Abstract
Although many of the genetic loci associated with breast cancer risk have been reported, there is a lack of systematic analysis of regulatory networks composed of different miRNAs and mRNAs on survival analysis in breast cancer. To reconstruct the microRNAs-genes regulatory network in breast cancer, we employed the expression data from The Cancer Genome Atlas (TCGA) related to five essential miRNAs including miR-21, miR-22, miR-210, miR-221, and miR-222, and their associated functional genomics data from the GEO database. Then, we performed an integration analysis to identify the essential target factors and interactions for the next survival analysis in breast cancer. Based on the results of our integrated analysis, we have identified significant common regulatory signatures including differentially expressed genes, enriched pathways, and transcriptional regulation such as interferon regulatory factors (IRFs) and signal transducer and activator of transcription 1 (STAT1). Finally, a reconstructed regulatory network of five miRNAs and 34 target factors was established and then applied to survival analysis in breast cancer. When we used expression data for individual miRNAs, only miR-21 and miR-22 were significantly associated with a survival change. However, we identified 45 significant miRNA-gene pairs that predict overall survival in breast cancer out of 170 one-on-one interactions in our reconstructed network covering all of five miRNAs, and several essential factors such as PSMB9, HLA-C, RARRES3, UBE2L6, and NMI. In our study, we reconstructed regulatory network of five essential microRNAs for survival analysis in breast cancer by integrating miRNA and mRNA expression datasets. These results may provide new insights into regulatory network-based precision medicine for breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that are approximately 21–25 nucleotides in length and mainly function in regulating gene expression in a variety of ways (Denli et al. 2004). Increasing evidence indicates that miRNAs are associated with a wide variety of human diseases, especially multiple cancers such as breast, lung, and colon cancer (Denli et al. 2004; Aaronson 1990). Compared to normal tissues, the miRNA expression has been revealed to be perturbed in tumors based on expression analysis studies (Meltzer 2005). As a consequence, miRNAs could be used as diagnostic biomarkers of human cancers in the future. The determination of miRNA binding characteristics with target messenger RNAs (mRNAs) is important to defining miRNA functions, but this task is complex and difficult due to the distinct sequence determinants of individual miRNAs that lead to diverse mRNA targeting. Distinct mRNA targeting “rules” have been frequently reported for individual miRNAs (Wang et al. 2010). Therefore, the identification of miRNA targets is thought to be critical to improving our understanding of the regulatory effects of miRNAs. In fact, both miRNA-mRNA and miRNA-miRNA interactions exist. Here, we reconstructed regulatory networks related to not only functional miRNA-mRNA interactions, but also potential miRNA-miRNA interactions involved in breast cancer.
Currently, several candidate miRNAs in human breast cancer biomarkers have been identified. Among them, there are five essential miRNAs, including miR-21, miR-22, miR-210, miR-221, and miR-222 associated with breast cancer. In at least three profiling studies by comparing miRNA expression level between breast cancer and normal tissues, miR-21 and miR-210 were consistently reported to be upregulated (Adhami et al. 2018). MiR-22 has been identified as a regulator of lipid and folate metabolism in breast cancer cells by systematic integration of molecular profiles (Koufaris et al. 2016). Based on high-throughput analysis and a human in vitro cell culture model of basal-like breast cancer, miR-221 and miR-222 were reported as regulators in multiple signaling pathways such as the oncogenic RAS-RAF-MEK pathway (Rao et al. 2011; Shah and Calin 2011; Stinson et al. 2011). However, most of previous studies have been based on the effects of individual miRNAs in breast cancer, which may not have sufficient evidence to show the prognostic roles of each miRNA. In this study, we have focused on these five essential miRNAs whose functional genomics data were available to reconstruct the regulatory network for further survival analysis to reveal the prognostic significance of miRNA-mRNA interactions in breast cancer.
Materials and methods
miRNA and gene expression data collection
RNA sequence expression data of breast invasive carcinoma miRNA were downloaded from GDC Data Portal (https://portal.gdc.cancer.gov/) in Dec. 2016, and the corresponding clinical data were downloaded from the cBioPortal database (http://www.cbioportal.org/). We downloaded and analyzed the “TCGA-BRCA” project. In total, we downloaded TCGA level 3 data that included 1096 primary breast cancer patients and 104 controls. All samples were tested by Illumina HiSeq 2000 RNA Sequencing Version 2 Analysis, and the reads per million mapped (RPMM) values were used for statistical analysis. To investigate the functions of these miRNAs, we searched for and downloaded several miRNAs treated breast cancer cell line expression data sets from the NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/) (Barrett et al. 2013; Edgar et al. 2002). After rigorous screening, we ultimately chose 4 datasets for re-analysis (Table 1). These datasets were GSE52674 (including 2 miR-21 knockdown samples and 2 controls), GSE25162 (including 2 miR-210 overexpression samples and 2 controls), GSE17508 (including 3 miR-22 knockout samples and 3 controls), and GSE19777 (including 3 miR-221 knockdown samples, 3 miR-222 knockdown samples, and 3 controls) (Rao et al. 2011). All four of these datasets used human breast cancer MCF-7 cell lines and were tested with an Affymetrix Human Genome U133 Plus 2.0 Array.
