Abstract
The molecular mechanisms of shell formation and pigmentation are issues of great interest in molluscan studies due to the unique physical and biological properties of shells. The Yesso scallop, Patinopecten yessoensis, is one of the most important maricultural bivalves in Asian countries, and its shell color shows polymorphism. To gain more information about the underlying mechanisms of shell formation and pigmentation, this study presents the first analyses of histological and transcriptional differences between different mantle regions of the Yesso scallop, which are thought to be responsible for the formation of different shell layers. The results showed major microstructural differences between the edge and central mantles, which were closely associated with their functions. Different biomineralization-related GO functions, which might participate in the formation of different shell layers, were significantly enriched in the different mantle regions, indicating the different molecular functions of the two mantle regions in shell formation. The melanogenesis pathway, which controls melanin biosynthesis, was the most significantly enriched pathway in the DEGs between the two mantle regions, indicating its important role in shell pigmentation. Tyr, the key and rate-limiting gene in melanogenesis, was expressed at a remarkably high level in the central mantle, while the upstream regulatory genes included in melanogenesis were mainly upregulated in the edge mantle, suggesting the different molecular functions of the two mantle regions in shell pigmentation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mollusca, a highly species-rich phylum, is the largest group in the marine realm. One of the most obvious features of mollusks is that a great majority of them are shelled, including nearly all bivalves, most gastropods, and some cephalopods (Williams 2016). Shells are multilayered structures composed of 95 to 99% calcium carbonate crystals and less than 5% organic macromolecules (Zhang and Zhang 2006), and they often occur in fabulous and diverse colors, generally due to the presence of biological pigments, such as melanins, carotenoids, and tetrapyrroles (Comfort 1949a, b, c, 1951; Hedegaard et al. 2006; Williams 2016; Sun et al. 2017; Zhao et al. 2017). In recent decades, shells have drawn much attention from scientists in biology, ecology, evolution, and material science due to their unique physical and biological properties (Zhang and Zhang 2006). There are two major hotspots in the study of shells, which are the underlying mechanisms of shell formation and pigmentation; these topics are still far from fully understood because of their high complexity.
Underlying the vast phenotypic diversity of molluscan shells is an evolutionarily homologous organ, the mantle, which is in direct contact with the shell and produces shell pigments (Williams 2016). The mantle tissue mainly consists of two sections, the edge mantle and the central mantle, and the different mantle regions are thought to be responsible for different shell layers, with the edge mantle secreting the periostracal and prismatic layers and the central mantle secreting the nacreous layer (Reindl and Haszprunar 1996; Audino et al. 2015). With the prevalence of next-generation sequencing, transcriptome analyses have been extensively carried out for the mantle tissues of many molluscan species to explore the molecular mechanisms of shell formation or pigmentation, and a number of related genes have been identified, such as the shell formation-related genes calmodulin (Sun et al. 2015; Fang et al. 2011), lustrin A (Zhao et al. 2012), collagen (Fang et al. 2011), perlucin (Bai et al. 2013; Joubert et al. 2010; Zhao et al. 2012), Pif (Joubert et al. 2010; Kinoshita et al. 2011; Bai et al. 2013; Lemer et al. 2015), and glycoprotein (Kinoshita et al. 2011), and the shell pigmentation-related genes tyrosinase (Sun et al. 2016; Feng et al. 2015; Ding et al. 2015), MITF (Yue et al. 2015; Sun et al. 2016), Notch (Yue et al. 2015; Feng et al. 2015), and so on. However, most of these studies were confined to whole mantle tissues, neglecting the different molecular functions of the different mantle regions. Transcriptome analysis performed separately on different mantle regions has been reported in only the pearl oyster (Pinctada fucata martensii), but it found divergent expression levels of many genes in the different mantle regions (Kinoshita et al. 2011). In our previous study, shell infestation by Polydora caused different transcriptional changes in the edge and central mantle (Mao et al. 2018). In addition, studies of individual shell-related genes or proteins have also found expression differences among the different mantle regions or shell layers, as reviewed by Zhang and Zhang (2006), indicating the different molecular functions of different mantle regions. Therefore, performing more molecular research on different mantle regions will help us better understand the genetic basis of shell formation and pigmentation.
The Yesso scallop (Patinopecten yessoensis) is a large and old (dating back to ~ 350 Ma) group naturally living on the cold and stable ocean bottoms of the northwestern Pacific Ocean. It is one of the most important maricultural shellfish in Asian countries due to its high economic value. Many studies have been conducted on various aspects of the Yesso scallop due to its evolutionarily and economically important position, including genetic breeding (Dou et al. 2016; Zhao et al. 2017), genomics (Wang et al. 2017), transcriptomics (Hou et al. 2011; Wang et al. 2013; Zhang et al. 2018), and epigenetics (Wang et al. 2015; Li et al. 2019), which have provided abundant genetic resources for studies of the Yesso scallop. As a bivalve, the Yesso scallop has two shell pieces, with the left valve slightly smaller and flatter than the right valve. The shell of the Yesso scallop mainly contains three layers: the periostracal layer, prismatic layer, and nacreous layer (Pang et al. 2015). The shell color of the Yesso scallop represents an unambiguous polymorphism. Typically, most of the left valves are brown and distinctly different from the white right valves. In addition, a small percentage of individuals in the natural population possess white or orange left valves. All of the above features make the Yesso scallop an ideal model for the study on shell pigmentation and biomineralization. In recent years, some transcriptome studies have been performed of the mantle tissue of the Yesso scallop (Sun et al. 2015, 2016; Ding et al. 2015), but none have focused on the expression differences among different mantle regions, limiting the mechanistic study of shell formation and pigmentation in Yesso scallops. In the present study, the histological structure of the edge and central mantles of the Yesso scallop was observed, and the transcriptional differences between the two mantle regions were first analyzed with RNA-seq data. Genes and pathways related to shell formation and pigmentation were screened. The aim of this study was to explore the potential molecular functions of different mantle regions of the Yesso scallop and provide more information for mechanistic studies of shell formation and pigmentation.
