR (low-temperature response), GT1-motif (light response), and MBS (MYB-binding site associated with drought response). This suggests that most SiMBR genes are controlled by many factors, including hormones and environmental stresses. For example, SiMBR2 gene had A-box, ABRE, AE-box, ARE, CGTCA-motif, GT1-motif, LTR, MBS, and other elements, showing it is part of a complex control system.
In silico transcript analysis of SiMBR genes in foxtail millet
An examination of publicly accessible RNA-seq data from foxtail millet (multi-omics database for Setaria italica; Li et al. 2023) disclosed the expression profiles of SiMBR genes across diverse tissues and developmental stages. Figure 5 illustrates that SiMBR5 (Seita.5G280100) is expressed in various tissues, with higher levels in the spikelet and root. The expression of SiMBR2 was observed in leaves. This suggests that SiMBR5 and SiMBR2 are important for foxtail millet's growth and function, probably because they are expressed differently in each tissue.
Expression analysis of SiMBR2 under different abiotic stresses and phytohormone treatments
In our previous investigation, we found that the transcription of SiMBR2 decreased in “Jingu21” seedlings under soil drought stress (Guo et al. 2022). We now explore how SiMBR2 affects plant responses to other stresses. The 14-day seedlings of “Jingu21” were subjected to 24 h abiotic stresses (cold, heat, NaCl, PEG, and NaHCO3). The transcription of SiMBR2 was repressed following 24 h of extreme temperature (both cold and heat), while its expression was induced under salt and osmotic stresses (Fig. 6a). Since we found many hormone-related cis-acting elements within SiMBR2, we tested its expression after 24 h of treatment with ABA (100 µmol/l), 6-BA (75 µmol/l), IAA (10 µmol/l), NAA (10 nmol/l), GA3 (1 mmol/l), MJ (100 µmol/l), and SA (10 mmol/l) (Fig. 6b). Among these, SA and GA3 increased SiMBR2 levels, while ABA suppressed it.
The relative transcript levels of SiMBR2 gene in foxtail millet under different abiotic stresses (a) and phytohormone treatments (b). The qRT-PCR analyses were used to access transcript levels of SiMBR2 gene from two-week-old foxtail millet seedlings with or without 12 and 24 h cold (4 ℃), hot (40 ℃ day/32 ℃ night), salt (200 mmol/l NaCl), osmotic stress (15% PEG), abscisic acid (ABA, 100 µmol/l), 6-benzylaminopurine (6-BA, 75 µmol/l), auxin (IAA, 10 µmol/l), 1-naphthaleneacetic acid (NAA, 10 nmol/l), gibberellic acid (GA3, 1 mmol/l), methyljasmonat (MJ, 100 µmol/l), and salicylic acid (SA, 10 mmol/l) treatments. Each bar represents the mean ± SE normalized to SiAct2 (Seita. 8G043100) and RNA POL II (Seita. 2G142700). All samples were run in three biological and three technical replicates. Asterisk indicates that the gene expression under stress has a significant difference compared with the control (*P < 0.05)
The osmotic stress from PEG mimics drought conditions that plants can tolerate. However, actual drought effects on plants are more complex (Kautz et al. 2015). In this study, we observed that the transcription level of SiMBR2 increased with PEG treatment. As previously reported, the level of SiMBR2 decreased after 14 days of drought stress induced by withholding water (Guo et al. 2022). To understand this difference, we monitored changes in the transcriptional levels of the SiMBR2 gene during a two-week drought process of water deficiency. As shown in Fig. S3, within the 14-day experimental period, SiMBR2 levels rose, then fell in both control and stress groups, peaking on day 1 and day 3 (the day when treatment group's soil water content below 10% was considered as day 0). After 14 days of water-deficit drought, the stress group had lower SiMBR2 levels than control, matching our earlier findings (Guo et al. 2022). This suggests that PEG-induced drought and water-deficit drought affect SiMBR2 gene expression differently.