Data preprocessing
R statistical software v3.3.3 (https://www.r-project.org/) was used to perform data preprocessing. The raw RPMM data of breast miRNA were log2 transformed, and miRNAs with a mean or median expression value of 0 were removed. After this step, we obtained expression data for 529 miRNAs in human primary breast cancers and controls. For these 4 miRNA-treated cell line datasets, we used the Robust Multichip Average (RMA) algorithm in the oligo package to normalize the raw expression data and generate a normalized gene expression matrix (Carvalho and Irizarry 2010). Gene annotation, integration, and renormalization of these 4 datasets were performed using our custom written Python code. The distributions of RMA-processed and globally renormalized gene expression values for these 4 studies are showed in Fig. S1. After the global renormalization, the distribution of gene expression values across all studies displayed a consistent range. Since we only collected 5 miRNA-treated cell line data sets, we therefore mainly focused on the functions of these 5 miRNAs (miR-21, miR-210, miR-22, miR-221, and miR-222). We divided these 4 datasets into 5 groups according to miRNA treatment.
miRNAs correlation and differentially expressed gene analysis
We used a heatmap to display the expression profiles of these 5 miRNAs in breast cancer patients and controls, and the unsupervised clustering method “ward.D2” was chosen. Univariate linear regression was used to explore the correlations among these 5 miRNAs, and an absolute value of the regression coefficient ≥ 0.5 and a P value ≤ 0.05 were considered as significant corrected. We constructed the interaction networks of these 5 miRNAs based on the regression coefficients and P values. The differentially expressed genes between the breast cancer cell line samples and controls in these 5 groups were identified using an empirical Bayes algorithm (function “eBayes”) in the limma package (Ritchie et al. 2015). Upregulated and downregulated genes were considered to be those with an absolute value of logarithmic transformed fold-change (log2(FC)) ≥ log2(1.5) and a P value ≤ 0.05.
Pathway annotation by gene set enrichment analysis
To understand the functions of these 5 miRNAs, we used the javaGSEA desktop application v2.2.4 to perform gene set enrichment analysis (GSEA) of KEGG pathways in these 5 groups of miRNA treatments (Subramanian et al. 2005). Gene sets containing less than 15 genes or more than 500 genes were excluded. The t-statistic mean of the genes was computed in each KEGG pathway using a permutation test with 1000 replications. Pathways with a normalized enrichment score (NES) > 0 were considered to be upregulated and pathways with a NES < 0 were considered to be downregulated. Statistical significance was identified as a P value ≤ 0.05.
miRNA and target gene regulatory network reconstruction
We used a Venn diagram to show the differently expressed genes in these 5 miRNA-treated groups. Shared deregulated genes in more than 3 groups were chosen to reconstruct the miRNA-mRNA regulatory network. We used Cytoscape v3.4.0 to display the results.
miRNA and miRNA-target gene interaction survival analysis
We used a Cox proportional hazards model to analyze the effect of miRNAs on overall patient survival. Each miRNA was divided into two groups according to the tertile expression levels. Samples with miRNA expression below the first tertile were classified in the low expression group, and those with miRNA expression above the second tertile were classified in the high expression group. To clarify the results, we did not include the samples with miRNAs expression between the first and second tertile in the analysis. The references for the associations between single miRNAs and overall survival were used in the low expression group. According to our groupings, highly expressed miRNAs with a hazard ratio (HR) > 1 were considered to be risk factors. Conversely, highly expressed miRNAs with a HR < 1 were considered to be correlated with better survival. For examination of the influences of miRNA-target gene interactions on overall survival, we divided these miRNA-target genes into two groups according to their tertile expression levels and then combined them with the above miRNA groups. Patients with both miRNAs and miRNA-target gene expression levels below the first tertile were defined as references. In addition, the other three groups were compared to the reference group. Statistical significance was considered to be a P value ≤ 0.05.