Materials and Methods
Scallop Collection
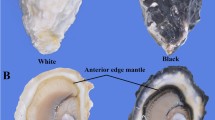
Two-year-old healthy Yesso scallops were collected from the Dalian Zhangzidao Sea area (Liaoning, China). The scallops were acclimated in the laboratory for one week prior to the experiments. Filtered and aerated seawater was maintained at approximately 8 °C, which is within the optimum temperature range for their growth. The edge and central mantles (EM and CM) of the left valves (as shown in Fig. 1) were separately sampled for histological sectioning and RNA extraction. No specific permits were required for the described field studies because the Yesso scallops used in the present study were cultured marine animals provided for market sale, and they are not part of an endangered or protected species. All experiments were conducted according to the regulations of the local and central government.
Histological Observation of the Edge and Central Mantles
Dissected specimens of the edge mantle and central mantle were fixed in Bouin’s solution. Then, the specimens were dehydrated in a graded ethanol series (80%, 95%, and 100%), cleared in xylene, and embedded in paraffin. Serial sections of 4 μm were produced on a Leica RM2255 microtome (Leica, Germany) and stained with hematoxylin and eosin (HE). Digital images were captured using a Leica DM4 B microscope equipped with a Leica DMC6200 camera.
Differential Gene Expression Analysis Between the Edge and Central Mantles
A total of 314,060,880 raw reads (156,785,500 for the edge mantle and 157,275,380 for the central mantle) of healthy Yesso scallops were downloaded from the NCBI database (accession numbers: SRR7287164, SRR7287165, SRR7287167, SRR7287170, SRR7287171, and SRR7287173); these reads were from a transcriptome project of the Yesso scallop in our previous study (Mao et al. 2018). Three biological replicates for each mantle region were included. The high-quality reads of each sample were mapped to the reference transcriptome constructed in our previous study by using Bowtie 2 software (http://bowtie-bio.sourceforge.net) (Langmead and Salzberg 2012), and the FPKM method (fragments per kilobase per million mapped reads) (Trapnell et al. 2010) was used to calculate the expression levels of the unigenes in each sample, which eliminated the effects of gene length and sequencing depth on the calculation of gene expression. A significance test for expression differences between the edge mantle and central mantle was conducted with negative binomial distribution hypothesis testing, and the false discovery rate (FDR) method was applied for multiple hypothesis testing to correct the significance levels (p values) and eliminate the influence of random fluctuations and errors. After calibration, a fold change over 2 and p ≤ 0.05 was set as the threshold for DEGs (differentially expressed genes).
Enrichment analyses were performed by mapping all DEGs to the GO and KEGG databases. GO functions or pathways with statistical significances (p values) lower than 0.05 as evaluated by the hypergeometric distribution test were considered to be enriched.
Sequence Identification and Characterization Analysis of the PyTyr Gene
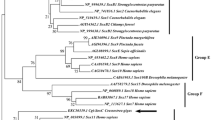
To identify the Tyr gene of the Yesso scallop, a BLASTN search was performed to align the cDNA sequence (CL14061Contig1) annotated with “putative tyrosinase-like protein tyr-3” in the transcriptome to the whole-genome sequences (Wang et al. 2017) with E value ≤ 1E−5. Gene structure was analyzed according to the genome sequence and annotation information. ORF (open reading frame) finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and DNAStar (version 7.1) were used to predict amino acid sequences. Then, the predicted amino acid sequence was confirmed by BLAST against the NCBI nonredundant protein sequence database. The conserved domains were identified by the simple modular architecture research tool (SMART) (http://smart.embl.de/). Multiple sequence alignment was performed using the ClustalW2 program (Larkin et al. 2007) with the Tyr protein sequences of P. yessoensis and other species, including Chlamys farreri, Crassostrea gigas, Hyriopsis cumingii, Pinctada margaritifera, Meretrix meretrix, Crassostrea virginica, Perna viridis, Homo sapiens, Mus musculus, Gallus gallus, and Danio rerio, which were downloaded from NCBI (http://www.ncbi.nlm.nih.gov) (the accession numbers of these sequences are listed in Supplementary Table S1), and pairwise sequence identity (percent identity) and genetic distance (divergence) among these species were calculated. A phylogenetic tree was constructed using MEGA 6 (Tamura et al. 2011) with the neighbor-joining method (Saitou and Nei 1987). Bootstrapping with 5000 replications was performed to evaluate the phylogenetic tree.