Over-expression of SiMBR2 in Arabidopsis seedlings contributes to abiotic stresses tolerance
To elucidate the physiological function of the SiMBR2 gene in plant stress responses, we introduced this gene into Arabidopsis for constitutive expression. After two weeks of soil water deprivation, plants over-expressing SiMBR2 were more tolerant to drought (Fig. 7a). We checked the levels of H2O2 and MDA, and the activity of antioxidant enzymes in leaf samples on day 10, when the leaves of plants were still green. Under drought stress, control plants exhibited elevated levels of H2O2 in their leaves, leading to an increase in MDA content and increased activities of antioxidant enzymes such as CAT, POD, and superoxide dismutase (SOD; Fig. 7b–f). In contrast, plants overexpressing SiMBR2 showed higher activities of these antioxidant enzymes, effectively reducing the rise in ROS and MDA levels. Additionally, we investigated the expression of representative antioxidant enzyme genes, including the superoxide dismutase isoform AtCSD1, peroxidase AtPOD1, and catalase AtCAT1 (Amoah and Seo 2021; Lan et al. 2022; Wang et al. 2023), in transgenic plants (Fig. 8). The data show that AtPOD1 and AtCAT1 were up-regulated in plants with extra SiMBR2 under drought. The transcription level of AtCSD1 remained consistently high in plants overexpressing SiMBR2, regardless of drought conditions (Fig. 8). This suggests that over-expression of the SiMBR2 gene in Arabidopsis improves their drought resistance, which is linked to increased gene activity and better ROS detoxification.
Comparison of drought tolerance of wild-type and transgenic Arabidopsis conferred by SiMBR2. a Growth phenotypes of 4-week-old soil-grown plants of wild-type and transgenic lines treated with natural drought for 2 weeks. Control plant kept water during the whole experiment. b H2O2 level. c MDA level. d CAT activity. e POD activity. f SOD activity. g The expression of SiMBR2 in Arabidopsis was confirmed by Western blot using anti-GFP (abcam). The leaf samples were harvested after 10 days of stop watering. The results were shown as means ± SE from three independent experiments. For b–f, different letters indicate significant differences (P < 0.05)
The relative transcript levels of AtCSD1, AtPOD1, and AtCAT1 in SiMBR2 overexpressed in Arabidopsis. The qRT-PCR analyses were used to access transcript levels of AtCSD1, AtPOD1, and AtCAT1 genes from two-week-old SiMBR2 overexpressing Arabidopsis with stopping watering treatment. AtACT2 (AT3G18780) and TIP41-like (TAP42 INTERACTING PROTEIN OF 41 kDa, AT4G34270) were used as the internal reference. Each bar represents the mean ± SE. All samples were run in three biological and three technical replicates. Asterisk indicates that the gene expression under stress has a significant difference compared with the transcript level at control plant (*P < 0.05)
Discussion
Foxtail millet, a C4 plant, exhibits remarkable resistance to various external stresses, and identifying the genes linked to its stress tolerance could help improve crop genetic engineering (Peng and Zhang 2020; Panchal et al. 2022; Zhang et al. 2023). With the full genome sequencing of foxtail millet now accessible (Bennetzen et al. 2012; Zhang et al. 2012), performing systematic analyses of millet genes using bioinformatics coupled with experimental methods has become feasible. The SiMBR genes belong to the RING proteins, which are part of the E3 ubiquitin ligase family. The functions of several RING proteins have been investigated, revealing their involvement in various plant processes such as responses to abiotic and biotic stresses, hormone signalling, leaf development, flowering, and more (Dong et al. 2006; Stone et al. 2006; Lazaro et al. 2012, 2015; Lee and Seo 2015; Zhang et al. 2017b; Sun et al. 2019). However, the functions of RING proteins are not yet fully elucidated, and the majority of research on these proteins’ function has been focused on model organisms like Arabidopsis, with limited investigation in crop species. Arabidopsis contains over 460 RING proteins (Stone et al. 2005; Sun et al. 2019), rice has 425 (Lim et al. 2010), and wheat has 1255 (Parveen et al. 2021). For the RING-H2 type alone, Arabidopsis has 291 and rice has 281. RING-type proteins play a role in plant growth, development, and responses to various abiotic stresses, including drought, salinity, temperature extremes, and toxic metals (Sun et al. 2019; Han et al. 2022). According to our initial transcriptome screening results (Guo et al. 2022), one SiMBR gene, Seita.7G226600, reacts to drought stress. Considering many members of the RING gene family and their complex roles, we chose to do a detailed analysis on the MBR family instead of the whole RING gene family.