Results and discussion
The correlation and interaction of five miRNAs revealed by TCGA data
Based on the gene expression matrices of our selected miRNAs (miR-21, miR-210, miR-22, miR-221, and miR-222) in 1200 samples including 1096 cases and 104 controls from the TCGA database, five miRNAs could be clustered into three groups (Fig. 1a). The miRNA-based clustering can also group patients according to subtypes (i.e., ER, PR, HER2 status). In short, miR-21 and miR-210 were clustered together as group 1, miR-221 and miR-222 were clustered together as group 2, and miR-22 was clustered by itself as group 3. To evaluate the transcriptional activity of miRNAs in breast cancer, the expression values of 1096 cases were compared with 104 controls for each miRNA. As a result, both miRNA21 and miRNA210 in group 1 were upregulated in primary breast cancer patients. In contrast, the other 3 miRNAs, including miR-22, miR-221, and miR-222, were downregulated in these patients. However, the expression values of miR-21 and miR-22 were higher than those of other miRNAs (Fig. 1b). To further explore the interactions of these five miRNAs, their pair-wise linear correlation coefficients were analyzed (Fig. 1c). A strong positive correlation was found between miR-221 and miR-222 (r = 0.83, P < 0.001). In fact, miR-221 and miR-222 have been frequently reported to regulate cell growth and cell cycle progression in various types of human malignancy (Li et al. 2016; Yoshimoto et al. 2011; Zhang et al. 2010). In addition, miR-22 was positively correlated with both miR-221 and miR-222 with correlation coefficients of 0.62 and 0.57, respectively. Although it was previously reported that miRNAs that directly target estrogen receptor (ER) α including miR-22 and miR-221/222 would have distinct roles in not only regulating ERα but also regulating other target genes in human breast cancer (Yoshimoto et al. 2011). Based on the correlation values, it can be suggested that there would be other miRNA-miRNA interactions exist, including miR-21/miR-22 (r = 0.39), miR-21/miR-221 (r = 0.37), miR-21/miR-210 (r = 0.22), and miR-210/miR-222 (r = 0.22). Potential interactions and correlations are summarized in Fig. 1d.
miRNAs correlations and interactions revealed by TCGA data. a Heatmap and clusters of expression of the five miRNAs (miR-21, miR-22, miR-210, miR-221, and miR-222) in 1200 samples including 1096 cases (red) and 104 controls (blue) from the TCGA database. It also showed the samples clustering according to subtypes (i.e., ER, PR, HER2 status). b Expression patterns of the five miRNAs between patients (red for miR-21, yellow for miR-210, green for miR-22, light-blue for miR-221, and dark-blue for miR-222) and control samples (gray). c The miRNA-miRNA correlation of the selected five miRNAs. d Hypothetical interactions of the selected five miRNAs revealed by TCGA data
Survival analysis of individual miRNAs
Survival analysis of individual miRNAs showed that the overexpression of both miR-21 (HR = 1.51, P = 0.031) and miR-22 (HR = 1.84, P = 0.004) could significantly decrease overall survival frequency in breast cancer patients, but no significant associations could be found for other miRNAs, namely, including miR-210, miR-221, or miR-222 (Fig. 2). Due to the poor overall survival associated with high levels of miR-21 and miR-22, both of these miRNAs have been recognized as prognostic indicators of breast cancer progression (Chen et al. 2015; Das and Lin, 2017; Jinling et al. 2016; Pandey et al. 2015; Wang et al. 2015). Furthermore, we have also performed survival analysis of each miRNA using the profiles of patient subtypes including ER, PR, and HER2 positive or negative (Fig. S2). As a result, miR-22 was significantly correlated with survival in subtypes of ER positive and PR positive or negative, miR-222 was significantly correlated with survival in subtype of ER positive and PR positive. In fact, the relationship between miRNAs alone and breast cancer prognosis has been widely reported, but the specific regulatory mechanism is still unknown. The use of miRNA information alone may be inefficient for survival prediction. In the view of systems biology, miRNA does not work alone. Both miRNA-mRNA and miRNA-miRNA interactions exist and function in diverse biological processes, especially in cancer regulation. It is necessary to integrate functional genomics data to reconstruct the network of miRNAs and target genes and apply it to additional studies on survival prediction.
Survival analysis of the five miRNAs. a The association of miR-21 with overall survival. b The association of miR-210 with overall survival. c The association of miR-22 with overall survival. d The association of miR-221 with overall survival. e The association of miR-222 with overall survival. The blue curves represent overall survival under low miRNA expression, the red curves represent overall survival under high miRNA expression
Gene expression profiling analysis of the five miRNAs
To further investigate the roles of the selected miRNAs in breast tumor development, we have employed four microarray datasets related to the above five miRNAs and performed gene expression profiling analysis (Fig. 3a and Fig. S1). Based on our analysis of differentially expressed genes (DEGs), we have significantly identified the respective significant DEGs in response to the regulation of each miRNA. As a result, there were 184 DEGs, including 151 upregulated genes and 33 downregulated genes, under miR-22 knockout; there were 68 DEGs, including only 2 upregulated genes and 66 downregulated genes, under miR-221 knockdown; there were 6 DEGs, including only one upregulated genes and 5 downregulated genes, under miR-222 knockdown; there were 1286 DEGs, including 984 upregulated genes and 302 downregulated genes, under miR-210 overexpression; and there were 448 DEGs, including 112 upregulated genes and 336 downregulated genes, under miR-21 knockdown. The details on all dysregulated genes for each miRNA are shown in Table S1.