Quantitative Real-time PCR Analysis of the PyTyr Gene
Total RNA of the edge and central mantle was extracted by using an RNAprep Pure Tissue Kit (Tiangen). The quantity and quality of the total RNA were determined by a Micro-Spectrophotometer NV3000 (Vastech) and agarose gel electrophoresis. First-strand cDNA was synthesized using a PrimeScript RT reagent Kit (TaKaRa) following the manufacturer’s protocol. All of the cDNA products were diluted to 200 ng/μl for use as qRT-PCR templates. The β-actin gene was selected as a reference gene. The qRT-PCR was performed in an ABI 7500 Real-time PCR System (Life Technologies) with a volume of 20 μl, which contained 1 × SYBR Green Master Mix (TaKaRa), 0.2 μM of each primer, 2.0 μl of cDNA template, and 0.4 μl of ROX Reference. The running program was as follows: 95 °C for 10 s, followed by 40 cycles of 95 °C for 10 s and 60 °C for 32 s. The primers used in the present study were designed by Primer Premier 5.0 software, and the sequences were as follows: β-actin-F: 5′-CCAAAGCCAACAGGGAAAAG-3′, β-actin-F: 5′-TAGATGGGGACGGTGTGAGTG-3′; Tyr-F: 5′-AGCGACAATGATGTGCGTAA-3′, Tyr-R: 5′-AGATGGCTCTATGGAGTTGA-3′. The specificity of the primers was assessed by alignment with the Yesso scallop genome. Melting curve analysis was also performed to verify that each primer set amplified a single product. Three technical replicates were performed for each reaction, and three individuals were used as biological replicates. The data from qRT-PCR were analyzed by the 2−ΔΔCT method (Livak and Schmittgen 2001) to evaluate the relative gene expression levels of Tyr in the different mantle regions of Yesso scallops. Statistical analysis of the data was performed with SPSS (version 19.0) software using the independent T test. Differences were considered significant at p ≤ 0.05.
Results
Histological Structure of the Edge and Central Mantles
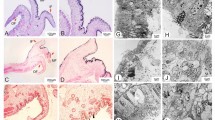
The mantle tissue of the Yesso scallop mainly consists of two parts: the edge mantle and the central mantle (Fig. 1). The microstructures of the edge mantle and central mantle were observed by histological study (Fig. 2). The edge mantle was a muscular, dissociative, and thick membrane (Fig. 1) composed of the secretory outer fold (of), the sensory middle fold (mf), and the muscular inner fold (if) (Fig. 2a, b). These folds were made up of the outer epithelium, inner epithelium, and connective tissues. The outer fold was the first finger-like protuberance close to the shell, and a periostracal groove (pg) was present between the outer fold and the middle fold (Fig. 2a). The middle fold was obviously composed of several protuberances, and most of them had abundant nerve fibers distributed between the two epithelia (Fig. 2d), while the inner fold was much larger than the other folds and had very developed muscle tissues (Fig. 2b). In addition, a large, rounded ganglion (ga) was observed in the region where the three folds converged (Fig. 2e). In contrast to the edge mantle, the central mantle was a transparent and thin membrane (Fig. 1) that adhered to the inner surface of the shell and was mainly composed of the outer epithelium, inner epithelium, and loose connective tissues (containing hemocytes, nerve fiber bundles, and a few muscle fibers) (Fig. 2c). Bushy and short cilia were distributed in the inner epithelium of the central mantle (Fig. 2g), while in the edge mantle, relatively sparse cilia were found in only the inner epithelium of the outer fold and outer epithelium of the middle fold (Fig. 2f), i.e., where the periostracal groove was located.
Histological sections of the edge mantle and the central mantle of P. yessoensis. a The edge mantle with outer fold, middle fold, and partial inner fold. b The inner fold of the edge mantle. c The central mantle. d The developed nerve fibers distributed in the middle fold. e The ganglion in the edge mantle. f The cilia distributed in the outer epithelium of the middle fold. g The cilia distributed in the inner epithelium of the central mantle. Abbreviations: of outer fold, mf middle fold, if inner fold, pg periostracal groove, nf nerve fiber, mf muscle fiber, he hemocytes
Differential Gene Expression Analysis Between the Edge and Central Mantles
Differential gene expression analysis was conducted between the edge mantle and the central mantle with RNA-seq data obtained from our previous Yesso scallop transcriptome project (Mao et al. 2018). A total of 3350 genes were differentially expressed between the two mantle regions, with 1730 upregulated in the edge mantle and 1620 in the central mantle (p ≤ 0.05) (Fig. 3).
To analyze the functions of these differentially expressed genes (DEGs), GO enrichment analysis was separately carried out for the up- and downregulated genes, the results of which are summarized in Table 1. For the upregulated genes in the edge mantle, one of the most enriched GO terms was related to nervous system constituent and neurotransmission, followed by terms related to muscular constituents, development, and activities. In addition, many GO functions related to responses to environmental stimuli, such as temperature, salt stress, wounding, fungi, and bacteria, were also significantly enriched. Most notably, many GO terms related to biomineralization were also significantly enriched for the upregulated genes in the edge mantle, e.g., “calcium ion binding,” “cellular calcium ion homeostasis,” “calcium ion transport,” “positive regulation of calcium ion transport,” “ossification,” “proteinaceous extracellular matrix,” “extracellular matrix,” “regulation of bone mineralization,” “collagen binding,” and “chitin binding.” Among these DEGs, many genes were related to shell formation, such as genes for “cartilage matrix protein,” “collagen,” “perlucin,” “pif,” “fibrinogen,” and “glycoprotein” (Table 3). For the upregulated genes in the central mantle (Table 2), the most enriched GO terms were those related to cilium components, assembly, and movement. Some nervous system–related terms were also enriched, but these were much fewer than in the edge mantle. In addition, the biomineralization-related terms were mainly related to calcium activity, such as “calcium ion binding,” “calmodulin binding,” “calcium channel activity,” and “positive regulation of cytosolic calcium ion concentration.”