The gene Seita.7G226600, which we named SiMBR2, exhibits similarity to the MBR genes found in Arabidopsis. Using the four MBR genes from Arabidopsis as a reference, we found that foxtail millet, as well as rice, barley, Brachypodium distachyon, and sorghum, has ten members of this gene family, and maize has 15 (Fig. 1 and Table S2). Phylogenetic analysis showed these MBR genes from different species fall into five distinct groups. Each group within foxtail millet shares comparable motifs and gene structures (Fig. 3). The SiMBR genes from foxtail millet show greater similarity to their orthologous counterparts in maize and sorghum than to those in Arabidopsis, indicating that the diversification of the MBR gene family predates the evolutionary split between monocots and dicots. Analysis of cis-acting elements (Fig. 4) indicates a regulatory network controlling SiMBR gene expression, involving multiple factors.
Using previously obtained RNA-seq data, we further analyzed the expression profiles of the SiMBR genes (Fig. 5). SiMBR5 was highly expressed in various tissues, with significantly elevated levels in the spikelet and root during the grain-filling stage. In contrast, SiMBR2 expression was highest in leaf tissue. Since SiMBR2 expression decreased under drought stress (Guo et al. 2022), we then focused on this gene to study its transcriptional regulation in leaves under different phytohormone treatments and abiotic stresses (Fig. 6). The findings indicated that SiMBR2 expression was up-regulated by PEG and salt stress, as well as by treatment with SA and GA3, while it was down-regulated under cold or heat stress, corroborating the presence of multiple cis-regulatory elements in the gene.
In two-week-old foxtail millet seedlings, a 24-h treatment with PEG6000 caused up-regulation of the SiMBR2 gene (Fig. 6). However, our earlier research showed that withdrawing water for 14 days led to down-regulation of the gene (Guo et al. 2022). To clarify this discrepancy, we further investigated the expression levels of SiMBR2 at various time points after stopping watering in two-week-old millet seedlings. We found that the gene up-regulated from day 0 to day 3, regardless of irrigation, matching RNA-seq data showing increased expression during leaf development (Fig. 5). After day 5, expression levels decreased in both the control and drought-stressed groups, with significantly lower expression in the drought-stressed group after 14 days, confirming our previous transcriptome analysis. Our research findings demonstrate that there are differences between PEG-simulated drought stress and water-deficit drought stress in regulating SiMBR2 expression. This distinction may be due to the osmotic stress induced by PEG-simulated drought is rapid, while plants experience a slower and more complex response to water-deficit stress. It also suggests that PEG-simulated drought and actual water-deficit drought are different and cannot be simply substituted in molecular mechanism research.
To elucidate the role of SiMBR2 in plant responses to drought stress, we introduced the gene into Arabidopsis. As shown in Fig. 7, expressing SiMBR2 in Arabidopsis conferred tolerance to drought conditions, indicated by improved antioxidant enzyme activity, lower ROS levels, and reduced MDA damage. Subsequent experiments involved quantifying the transcript levels of key antioxidant enzyme genes AtCSD1, AtPOD1, and AtCAT1 in Arabidopsis (Amoah and Seo 2021; Lan et al. 2022; Wang et al. 2023). Under drought stress, we found increased transcription levels of AtPOD1 and AtCAT1 in plants expressing SiMBR2. In contrast, AtCSD1 levels remained consistently elevated in these plants, irrespective of stress conditions (Fig. 8). These results suggest that expressing SiMBR2 in Arabidopsis enhances antioxidant enzyme gene expression, improving ROS scavenging and plant tolerance to drought.
In Arabidopsis, MBR binds to PFT1 and promotes its degradation, ultimately promoting flowering (Iñigo et al. 2012). PFT1 is a subunit of the plant Mediator complex, which links the transcription factor to the RNA Polymerase II and regulates gene expression (Chen et al. 2022). Zhang et al. (2017b) reported that MBR could also interact with the transcription factor repressor TIE1, promote its degradation and control leaf defects. AtCSD1's consistent up-regulation in SiMBR2 overexpressing plants, even without drought stress, suggests a complex regulation of gene expression by SiMBR2, as AtPOD1 and AtCAT1 levels increased only under stress. Whether this involves SiMBR2-mediated degradation of the Mediator complex or transcription regulators requires further investigation.