Overview of gene expression profiles analysis of the five miRNAs. a Heatmap of differentially expressed genes associated with the regulation of the five miRNAs, including miR-21 knockdown (red), miR-210 overexpression (yellow), miR-22 knockout (green), miR-221 knockdown (light blue), and miR-222 knockdown (dark blue). The control samples are shown in gray. b Summary of the overlapping differentially expressed genes associated with the regulation of these five miRNAs
In comparison, the overlapping genes among correlated miRNAs have been identified. We focused on commonly dysregulated genes associated with at least 3 miRNAs and ultimately obtained 34 target genes that overlapped in more than two miRNAs, which are described in Table 2 (Fig. 3b). There were 4 overlapping genes among miR-22, miR-21, and miR-210, including OAS3, DDX60, IRF9, and IFI6, which were upregulated by the regulation of both miR-22 and miR-21 but downregulated by the regulation of miR-210. Four other overlapping genes, including IRF7, CMPK2, HLA-C, and NMI, were identified among miR-22, miR-21, and miR-221, and the first three genes (IRF7, CMPK2, HLA-C) were upregulated by the regulation of both miR-22 and miR-21 but downregulated by the regulation of miR-221, whereas NMI was upregulated by the regulation of miR-22 but downregulated by the regulation of both miR-21 and miR-221. Only one gene, AKR1C3, was commonly identified among miR-21, miR-221, and miR-222, and it was upregulated by the regulation of miR-21 but downregulated by the regulation of both miR-221 and miR-222. A total of 17 overlapping genes were identified among miR-22, miR-210, and miR-221. Most of these genes were upregulated by the regulation of miR-22 but downregulated by the regulation of both miR-210 and miR-221, including GBP3, TAP1, HCP5, BST2, RARRES3, SAMHD1, UBE2L6, IFITM1, OASL, LGALS3BP, TRIM22, OAS2, IFI44L, IFI44, RSAD2, and DDX60L. Only one of these genes, PSMB10, was upregulated by the regulation of both miR-22 and miR-210 but downregulated by the regulation of miR-221. There were 8 overlapping genes among 4 miRNAs (miR-22, miR-21, miR-210, and miR-221). They were IFI27, ISG15, OAS1, PSMB9, IFIT1, IFIT2, IFIT3, and HLA-B, all of which were upregulated by the regulation of both miR-22 and miR-21 but downregulated by the regulation of both miR-210 and miR-221.
Functional enrichment analysis of the five miRNAs
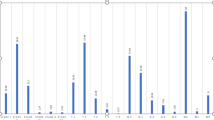
Based on the GSEA of gene expression profiles in these 5 groups of miRNA treatments, there were 86 enriched KEGG pathways involved in at least one miRNA-treated group (Fig. 4a). Among them, 3 pathways, including the RIG – I-like receptor signaling pathway, Toll-like receptor signaling pathway, and Cytokine − cytokine receptor interaction, were enriched by the regulation of all five miRNAs, which were associated with immune system and signaling molecule interactions. Three immune diseases-related pathways, including allograft rejection, graft-versus-host disease, and autoimmune thyroid disease, and the immune system-related pathway of antigen processing and presentation, and the folding, sorting and degradation-related pathway of the proteasome were enriched in the regulatory network of four miRNAs. Functional enrichment analysis indicated that dysregulated KEGG pathways in response to diverse miRNAs regulation were mainly related to immune functions. The immune response has been reported to be prominent demonstrating an important modulatory role of miRNAs in the biology of breast tumors devoid of somatic copy-number aberrations (CNA-devoid) (Dvinge et al. 2013). It has been suggested that some miRNAs could regulate the function of several types of immune cells as a novel regulator of autoimmune diseases (Arora et al. 2017; Garo and Murugaiyan 2016; Junker et al. 2015; Wang et al. 2017, 2016). Furthermore, we have identified 88 immune response mediators whose gene expression values significantly changed across diverse stages (from stage I to stage IV) of breast cancer. The gene expression patterns of these mediators are shown in Fig. 4b. As a whole, most of these genes showed little difference in their expression from stage I to stage III, but their expression differed significantly in stage IV. In the last stage, more genes were significantly upregulated, such as CXCL13 and PIGR, and fewer genes were significantly downregulated, such as CRIP1 and FGFR3, compared to the other stages.