KEGG Enrichment Analysis of DEGs Between the Edge and Central Mantles
First, KEGG enrichment analysis was performed for all the differentially expressed genes between the edge and central mantle. A total of 54 pathways were significantly enriched (p ≤ 0.05), and those ranked in the top 20 are shown in Fig. 4a. Among these pathways, “melanogenesis” was notably the most significantly enriched pathway (marked with a red star); this pathway controls the synthesis of melanins. Then, KEGG enrichment analysis was separately carried out for the upregulated genes in the edge and central mantles, and 63 and 36 pathways were separately enriched (Fig. 4b, c, marked with red stars). Notably, the pathway of melanogenesis was significantly enriched in both mantle regions. The genes included in the melanogenesis pathway and the genes detected to have altered regulation are marked in the pathway map (map04916) (Fig. 5). Among these genes, Wnt (CL2260Contig1, CL5293Contig1, CL33407Contig1, CL39128Contig1, CL15607Contig1, CL3041Contig1, CL58260Contig1), GNAS (guanine nucleotide-binding protein G(s) subunit alpha, CL54338Contig1), GSK-3β (glycogen synthase kinase-3 beta, CL33363Contig1), and MAPK (Mitogen-activated protein kinase, CL45618Contig1) were upregulated in the edge mantle (marked with red); Tyr (tyrosinase, CL14061Contig1) was upregulated in the central mantle (marked with green), and FZD (Frizzled: CL34951Contig1, upregulated in the edge mantle, and CL26136Contig1 and CL3646Contig1, upregulated in the central mantle) and CAM (calmodulin: CL33708Contig1, CL56040Contig1, CL32987Contig1, CL34373Contig1, comp134620_c1_seq1_3, CL47922Contig1, CL12604Contig1, CL14037Contig1, CL62949Contig1, CL28607Contig1 and comp127217_c0_seq1_2, upregulated in the edge mantle, and CL11292Contig1, CL26634Contig1, CL30465Contig1, CL36393Contig1, and CL50141Contig1, upregulated in the central mantle) both showed altered regulation in both mantle regions (marked with yellow). Additionally, another pathway, “tyrosine metabolism,” which was also related to melanin synthesis, was significantly enriched among the upregulated genes in the central mantle (Fig. 4c, marked with a blue star). In addition, “calcium signaling pathway” was enriched in both mantle regions, and “mineral absorption” was enriched in the central mantle (Fig. 4, marked with a green star), and these pathways were probably related to shell formation.
Sequence and Expression Characterization of the Gene PyTyr
Tyrosinase (Tyr) is the key enzyme during the synthesis of melanins, which has been widely studied in vertebrates. In this study, a gene for Tyr (CL14061Contig1, named PyTyr) was detected as significantly differentially expressed between the edge and central mantle using RNA-seq data. To explore the molecular function of PyTyr in scallop pigmentation and test its expression level in different mantle regions of Yesso scallop, sequence characterization of PyTyr was analyzed aided by the whole-genome sequence of Yesso scallop (Wang et al. 2017), and its expression level was validated separately in the edge and central mantle by qRT-PCR.
PyTyr was confirmed to be present in the genome of Yesso scallop by BLAST search of the transcript against the genome sequence, and the complete DNA sequence of PyTyr was 6667 bp, including 3 exons and 2 introns with an average length of 647 bp and 2201 bp. The GT-AG rule was used to distinguish intron-exon boundaries. The full-length cDNA of PyTyr was 2265 bp with a 137 bp 5′UTR, 187 bp 3′UTR, and 1941 bp ORF encoding 646 amino acids. Two unambiguous functional domains, a signal peptide domain and a tyrosinase domain, were recognized in the amino acid sequence of PyTyr in the 1–20 and 124–294 residue regions. The gene structure of PyTyr is shown in Fig. 6a. Two highly conserved copper-binding sites, the Cu(A)-binding site and the Cu(B)-binding site, which are characterized by the H1(n)-H2(8)-H3 and H1(3)-H2(n)-H3 motifs, were found in the tyrosinase domain, and six conserved histidine residues were detected in PyTyr (Fig. 6b), which confirmed the identification of PyTyr in the present study. Multiple sequence alignment analysis (Supplementary Table S2) showed that the amino acid identity of Tyr between Yesso scallop and other mollusks ranged from 16.9% (P. yessoensis and M. meretrix) to 26.4% (P. yessoensis and C. farreri), and the conservation of Tyr among molluscan species (16.9–59.4%) was much lower than that among vertebrates (60.5–86.2%). These results were consistent with the phylogenetic analysis, the tree of which clustered into two groups representing vertebrates and Mollusca (Fig. 6c). The expression levels of PyTyr in different mantle regions as detected by qRT-PCR were consistent with the RNA-seq data; significantly more PyTyr mRNAs were detected in the central mantle than in the edge mantle (p < 0.01).