It is also important to note that our current method involves rapidly screening and evaluating gene functions through heterologous expression in Arabidopsis. However, given the substantial differences between Arabidopsis and foxtail millet, further functional characterization of the gene must be performed within the millet system.
Conclusion
We identified 10 SiMBR genes in the foxtail millet genome. We analyzed their phylogenetic relationships, conserved motifs, chromosomal locations, gene structures, promoter regions, and predicted expression profiles in different tissues and developmental stages, as well as responses to plant hormones and abiotic stresses. We found that SiMBR2 is regulated by various stress factors. Its heterologous expression in Arabidopsis improved drought tolerance by reducing ROS. This research opens avenues for further exploration into genetically enhancing foxtail millet and other crops for better resistance to environmental stressors.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- CAT:
-
Catalase
- MBR:
-
MED25-BINDING RING-H2 PROTEIN
- MDA:
-
Malondialdehyde
- POD:
-
Peroxidase
- SA:
-
Salicylic acid
References
Amoah JN, Seo YW (2021) Effect of progressive drought stress on physio-biochemical responses and gene expression patterns in wheat. 3 Biotech 11:1–18. https://doi.org/10.1007/s13205-021-02991-6
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME suite: tools for motif discovery and searching. Nucleic Acids Res 37:202–208. https://doi.org/10.1093/nar/gkp335
Bennetzen JL, Schmutz J, Wang H et al (2012) Reference genome sequence of the model plant Setaria. Nat Biotechnol 30:555–561. https://doi.org/10.1038/nbt.2196
Chen L, Hellmann H (2013) Plant E3 ligases: flexible enzymes in a sessile world. Mol Plant 6:1388–1404. https://doi.org/10.1093/mp/sst005
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Chen J, Yang S, Fan B, Zhu C, Chen Z (2022) The Mediator complex: a central coordinator of plant adaptive responses to environmental stresses. Int J Mol Sci 23:6170. https://doi.org/10.3390/ijms23116170
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313X.1998.00343.x
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17. https://doi.org/10.1104/pp.105.063743
Diao X, Schnable J, Bennetzen JL, Li J (2014) Initiation of Setaria as a model plant. Front Agric Sci Eng. 1:16–20. https://doi.org/10.15302/J-FASE-2014011
Dong CH, Agarwal M, Zhang Y, Xie Q, Zhu JK (2006) The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA 103:8281–8286. https://doi.org/10.1073/pnas.0602874103
Fan Y, Lai D, Yang H et al (2021) Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.). BMC Genomics 22:778. https://doi.org/10.1186/s12864-021-08095-y
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. https://doi.org/10.1093/nar/gkg563
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:1178–1186. https://doi.org/10.1093/nar/gkr944
Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64:355–365. https://doi.org/10.1111/j.1365-313X.2010.04322.x
Guo Y, Hao D, Wang X, Wang H, Wu Z, Yang P, Zhang B (2022) Comparative transcriptomics reveals key genes contributing to the differences in drought tolerance among three cultivars of foxtail millet (Setaria italica). Plant Growth Regul 99:45–64. https://doi.org/10.1007/s10725-022-00875-0
Han G, Qiao Z, Li Y, Yang Z, Wang C, Zhang Y, Liu L, Wang B (2022) RING zinc finger proteins in plant abiotic stress tolerance. Front Plant Sci 13:877011. https://doi.org/10.3389/fpls.2022.877011
He L, Zhang B, Wang X, Li H, Han Y (2015) Foxtail millet: nutritional and eating quality, and prospects for genetic improvement. Front Agric Sci Eng. 2:124–133. https://doi.org/10.15302/J-FASE-2015054
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:585–587. https://doi.org/10.1093/nar/gkm259