Functional enrichment associated with regulation of the five miRNAs. Summary of the enriched pathways significantly associated with regulation of the five miRNAs (red column for miR-21, yellow column for miR-210, green column for miR-22, light-blue column for miR-221, and dark-blue column for miR-222). The size of the circles represents the significance level, and the color of the squares represents the normalized enrichment scores (NES) of each pathway. Heatmap showing the expression patterns of immune-related genes that are significantly differentially expressed in diverse stages of breast cancer. The color bar represents the value of logFC compared to the control sample
Transcriptional regulation relationship among the five miRNAs
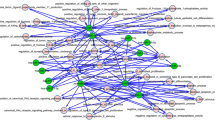
To determine the specific regulatory functions of each miRNA, gene co-expression data and profiles of transcription factor binding sites (TFBSs) were integrated to predict the significantly associated transcription factors (TFs) of individual miRNAs. As a result, there were 21 TFs for miR-21, 18 TFs for miR-22, 84 TFs for miR-210, 2 TFs for miR-221, and one TF for miR-222, respectively (Table S3). These TFs are mainly involved in several transcription factor families, such as the basic helix-loop-helix (bHLH) family, the interferon regulatory factors (IRFs) family, the basic leucine (Leu) zipper (bZIP) family, and the zinc finger zf-C2H2 family. Among them, the activation of the interferon-stimulated genes, such as IRF-1 and IRF-5, has been previously linked to tumor immune rejection (Koelzer et al. 2017). In our study, co-expression analysis of transcriptional regulation revealed that diverse expression patterns and novel potential regulatory genes were induced by the deregulation of IRF1 in response to the altered expression of miRNAs (Fig. 5). IRF1 was significantly downregulated by miR-21 and miR-221 knockdown; it is the activator of target genes with lower expressions, such as IFIT3 and PSMB9, and the repressor of other genes with higher expressions, such as E2F1 and MYB. Instead, IRF1 was significantly upregulated by miR-22, and the activated genes of IRF1, such as IFIT3 and PSMB9, were also over-expressed. The expression of IRF1 was not significantly altered by the overexpression of miR-210 or the knockdown of miR-222, whereas the expression of most IRF1 target genes was not changed significantly, except for the upregulation of IFIT3 and the downregulation of SOCS2. Recent comparative structure function analysis and in silico modeling have demonstrated that DNA-binding domain (DBD) monoubiquitination may play an essential role in IRF-1 activation (Landre et al. 2017). Besides IRF1, we identified signal transducer and activator of transcription 1 (STAT1) as another important TF in the transcriptional regulation among the five miRNAs. The regulatory patterns of STAT1 were shown in Fig. S3. Our findings may provide insights into a new level of transcriptional regulation of IRF-1 and STAT1 in breast cancer.
IRF transcriptional regulations among the five miRNAs. Diverse expression patterns and novel potential regulatory genes induced by the deregulation of IRF1 in response to the altered expression of the 5 miRNAs are revealed by co-expression analysis of transcriptional regulation, including miR-21 knockdown, miR-210 overexpression, miR-22 knockout, miR-221 knockdown, and miR-222 knockdown
Application of the reconstructed miRNAs regulatory network for survival analysis
Based on the above TFs and target genes information, a miRNA-TF-gene regulatory network was reconstructed (Fig. 6). We combined the expression signatures of miRNAs with their target genes according to the reconstructed miRNA regulatory network in our study and applied the results to survival prediction. Out of 170 one-on-one interactions of 5 miRNAs with 34 genes, we identified 45 significant miRNA-gene interactions for predicting survival in breast cancer (Table 3 and Fig. S4). Due to the lack of relevant data, we did not perform multi-factor correction. The six most significant effects of miRNA-target gene interactions on overall survival, including miR-21/proteasome subunit beta 9 (PSMB9), miR-21/human leukocyte antigen C (HLA-C), miR-210/retinoic acid receptor responder 3 (RARRES3), miR-22/HLA-C, miR-221/ubiquitin conjugating enzyme E2 L6 (UBE2L6), and miR-222/N-myc interactor (NMI), are shown in Fig. 7.
Most significant effect of miRNA-target gene interactions on overall survival. a Interaction of miR-21 and PSMB9 on overall survival. b Interaction of miR-21 and HLA-C on overall survival. c Interaction of miR-210 and RARRES3 on overall survival. d Interaction of miR-22 and HLA-C on overall survival. e Interaction of miR-221 and UBE2L6 on overall survival. f Interaction of miR-222 and NMI on overall survival. The red curves represent overall survival under low expression of both miRNAs and genes. The green curves represent overall survival under low expression of miRNAs but high expression of genes. The blue curves represent overall survival under high expression of miRNAs but low expression of genes. The purple curves represent overall survival under high expression of both miRNAs and genes
In particular, patients with miR-21 expression > 18.06 and PSMB9 expression < 10.32 showed significantly poor survival (HR = 1.36, 95% CI 1.10–1.70, P = 0.005) compared to the reference group. However, patients with miR-21 expression < 17.55 and PSMB9 expression > 9.26 or miR-21 expression > 18.06 and PSMB9 expression > 10.32 all showed no significant difference in overall survival (Fig. 7a). The association between high expression levels of one of the immunoproteasome (IP) genes, PSMB9, with longer survival depends on regulation by cell-intrinsic and -extrinsic factors in human cancers (Rouette et al. 2016). The oncogenic miR-21 signaling pathway may play important roles in cancer initiation and progression as an environmental driver induced by intrinsic and extrinsic stimuli (Melnik 2015). In this sense, the correlation between miR-21 and PSMB9 may primarily affect the survival of breast tumor patients.