Sequence characterization and expression analysis of the gene PyTyr. a The gene structure of PyTyr. b Multiple sequence alignment analysis of the Cu(A)-binding site and Cu(B)-binding site among different mollusks. Red triangles represent the six conserved histidine residues. c Phylogenetic analysis of Tyr among different species. d Expression levels of PyTyr in the edge and central mantles as measured by RNA-seq and qRT-PCR
Discussion
The Yesso scallop has a typical mantle structure, which is mainly composed of the edge mantle and the central mantle. The morphological and histological structure of the edge mantle is more complex than that of the central mantle, and their structures are closely associated with their functions. At the molecular level, differential gene expression analysis with the RNA-seq data showed that many differentially expressed genes were related to tissue composition, which was consistent with the histological observations. For example, we found that the edge mantle possessed a much more developed nerve distribution than the central mantle based on histological observations. First, a large, rounded ganglion was observed in the edge mantle (Fig. 2e). It is the most significant nerve in the scallop mantle margin and is responsible for the innervation of the mantle folds and organs (Audino et al. 2015). Second, developed nerve fibers were found in the middle fold, which extended to the epithelial cells (Fig. 2d), allowing for sensory functions. Meanwhile, in the differential gene expression analysis between the two mantle regions, among the most enriched GO functions of the upregulated genes in the edge mantle were those related to the nervous system (Table 1), such as “postsynaptic membrane,” “synaptic transmission,” “neuropeptide signaling pathway,” “axon guidance,” and “synapse.”. Most likely because of the developed nerves in the edge mantle, especially in the middle fold, it is more sensitive to environmental changes and stimulations, and as a result, many functions related to response reaction were also significantly enriched among the upregulated genes in the edge mantle (Table 1). In addition, it has been reported that both shell growth and pigmentation were under neurosecretory control (Boettiger et al. 2009; Budd et al. 2014); thus, the developed nerve system in the Yesso scallop mantle may also play an important role in shell formation and pigmentation. In the central mantle, the most enriched GO functions among the upregulated genes were those associated with cilium activity (Table 2). Histologically, bushy cilia were found in the outer epithelium of the central mantle (Fig. 2g), which probably took part in substance (e.g., shell-related) absorption, secretion, and transportation, as many secretory cells were discovered under the cilia layer and some related signaling pathways, such as “salivary secretion” and “mineral absorption,” were significantly enriched in the KEGG analysis; in contrast, in the edge mantle, relatively sparse cilia were mainly observed in the periostracal groove region, which was responsible for periostracum secretion (Wilbur and Saleuddin 1983; Audino et al. 2015), so the cilia here were probably related to the secretion and transportation of the shell components.
Functions related to shell formation were of special interest. In the present study, biomineralization-associated GO functions and KEGG pathways were significantly enriched in both the edge and central mantles, especially in the edge mantle (Tables 1 and 2). Among these functions, two aspects were included, one of which was related to “calcium” while the other was related to “organic shell matrix.” Calcium is an important component of the shell, and the calcium metabolic process plays an important role in regulating shell formation (Liu et al. 2007; Huang et al. 2007; Kinoshita et al. 2011; Sun et al. 2015). In our KEGG enrichment analysis, “calcium signaling pathway” was significantly enriched in both mantle regions (Fig. 4), indicating its important role in all shell layer formation. In the edge mantle, the GO functions “calcium ion binding,” “cellular calcium ion homeostasis,” “calcium ion transport,” and “positive regulation of calcium ion transport” were significantly enriched, and all have strong relationships with calcium ion transport. In the central mantle, in addition to “calcium ion binding,” the additional functions “calmodulin binding,” “calcium channel activity,” and “positive regulation of cytosolic calcium ion concentration” were significantly enriched; these pathways were probably related to the processes of calcium ion absorption and storage, as the pathway “mineral absorption” was also enriched in the central mantle (Fig. 4c). That “calcium ion binding” was obviously enriched in both the mantle regions is probably because it was the basic process for other calcium ion activities, and the differences in calcium ion activities between the two mantle regions probably suggested their different molecular roles in shell formation.
Despite its low content in the shell, the organic shell matrix, including proteins, glycoproteins, polysaccharides, and lipids, is thought to control the formation of calcium carbonate crystals, such as the nucleation, growth, and spatial orientation of calcium carbonate crystals, and thus to be responsible for their extraordinary properties (Wheeler and Sikes 1984; Belcher et al. 1996; Naka and Chujo 2001; Blank et al. 2003; Fu et al. 2005; Addadi et al. 2006). The different crystal polymorphisms and microstructures of the different shell layers are controlled by proteins secreted from different mantle regions, with the outer calcite prismatic layer formed by the edge mantle and the inner aragonite nacreous layer formed by the central mantle (Zhang and Zhang 2006). In the present study, GO functions related to the organic shell matrix and biomineralization process were significantly enriched mainly in the edge region of the mantle, such as “proteinaceous extracellular matrix,” “extracellular matrix,” “ossification,” “regulation of bone mineralization,” “collagen binding,” and “chitin binding,” consistent with the fact that shell formation is more strongly activated in the edge mantle than in the central mantle (Lydie et al. 2001; Kinoshita et al. 2011). Moreover, some genes for shell matrix proteins were included in these functions, such as “cartilage matrix protein,” “collagen,” “perlucin,” “pif,” “fibrinogen,” and “glycoprotein,” which have also been reported in other studies of shell formation (Miyamoto et al. 2002; Weiss et al. 2000; Kröger 2009; Suzuki et al. 2013; Kinoshita et al. 2011; Areva et al. 2002; Luo et al. 2015). The expression levels of these genes were significantly higher in the edge mantle than in the central mantle (fold change > 2 and p ≤ 0.05, Table 3), which probably indicated that they might be involved in regulation the formation of the prismatic layer in Yesso scallop. However, “perlucin” and “pif” have been reported as important nacre-specific genes (Weiss et al. 2000; Kröger 2009), in contrast to the present study. The likely reasons are that functional differences in these genes may exist among different mollusks due to gene differentiation or that these genes can participate in the formation of both the prismatic and nacreous layers. A similar situation has been found in other studies. Some prismatic genes, including MSI31 (Sudo et al. 1997), aspein (Tsukamoto et al. 2004), and prismalin-14 (Suzuki et al. 2004), were expressed at similar levels in the edge mantle and central mantle according to Kinoshita et al. (2011). Gene duplication often causes divergent expression of paralogous genes (Force et al. 1999). In Pinctada fucata martensii, different isoforms of N16, another nacreous gene (Samata et al. 1999), were specifically expressed in different mantle regions, with N16.6 specific to the mantle edge and N16.7 specific to the central mantle (Kinoshita et al. 2011). The functions of the shell formation-related genes detected in our study require further investigation.