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Iñigo S, Giraldez AN, Chory J, Cerdán PD (2012) Proteasome-mediated turnover of Arabidopsis MED25 is coupled to the activation of FLOWERING LOCUS T transcription. Plant Physiol 160:1662–1673. https://doi.org/10.1104/pp.112.205500
Kautz B, Noga G, Hunsche M (2015) PEG and drought cause distinct changes in biochemical, physiological and morphological parameters of apple seedlings. Acta Physiol Plant 37:1–6. https://doi.org/10.1007/s11738-015-1914-8
Kumar K, Muthamilarasan M, Prasad M (2013) Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell Tissue Organ Cult 115:13–22. https://doi.org/10.1007/s11240-013-0335-x
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lan Y, Zhang K, He T, Wang H, Jiang C, Yan H, Xiang Y (2022) Systematic analysis of the serine/arginine-rich protein splicing factors (SRs) and focus on salt tolerance of PtSC27 in Populus trichocarpa. Plant Physiol Biochem 173:97–109. https://doi.org/10.1016/j.plaphy.2022.01.015
Lazaro A, Valverde F, Piñeiro M, Jarillo JA (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24:982–999. https://doi.org/10.1105/tpc.110.081885
Lazaro A, Mouriz A, Piñeiro M, Jarillo JA (2015) Red light-mediated degradation of constans by the e3 ubiquitin ligase hos1 regulates photoperiodic flowering in Arabidopsis. Plant Cell 27:2437–2454. https://doi.org/10.1105/tpc.15.00529
Lee K, Seo PJ (2015) The E3 ubiquitin ligase HOS1 is involved in ethylene regulation of leaf expansion in Arabidopsis. Plant Signal Behav 10:1–4. https://doi.org/10.1080/15592324.2014.1003755
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van De Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. https://doi.org/10.1093/nar/30.1.325
Li Y, Yu S, Zhang Q et al (2022) Genome-wide identification and characterization of the CCT gene family in foxtail millet (Setaria italica) response to diurnal rhythm and abiotic stress. Genes (Basel) 13:1829. https://doi.org/10.3390/genes13101829
Li X, Hou S, Feng M, Xia R, Li J, Tang S, Han Y, Gao J, Wang X (2023) MDSi: multi-omics database for Setaria italica. BMC Plant Biol 23:223. https://doi.org/10.1186/s12870-023-04238-3
Lim SD, Yim WC, Moon JC, Kim DS, Lee BM, Jang CS (2010) A gene family encoding RING finger proteins in rice: their expansion, expression diversity, and co-expressed genes. Plant Mol Biol 72:369–380. https://doi.org/10.1007/s11103-009-9576-9
Liu R, Xia R, Xie Q, Wu Y (2021) Endoplasmic reticulum-related E3 ubiquitin ligases: key regulators of plant growth and stress responses. Plant Commun 2:100186. https://doi.org/10.1016/j.xplc.2021.100186
Liu M, Li C, Li Y et al (2023) Genome-wide identification and characterization of the VQ motif-containing gene family based on their evolution and expression analysis under abiotic stress and hormone treatments in foxtail millet (Setaria italica L.). Genes (Basel) 14:1032. https://doi.org/10.3390/genes14051032
Mandal A, Sharma N, Muthamilarasan M, Prasad M (2018) Ubiquitination: a tool for plant adaptation to changing environments. Nucleus 61:253–260. https://doi.org/10.1007/s13237-018-0255-6
Panchal A, Singh RK, Prasad M (2022) Recent advancements and future perspectives of foxtail millet genomics. Plant Growth Regul 99:11–23. https://doi.org/10.1007/s10725-022-00858-1
Parveen A, Rahim MS, Sharma A, Mishra A, Kumar P, Fandade V, Kumar P, Bhandawat A, Verma SK, Roy J (2021) Genome-wide analysis of RING-type E3 ligase family identifies potential candidates regulating high amylose starch biosynthesis in wheat (Triticum aestivum L.). Sci Rep 11:11461. https://doi.org/10.1038/s41598-021-90685-7
Peng R, Zhang B (2020) Foxtail millet: a new model for C4 plants. Trends Plant Sci 26:199–201. https://doi.org/10.1016/j.tplants.2020.12.003
Škiljaica A, Jagić M, Vuk T, Leljak Levanić D, Bauer N, Markulin L (2022) Evaluation of reference genes for RT-qPCR gene expression analysis in Arabidopsis thaliana exposed to elevated temperatures. Plant Biol (stuttg) 24:367–379. https://doi.org/10.1111/plb.13382
Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137:13–30. https://doi.org/10.1104/pp.104.052423
Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18:3415–3428. https://doi.org/10.1105/tpc.106.046532
Sun J, Sun Y, Ahmed RI, Ren A, Xie M (2019) Research progress on plant RING-finger proteins. Genes (Basel) 10:973. https://doi.org/10.3390/genes10120973
Wang J, Moeen-ud-din M, Yin R, Yang S (2023) ROS homeostasis involved in dose-dependent responses of Arabidopsis seedlings to copper toxicity. Genes (Basel). https://doi.org/10.3390/genes14010011
Wen Y, Zhao Z, Cheng L et al (2024) Genome-wide identification and expression profiling of the ABI5 gene family in foxtail millet (Setaria italica). BMC Plant Biol 24:164. https://doi.org/10.1186/s12870-024-04865-4
Xu FQ, Xue HW (2019) The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ 42:2931–2944. https://doi.org/10.1111/pce.13633
Zhang G, Liu X, Quan Z et al (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotechnol 30:549–554. https://doi.org/10.1038/nbt.2195
Zhang B, Karnik R, Wang Y, Wallmeroth N, Blatt MR, Grefen C (2015) The Arabidopsis R-SNARE VAMP721 interacts with KAT1 and KC1 K+ channels to moderate K+ current at the plasma membrane. Plant Cell 27:1697–1717. https://doi.org/10.1105/tpc.15.00305
Zhang B, Karnik R, Waghmare S, Donald N, Blatt MR (2017a) VAMP721 conformations unmask an extended motif for K+ channel binding and gating control. Plant Physiol 173:536–551. https://doi.org/10.1104/pp.16.01549
Zhang J, Wei B, Yuan R, Wang J, Ding M, Chen Z, Yu H, Qina G (2017b) The Arabidopsis ring-type E3 ligase TEAR1 controls leaf development by targeting the TIE1 transcriptional repressor for degradationopen. Plant Cell 29:243–259. https://doi.org/10.1105/tpc.16.00771
Zhang B, Guo Y, Wang H, Wang X, Lv M, Yang P, Zhang L (2022) Identification and characterization of shaker K+ channel gene family in foxtail millet (Setaria italica) and their role in stress response. Front Plant Sci 13:907635. https://doi.org/10.3389/fpls.2022.907635
Zhang B, Liang Z, Wang X (2023) Genetic and genomic research in foxtail millet: preface. Plant Growth Regul 99:1–2. https://doi.org/10.1007/s10725-022-00950-6
Zhou X, Li Y, Wang J et al (2024) Genome-wide identification of U-box gene family and expression analysis in response to saline-alkali stress in foxtail millet (Setaria italica L. Beauv). Front Genet 15:1356807. https://doi.org/10.3389/fgene.2024.1356807
Acknowledgements
We thank all the colleagues in our laboratory for providing useful discussions and technical assistance. This work was supported by the National Natural Science Foundation of China (32272012), Research Project Supported by Shanxi Scholarship Council of China (No. 2021-015), the open funding of State Key Laboratory of Sustainable Dryland Agriculture, Shanxi Agricultural University (No. YJHZKF2108), and Fund Program for the Scientific Activities of Selected Returned Overseas Professionals in Shanxi Province (No. 20230003) to BZ. The Doctoral Research project of Shanxi Agricultural University (No. 2021BQ40) to HZ. The Science and Technology Innovation Young Talent Team of Shanxi Province (No. 202204051001019) to ZL and BZ. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
Experiments were designed by BZ, PY, and LZ. Bioinformatics analysis was performed by YF, YG, ZL, and BZ. Plant physiology experiments were performed by YF, RH, HZ, PY, and BZ. Plant molecular experiments were performed by YF, RH, and PY. BZ, ZL, and LZ analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fan, Y., Guo, Y., Zhang, H. et al. Genome-wide identification of the MED25 BINDING RING-H2 PROTEIN gene family in foxtail millet (Setaria italica L.) and the role of SiMBR2 in resistance to abiotic stress in Arabidopsis. Planta 260, 22 (2024). https://doi.org/10.1007/s00425-024-04455-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04455-6