Patients with miR-21 expression > 18.06 and HLA-C expression < 15.41 showed significantly poor survival (HR = 1.32, 95% CI 1.03–1.68, P = 0.026), but patients with miR-21 expression < 17.55 and HLA-C expression > 14.62 or miR-21 expression > 18.06 and HLA-C expression > 15.41 all showed no significant difference in overall survival (Fig. 7b). Patients with miR-22 expression > 16.14 and HLA-C expression < 15.41 (HR = 1.33, 95% CI 1.04–1.70, P = 0.023), as well as patients with miR-22 expression > 16.14 and HLA-C expression > 15.41 (HR = 1.21, 95% CI 1.03–1.41, P = 0.022), showed significantly poor survival, but patients with miR-22 expression < 15.70 and HLA-C expression > 14.62 showed no significant difference in overall survival (Fig. 7d). Recently, HLA-C has been identified to be associated with favorable clinical outcomes in basal-like breast tumors by transcriptomic studies on immunologic signatures (Martinez-Canales et al. 2017). Although there was no direct evidence regarding the relation between miR-21 and HLA-C, the binding of another microRNA, miR-148a, may regulate the expression of human HLA-C allotypes, which suggests its indispensable roles in controlling miRNA regulation in cancers (O'Huigin et al. 2011).
Patients with miR-210 expression < 7.64 and RARRES3 expression > 11.13 showed significantly high survival (HR = 0.61, 95% CI 0.38–1.00, P = 0.050), but patients with miR-210 expression > 9.39 and RARRES3 expression < 12.29 or miR-210 expression > 9.39 and RARRES3 expression > 12.29 all showed no significant difference in overall survival (Fig. 7c). In a recent study, the metastasis suppressor RARRES3 was identified as an endogenous inhibitor of immunoproteasome expression in breast cancer cells, the expression of which is controlled by the interferon regulatory factor IRF1 (Anderson et al. 2017). Moreover, RARRES3 plays a pivotal role in modulating Wnt protein acylation and signaling activities in human breast cancer cells (Hsu et al. 2015).
Patients with miR-221 expression < 6.00 and UBE2L6 expression > 11.61 showed significantly high survival (HR = 0.54, 95% CI 0.33–0.89, P = 0.015), but patients with miR-221 expression > 6.76 and UBE2L6 expression < 12.38 or miR-221 expression > 6.76 and UBE2L6 expression > 12.38 all showed no significant difference in overall survival (Fig. 7e). The expression of UBE2L6 has been confirmed to be downregulated in human breast cells with BRCA2 knockdown (Tripathi and Chaudhuri 2005). Patients with miR-222 expression > 5.57 and NMI expression < 10.09 showed significantly poor survival (HR = 1.26, 95% CI 1.01–1.58, P = 0.040), but patients with miR-222 expression < 4.75 and NMI expression > 9.47 or miR-222 expression > 5.57 and NMI expression > 10.09 all showed no significant difference in overall survival (Fig. 7f). NMI has been reported to function in the regulation of autophagy and chemosensitivity in breast cancer cells has been reported, and it may be negatively regulated by miR-29 (Metge et al. 2015; Rostas 3rd et al. 2014).
Conclusions
miRNAs play crucial roles in the regulation of multiple biological functions, and they may be used as diagnostic markers in breast cancer for patient prognosis analysis. However, due to the inefficiency of survival prediction using miRNA alone, the reconstruction of a miRNA regulatory network and its application in survival analysis was necessary and sufficient to systematically reveal the direct and indirect relationships between miRNAs and survival. In our study, this strategy was used to evaluate five essential miRNAs related to breast cancer, including miR-21, miR-22, miR-210, miR-221, and miR-222. The final results based on the integration of functional genomics data showed that in addition to the identification of two distinct miRNAs, we identified 45 significant miRNA-gene interactions that could be used for survival analysis. Further validation experiments are certainly needed to reveal the involved genetic mechanisms.