The coloration of shells is most commonly due to the presence and distribution of biological pigments (Williams 2016). Melanins, as one of the most widespread pigments in nature, have also been detected in molluscan shells, including that of the Yesso scallop (Sun et al. 2017). The molecular mechanism of melanin biosynthesis is well understood in vertebrates, but understanding remains limited in mollusks. Melanin biosynthesis is initiated by the oxidation of tyrosine to dopaquinone, catalyzed by the key and rate-limiting enzyme tyrosinase (Tyr) (Chang 2012), which is directly and positively regulated by the transcription factor MITF, the master regulator of melanogenesis (Vance and Goding 2004). MITF is the target of many regulatory pathways involved in melanogenesis, such as the cAMP signaling pathway, Wnt signaling pathway, and ERK (MAPK) signaling pathway (Fig. 5). In the present study, melanogenesis was the most significantly enriched pathway in the DEGs between the edge and central mantles, illustrating its important role in shell pigmentation. Genes included in melanogenesis and highly expressed in the edge mantle were mainly upstream regulatory genes in the cAMP, Wnt, and ERK signaling pathways. However, Tyr, the key gene for melanogenesis, was significantly upregulated in the central mantle, and this finding was validated by sequence analysis and qRT-PCR. In our previous study, PyMITF had the same expression trend as PyTyr, being expressed at a significantly higher level in the central mantle than in the edge mantle (Mao et al. 2019). In addition, the pathway of tyrosine metabolism was significantly enriched in the central mantle (Fig. 4c), which provided the raw material tyrosine for melanin biosynthesis. Therefore, these findings probably suggest the different molecular roles of different mantle regions in shell pigmentation; the main melanin synthesis process probably occurs in the central mantle cells and is regulated by products from the edge mantle, although more studies are required to validate the conclusion.
In conclusion, histological and transcriptional difference analyses among different mantle regions were first conducted in the Yesso scallop. The results showed that large microstructural differences existed between the edge and central mantles, which were closely associated with their functions. Different biomineralization-related GO functions were significantly enriched in the different mantle regions, which might participate in the formation of different shell layers, and indicated the different molecular functions of the two mantle regions in shell formation. The pathway of melanogenesis, which controls melanin biosynthesis, was the most significantly enriched pathway among the DEGs between the two mantle regions, indicating its important role in shell pigmentation. Tyr, the key and rate-limiting gene in melanogenesis, was expressed at a remarkably higher level in the central mantle, while the upstream regulatory genes included in melanogenesis were mainly upregulated in the edge mantle, suggesting the different molecular functions of the two mantle regions in shell pigmentation. The present study provides valuable information for mechanistic molecular studies of shell formation and pigmentation.
References
Addadi L, Joester D, Nudelman F, Weiner S (2006) Mollusk shell formation: a source of new concepts for understanding biomineralization processes. Chem Eur J 12:980–987
Areva S, Peltola T, Säilynoja E, Laajalehto K, Linden M, Rosenholm JB (2002) Effect of albumin and fibrinogen on calcium phosphate formation on sol-gel-derived titania coatings in vitro. Chem Mater 14(4):1614–1621
Audino JA, Marian JEA, Wanninger A, Lopes SG (2015) Mantle margin morphogenesis in Nodipecten nodosus (Mollusca: Bivalvia): new insights into the development and the roles of bivalve pallial folds. BMC Dev Biol 15:22
Bai Z, Zheng H, Lin J, Wang G, Li J (2013) Comparative analysis of the transcriptome in tissues secreting purple and white nacre in the pearl mussel Hyriopsis cumingii. PLoS One 8:e53617
Belcher AM, Wu XH, Christensen RJ, Hansma PK, Stucky GD, Morse DE (1996) Control of crystal phase switching and orientation by soluble mollusc-shell proteins. Nature 381:56–58
Blank S, Arnoldi M, Khoshnavaz S, Treccani L, Kuntz M, Mann K, Grathwohl G, Fritz M (2003) The nacre protein perlucin nucleates growth of calcium carbonate crystals. J Microsc 212(3):280–291
Boettiger A, Ermentrout B, Oster G (2009) The neural origins of shell structure and pattern in aquatic mollusks. Proc Natl Acad Sci 106:6837–6842
Budd A, McDougall C, Green K, Degnan BM (2014) Control of shell pigmentation by secretory tubules in the abalone mantle. Front Zool 11:62
Chang TS (2012) Natural melanogenesis inhibitors acting through the down-regulation of tyrosinase activity. Materials 5:1661–1685
Comfort A (1949a) Acid-soluble pigments of shells. 1. The distribution of porphyrin fluorescence in molluscan shells. Biochem J 44:111–117
Comfort A (1949b) Acid-soluble pigments of molluscan shells. 2. Pigments other than porphyrins. Biochem J 45:199–204
Comfort A (1949c) Acid soluble pigments of molluscan shells. 3. The indigoid character of the blue pigment of Haliotis cracherodii Leach. Biochem J 45:204–208
Comfort A (1951) The pigmentation of molluscan shells. Biol Rev 26:285–301
Ding J, Zhao L, Chang Y, Zhao W, Du Z, Hao Z (2015) Transcriptome sequencing and characterization of Japanese scallop Patinopecten yessoensis from different shell color lines. PLoS One 10:e0116406
Dou J, Li X, Fu Q, Jiao W, Li Y, Li T, Wang Y, Hu X, Wang S, Bao Z (2016) Evaluation of the 2b-RAD method for genomic selection in scallop breeding. Sci Rep 6:19244
Fang D, Xu G, Hu Y, Pan C, Xie L, Zhang R (2011) Identification of genes directly involved in shell formation and their functions in pearl oyster, Pinctada fucata. PLoS One 6:e21860
Feng D, Li Q, Yu H, Zhao X, Kong L (2015) Comparative transcriptome analysis of the Pacific oyster Crassostrea gigas characterized by shell colors: identification of genetic bases potentially involved in pigmentation. PLoS One 10:e0145257
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545
Fu G, Valiyaveettil S, Wopenka B, Morse DE (2005) CaCO3 biomineralization: acidic 8-kDa proteins isolated from aragonitic abalone shell nacre can specifically modify calcite crystal morphology. Biomacromolecules 6:1289–1298
Hedegaard C, Bardeau J-F, Chateigner D (2006) Molluscan shell pigments: an in situ resonance Raman study. J Molluscan Stud 72:157–162
Hou R, Bao Z, Wang S, Su H, Li Y, Du H, Hu J, Wang S, Hu X (2011) Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PLoS One 6:e21560
Huang J, Zhang C, Ma Z, Xie L, Zhang R (2007) A novel extracellular EF-hand protein involved in the shell formation of pearl oyster. Biochim Biophys Acta Gen Subj 1770:1037–1044
Joubert C, Piquemal D, Marie B, Manchon L, Pierrat F, Zanella-Cléon I, Cochennec-Laureau N, Montagnani C (2010) Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics 11:613
Kinoshita S, Wang N, Inoue H, Maeyama K, Okamoto K, Nagai K, Kondo H, Hirono I, Asakawa S, Watabe S (2011) Deep sequencing of ESTs from nacreous and prismatic layer producing tissues and a screen for novel shell formation-related genes in the pearl oyster. PLoS One 6:e21238
Kröger N (2009) The molecular basis of nacre formation. Science 325:1351–1352
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lemer S, Saulnier D, Gueguen Y, Planes S (2015) Identification of genes associated with shell color in the black-lipped pearl oyster, Pinctada margaritifera. BMC Genomics 16:568
Li Y, Zhang L, Li Y, Li W, Guo Z, Li R, Hu X, Bao Z, Wang S (2019) Dynamics of DNA methylation and DNMT expression during gametogenesis and early development of scallop Patinopecten yessoensis. Mar Biotechnol 21:196–205
Liu H, Liu S, Ge Y, Liu J, Wang X, Xie L, Zhang R, Wang Z (2007) Identification and characterization of a biomineralization related gene PFMG1 highly expressed in the mantle of Pinctada fucata. Biochemistry 46:844–851
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408
Luo YJ, Takeuchi T, Koyanagi R, Yamada L, Kanda M, Khalturina M, Fujie M, S-i Y, Endo K, Satoh N (2015) The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat Commun 6:8301
Lydie MAO, Golubic S, Le Campion-Alsumard T, Payri C (2001) Developmental aspects of biomineralisation in the Polynesian pearl oyster Pinctada margaritifera var. cumingii. Oceanol Acta 24:37–49
Mao J, Zhang W, Zhang X, Tian Y, Wang X, Hao Z, Chang Y (2018) Transcriptional changes in the Japanese scallop (Mizuhopecten yessoensis) shellinfested by Polydora provide insights into the molecular mechanism of shell formation and immunomodulation. Sci Rep 8:17664
Mao J, Zhang X, Zhang W, Tian Y, Wang X, Hao Z, Chang Y (2019) Genome-wide identification, characterization and expression analysis of the MITF gene in Yesso scallops (Patinopecten yessoensis) with different shell colors. Gene 688:155–162
Miyamoto H, Hamaguchi M, Okoshi K (2002) Analysis of genes expressed in the mantle of oyster Crassostrea gigas. Fish Sci 68:651–658
Naka K, Chujo Y (2001) Control of crystal nucleation and growth of calcium carbonate by synthetic substrates. Chem Mater 13:3245–3259
Pang Y, Ding J, Tian Y, Hao Z, Chang Y (2015) Analysis of shell microstructure and five surface elements of Patinopecten yessoensis at different ages. Mar Sci 39:28–34
Reindl S, Haszprunar G (1996) Fine structure of caeca and mantle of arcoid and limopsoid bivalves (Mollusca: Pteriomorpha). Veliger 39:101–116
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Samata T, Hayashi N, Kono M, Hasegawa K, Horita C, Akera S (1999) A new matrix protein family related to the nacreous layer formation of Pinctada fucata. FEBS Lett 462:225–229
Sudo S, Fujikawa T, Nagakura T, Ohkubo T, Sakaguchi K, Tanaka M, Nakashima K, Takahashi T (1997) Structures of mollusc shell framework proteins. Nature 387:563–564