Abbreviations
- TCGA:
-
The Cancer Genome Atlas
- IRFs:
-
interferon regulatory factors
- STAT1:
-
signal transducer and activator of transcription 1
- miRNAs:
-
microRNAs
- mRNAs:
-
messenger RNAs
- RPMM:
-
reads per million mapped
- RMA:
-
Robust Multichip Average
- GSEA:
-
gene set enrichment analysis
- NES:
-
normalized enrichment score
- HR:
-
hazard ratio
- ER:
-
estrogen receptor
- DEGs:
-
differentially expressed genes
- TFBSs:
-
transcription factor binding sites
- bHLH:
-
the basic helix-loop-helix
- DBD:
-
DNA-binding domain
- PSMB9:
-
proteasome subunit beta 9
- HLA-C:
-
human leukocyte antigen C
- RARRES3:
-
retinoic acid receptor responder 3
- UBE2L6:
-
ubiquitin conjugating enzyme E2 L6
- NMI:
-
N-myc interactor
References
Aaronson SA (1990) Oncogenes and the molecular origins of Cancer. Robert A. Weinberg, Ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989. xii, 367 pp., illus. Paper, $55. Cold Spring Harbor Monograph 18. Science 249:1177–1178
Adhami M, Haghdoost AA, Sadeghi B, Malekpour Afshar R (2018) Candidate miRNAs in human breast cancer biomarkers: a systematic review. Breast Cancer 25:198–205
Anderson AM, Kalimutho M, Harten S, Nanayakkara DM, Khanna KK, Ragan MA (2017) The metastasis suppressor RARRES3 as an endogenous inhibitor of the immunoproteasome expression in breast cancer cells. Sci Rep 7:39873
Arora N, Tripathi S, Singh AK, Mondal P, Mishra A, Prasad A (2017) Micromanagement of immune system: role of miRNAs in helminthic infections. Front Microbiol 8:586
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M et al (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41:D991–D995
Carvalho BS, Irizarry RA (2010) A framework for oligonucleotide microarray preprocessing. Bioinformatics 26:2363–2367
Chen B, Tang H, Liu X, Liu P, Yang L, Xie X, Ye F, Song C, Xie X, Wei W (2015) miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett 356:410–417
Das S, Lin TS (2017) The role of microRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr Pharm Des 23:1845-1859
Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ (2004) Processing of primary microRNAs by the microprocessor complex. Nature 432:231–235
Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A et al (2013) The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 497:378–382
Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210
Garo LP, Murugaiyan G (2016) Contribution of microRNAs to autoimmune diseases. Cell Mol Life Sci 73:2041–2051
Hsu TH, Jiang SY, Chang WL, Eckert RL, Scharadin TM, Chang TC (2015) Involvement of RARRES3 in the regulation of Wnt proteins acylation and signaling activities in human breast cancer cells. Cell Death Differ 22:801–814
Jinling W, Sijing S, Jie Z, Guinian W (2016) Prognostic value of circulating microRNA-21 for breast cancer: a systematic review and meta-analysis. Artif Cells Nanomed Biotechnol:1–6
Junker F, Chabloz A, Koch U, Radtke F (2015) Dicer1 imparts essential survival cues in Notch-driven T-ALL via miR-21-mediated tumor suppressor Pdcd4 repression. Blood 126:993–1004
Koelzer VH, Sokol L, Zahnd S, Christe L, Dawson H, Berger MD, Inderbitzin D, Zlobec I, Lugli A (2017) Digital analysis and epigenetic regulation of the signature of rejection in colorectal cancer. Oncoimmunology 6:e1288330
Koufaris C, Valbuena GN, Pomyen Y, Tredwell GD, Nevedomskaya E, Lau CH et al (2016) Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene 35:2766–2776
Landre V, Revi B, Mir MG, Verma C, Hupp TR, Gilbert N et al (2017) Regulation of transcriptional activators by DNA-binding domain ubiquitination. Cell Death Differ 24:903–916
Li X, Zheng J, Chen L, Diao H, Liu Y (2016) Predictive and prognostic roles of abnormal expression of tissue miR-125b, miR-221, and miR-222 in glioma. Mol Neurobiol 53:577–583
Martinez-Canales S, Cifuentes F, Lopez De Rodas Gregorio M, Serrano-Oviedo L, Galan-Moya EM, Amir E et al (2017) Transcriptomic immunologic signature associated with favorable clinical outcome in basal-like breast tumors. PLoS One 12:e0175128
Melnik BC (2015) MiR-21: an environmental driver of malignant melanoma? J Transl Med 13:202
Meltzer PS (2005) Cancer genomics: small RNAs with big impacts. Nature 435:745–746
Metge BJ, Mitra A, Chen D, Shevde LA, Samant RS (2015) N-Myc and STAT interactor regulates autophagy and chemosensitivity in breast cancer cells. Sci Rep 5:11995
O'Huigin C, Kulkarni S, Xu Y, Deng Z, Kidd J, Kidd K et al (2011) The molecular origin and consequences of escape from miRNA regulation by HLA-C alleles. Am J Hum Genet 89:424–431
Pandey AK, Zhang Y, Zhang S, Li Y, Tucker-Kellogg G, Yang H, Jha S (2015) TIP60-miR-22 axis as a prognostic marker of breast cancer progression. Oncotarget 6:41290–41306
Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C et al (2011) MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 30:1082–1097
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47
Rostas JW 3rd, Pruitt HC, Metge BJ, Mitra A, Bailey SK, Bae S et al (2014) microRNA-29 negatively regulates EMT regulator N-myc interactor in breast cancer. Mol Cancer 13:200
Rouette A, Trofimov A, Haberl D, Boucher G, Lavallee VP, D'Angelo G et al (2016) Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci Rep 6:34019
Shah MY, Calin GA (2011) MicroRNAs miR-221 and miR-222: a new level of regulation in aggressive breast cancer. Genome Med 3:56
Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O'Brien C et al (2011) TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal 4:ra41
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550
Tripathi MK, Chaudhuri G (2005) Down-regulation of UCRP and UBE2L6 in BRCA2 knocked-down human breast cells. Biochem Biophys Res Commun 328:43–48
Wang WX, Wilfred BR, Xie K, Jennings MH, Hu YH, Stromberg AJ, Nelson P (2010) Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. RNA Biol 7:373–380
Wang Y, Zhang Y, Pan C, Ma F, Zhang S (2015) Prediction of poor prognosis in breast cancer patients based on microRNA-21 expression: a meta-analysis. PLoS One 10:e0118647
Wang S, Wan X, Ruan Q (2016) The MicroRNA-21 in autoimmune diseases. Int J Mol Sci 17
Wang B, Yao Q, Xu D, Zhang JA (2017) MicroRNA-22-3p as a novel regulator and therapeutic target for autoimmune diseases. Int Rev Immunol:1–6
Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, Fujii Y, Yamashita H (2011) Distinct expressions of microRNAs that directly target estrogen receptor alpha in human breast cancer. Breast Cancer Res Treat 130:331–339
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, Pu PY, Cheng JQ, Kang CS (2010) MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9:229
Acknowledgments
Many thanks to the National Postdoctoral office and the Hongkong Scholars Association.
Funding
We acknowledge financial support from the Natural Science Foundation Project of Anhui Province (1508085QC63), and Key University Science Research Project of Anhui Province (KJ2017A021), and the Scientific Research Foundation and Academic & Technology Leaders Introduction Project (the 211 Project) of Anhui University (10117700023). Our work was also supported by the Hong Kong Scholars Program 2016 (XJ2016062) and National Basic Research Program of China (Grant No. 2013CB835100). Financial support by the Hong Kong Baptist University Strategic Development Fund (SDF) (SDF15-0324-P02(b) to A.L.) should also be acknowledged.
Author information
Authors and Affiliations
Contributions
KH, JFH, and APL designed the study. WXL, DGG, MTG, SDY, and ZF performed the experiments and/or data analysis. KH, WXL, DGG, and APL wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
Expression value distributions of the normalized datasets. The boxplots show the expression value distributions of the normalized GEO datasets (GSE17508, GSE19777, GSE19777, and GSE52674) that were selected for our integrated analysis associated with the five miRNAs. The red box represents case samples and the green box represents control samples. (PNG 7 kb)
Figure S2
Survival analysis of individual miRNAs based on subtypes. It showed the survival analysis of each miRNA using the profiles of patient subtypes including ER, PR, and HER2 positive or negative. (PDF 690 kb)
Figure S3
Transcriptional regulation of STAT1 among the five miRNAs. It shows the transcriptional regulation of STAT1 among the five miRNAs, including miR-21 knockdown, miR-210 overexpression, miR-22 knockout, miR-221 knockdown, and miR-222 knockdown. (PNG 5866 kb)
Figure S4
Survival analysis of 45 significant miRNA-gene interactions. Survival analysis of 45 significant miRNA-gene interactions. The red curves represent overall survival under low expression of both miRNAs and genes. The green curves represent overall survival under low expression of miRNAs but high expression of genes. The blue curves represent overall survival under high expression of miRNAs but low expression of genes. The purple curves represent overall survival under high expression of both miRNAs and genes. (PDF 694 kb)
Table S1
Differentially expressed genes in response to the regulation of the five miRNAs. Significantly upregulated or downregulated genes in response to the regulation of the five miRNAs, including miR-21 knockdown, miR-210 overexpression, miR-22 knockout, miR-221 knockdown, and miR-222 knockdown. (XLS 332 kb)
Table S2
KEGG pathway enrichment analysis for the five miRNAs. Details of the significantly enriched KEGG pathways associated with the regulation of the five miRNAs, including miR-21 knockdown, miR-210 overexpression, miR-22 knockout, miR-221 knockdown, and miR-222 knockdown. (XLS 31 kb)
Table S3
Transcription factors significantly related to the five mRNAs. Transcription factors significant related to the regulation of the five miRNAs, including miR-21 knockdown, miR-210 overexpression, miR-22 knockout, miR-221 knockdown, and miR-222 knockdown. (XLS 50 kb)
Rights and permissions
About this article
Cite this article
He, K., Li, WX., Guan, D. et al. Regulatory network reconstruction of five essential microRNAs for survival analysis in breast cancer by integrating miRNA and mRNA expression datasets. Funct Integr Genomics 19, 645–658 (2019). https://doi.org/10.1007/s10142-019-00670-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-019-00670-7