Sun X, Yang A, Wu B, Zhou L, Liu Z (2015) Characterization of the mantle transcriptome of Yesso scallop (Patinopecten yessoensis): identification of genes potentially involved in biomineralization and pigmentation. PLoS One 10:e0122967
Sun X, Liu Z, Zhou L, Wu B, Dong Y, Yang A (2016) Integration of next generation sequencing and EPR analysis to uncover molecular mechanism underlying shell color variation in scallops. PLoS One 11:e0161876
Sun X, Wu B, Zhou L, Liu Z, Dong Y, Yang A (2017) Isolation and characterization of melanin pigment from Yesso scallop Patinopecten yessoensis. J Ocean Univ China 16:279–284
Suzuki M, Murayama E, Inoue H, Ozaki N, Tohse H, Kogure T, Nagasawa H (2004) Characterization of Prismalin-14, a novel matrix protein from the prismatic layer of the Japanese pearl oyster (Pinctada fucata). Biochem J 382:205–213
Suzuki M, Iwashima A, Kimura M, Kogure T, Nagasawa H (2013) The molecular evolution of the Pif family proteins in various species of mollusks. Mar Biotechnol 15:145–158
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg S, Wold B, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Tsukamoto D, Sarashina I, Endo K (2004) Structure and expression of an unusually acidic matrix protein of pearl oyster shells. Biochem Biophys Res Commun 320:1175–1180
Vance KW, Goding CR (2004) The transcription network regulating melanocyte development and melanoma. Pigment Cell Res 17:318–325
Wang S, Hou R, Bao Z, Du H, He Y, Su H, Zhang Y, Fu X, Jiao W, Li Y, Zhang L, Wang S, Zhang L (2013) Transcriptome sequencing of Zhikong scallop (Chlamys farreri) and comparative transcriptomic analysis with Yesso scallop (Patinopecten yessoensis). PLoS One 8:e63927
Wang S, Lv J, Zhang L, Dou J, Sun Y, Li X, Fu X, Dou H, Mao J, Hu X, Bao Z (2015) MethylRAD: a simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes. Open Biol 5:150130
Wang S, Zhang J, Jiao W, Li J, Xun X, Sun Y, Guo X, Huan P, Dong B, Zhang L, Hu X, Sun X, Wang J, Zhao C, Wang Y, Wang D, Huang X, Wang R, Lv J, Li Y, Zhang Z, Liu B, Lu W, Hui Y, Liang J, Zhou Z, Hou R, Li X, Liu Y, Li H, Ning X, Lin Y, Zhao L, Xing Q, Dou J, Li Y, Mao J, Guo H, Dou H, Li T, Mu C, Jiang W, Fu Q, Fu X, Miao Y, Liu J, Yu Q, Li R, Liao H, Li X, Kong Y, Jiang Z, Chourrout D, Li R, Bao Z (2017) Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat Ecol Evol 1:0120
Weiss IM, Kaufmann S, Mann K, Fritz M (2000) Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusc Haliotis laevigata. Biochem Biophys Res Commun 267:17–21
Wheeler AP, Sikes CS (1984) Regulation of carbonate calcification by organic matrix. Am Zool 24:933–944
Wilbur KM, Saleuddin ASM (1983) Shell formation. In: Saleuddin ASM, Wilbur KM (eds) The mollusca, vol 4. Academic Press, New York, pp 235–287
Williams ST (2016) Molluscan shell colour. Biol Rev 92(2):1039–1058
Yue X, Nie Q, Xiao G, Liu B (2015) Transcriptome analysis of shell color-related genes in the clam Meretrix meretrix. Mar Biotechnol 17:364–374
Zhang C, Zhang R (2006) Matrix proteins in the outer shells of molluscs. Mar Biotechnol 8:572–586
Zhang M, Wang Y, Li Y, Li W, Li R, Xie X, Wang S, Hu X, Zhang L, Bao Z (2018) Identification and characterization of neuropeptides by transcriptome and proteome analyses in a bivalve mollusc Patinopecten yessoensis. Front Genet 9:197
Zhao X, Wang Q, Jiao Y, Huang R, Deng Y, Wang H, Du X (2012) Identification of genes potentially related to biomineralization and immunity by transcriptome analysis of pearl sac in pearl oyster Pinctada martensii. Mar Biotechnol 14:730–739
Zhao L, Li Y, Li Y, Yu J, Liao H, Wang S, Lv J, Liang J, Huang X, Bao Z (2017) A genome-wide association study identifies the genomic region associated with shell color in yesso scallop, Patinopecten yessoensis. Mar Biotechnol 19:301–309
Acknowledgments
We thank Dalian Zhangzidao Fishery Group Co., Ltd. (Dalian, China) for providing the scallop materials.
Funding
This project was supported by the National Natural Science Foundation of China (31702342), the Major Science and Technology Research Project of Liaoning Province (2017203003), and the Doctoral Startup Foundation of Liaoning Province (20170520095).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: JM and YC. Prepared the samples: XW, JS, and DY. Performed the experiments: JM, WZ, and YT. Analyzed the data: JM, WZ, ZH, and BH. Wrote the paper: JM and YC.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 80 kb)
Rights and permissions
About this article
Cite this article
Mao, J., Zhang, W., Wang, X. et al. Histological and Expression Differences Among Different Mantle Regions of the Yesso Scallop (Patinopecten yessoensis) Provide Insights into the Molecular Mechanisms of Biomineralization and Pigmentation. Mar Biotechnol 21, 683–696 (2019). https://doi.org/10.1007/s10126-019-09913-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-019-09913-x