Abstract
All-female lines of fish are created by crossing sex reversed (XX genotype) males with normal females. All-female lines avoid the deleterious phenotypic effects that are typical of precocious maturation in males. To determine whether all-female and mixed sex populations of rainbow trout (Oncorhynchus mykiss) differ in performance, we compared the growth and gene expression profiles in progeny groups produced by crossing a XX male and a XY male to the same five females. Body weight and length were measured in the resulting all-female (XX) and mixed sex (XX/XY) offspring groups. Microarray experiments with liver and white muscle were used to determine if the gene expression profiles of large and small XX offspring differ from those in large and small XX/XY offspring. We detected no significant differences in body length and weight between offspring groups but XX offspring were significantly less variable in the value of these traits. A large number of upregulated genes were shared between the large XX and large XX/XY offspring; the small XX and small XX/XY offspring also shared similar expression profiles. No GO category differences were seen in the liver or between the large XX and large XX/XY offspring in the muscle. The greatest differences between the small XX and small XX/XY offspring were in the genes assigned to the “small molecule metabolic process” and “cellular metabolic process” GO level 3 categories. Similarly, genes within these categories as well as the category “macromolecule metabolic process” were more highly expressed in small compared to large XX fish.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In fishes, differences in growth between the sexes at the onset of sexual maturation can be the result of differential allocation of energy into gametic tissue (Ji et al. 2011; Liu et al. 2012). In salmonids, early rapid growth is associated with an increased rate of early maturation, particularly in males (Berejikian et al. 2010; Guiry et al. 2010). Early maturation is undesirable in aquaculture as the energy demands of gamete production result in muscle deterioration (Salem et al. 2006a, b). One strategy to reduce the frequency of early maturation has been to produce all-female lines. Within XY sex-determining species of fishes, all-female lines can be created by crossing a genotypic female that has been made into a phenotypic male (neomale) by androgen exposure (XX genotype) with a normal female (Nynca et al. 2012a).
The potential use of neomales in aquaculture has prompted studies of the performance of their progeny (all-females) relative to that of typical XY males (mixed sex progeny). The offspring of sex-reversed males and mixed sex control populations appear to show similar fertility and early survival based on studies with crucian carp, Carassius carassius (Luo et al. 2011), Eurasian perch, Perca fluviatilis (Stejskal et al. 2009) and Chinook salmon, Oncorhynchus tshawytscha (Hopkins and Todd 1991). However, the relative growth of all-female and mixed sex offspring can vary in magnitude and direction depending on species and strain. For instance, all-female offspring of Chinook salmon had slower growth compared to mixed sex controls (Hopkins and Todd 1991) while the opposite relationship was detected in Eurasian perch (Stejskal et al. 2009). In common carp Cyprinus curpio L., all-female offspring from one strain showed a higher growth rate compared to mixed sex controls while no differences were detected in another strain (Kocour et al. 2005). Although rainbow trout males often have elevated growth rates compared to females, growth rates can vary among families leading to similar performance in the sexes (Naevdal et al. 1979). Therefore, all-female lots of rainbow trout could have lower or possibly comparable growth rates compared to mixed sex family lots.
Comparisons of the biochemical and physiological profiles and the growth of XY males and XX males from different species have yielded varied results. Studies with salmonids report that XX males and XY males have similar androgenic and oestrogenic profiles, fertilization rates (Nynca et al. 2012b), and weights (Hopkins and Todd 1991). In contrast, XX males have more concentrated sperm, with a slower swimming speed (Nynca et al. 2012a), lower 20β-dihydroxy-4-pregnen-3-one levels (Espinosa et al. 2011), and higher antitrypsin activity, lactate dehydrogenase levels, protein concentration, and osmolality than XY males (Nynca et al. 2012a, b). Studies with non-salmonids suggest that XY males and XX males may not show consistent differences in performance. No significant differences were found between normal male and XX male Eurasian perch when gonadosomatic index, fertilization rate, sex steroid levels, and sperm concentration and motility were compared (Rougeot et al. 2004). However, Nile tilapia, Oreochromis niloticus, XY males and XX males showed comparable growth but XY males had higher hemoglobin levels than XX males (Phumyu et al. 2012).
Gene expression analysis is a powerful approach to detect the genetic basis of physiological and performance differences between fish with different phenotypes (Rescan et al. 2007). Growth involves an increase in skeletal muscle mass (Carpio et al. 2009) coupled with physiological processes occurring in the liver (Devlin et al. 2013). Therefore, expression studies are often based on examination of those tissues. Studies of salmonid liver and muscle gene expression indicate that the major GO categories associated with growth differences contain genes central to energy production, carbohydrate and lipid metabolism, and cytoskeletal components (Tymchuk et al. 2009; Devlin et al. 2013). In rainbow trout experiencing muscle growth after a period of starvation, genes related to protein production, cell proliferation, mitochondrial bioenergetics, and muscle remodeling were upregulated in the white muscle compared to normally growing controls (Rescan et al. 2007). In the muscle of rainbow trout experiencing muscle wastage, genes related to the catheptic and collagenase proteolytic pathways, muscle components, protein production, and cellular respiration were downregulated compared to the control fish (Salem et al. 2006a). However, rainbow trout grown under normal aquaculture conditions showed genes related to cytoskeleton components and the stress responses upregulated in the larger fish and genes related to the myofibril component of muscle tissue and apoptosis were upregulated in the smaller fish in the white muscle (Kocmarek et al. 2014). In the liver, small fish showed upregulation of insulin-like growth factor binding protein 1 (IGFBP-1) compared to large fish. Thus, comparisons across studies suggest that different genes might be involved in normal and altered growth.
In this study, we examined the gene expression profiles in the liver and white muscle of large and small rainbow trout progeny produced by crossing a XX male (neomale) with multiple females using a 44 K gene Atlantic salmon Salmo salar microarray. We then compared their expression profiles and growth-related traits to progeny of the same females crossed to an XY male (data from Kocmarek et al. 2014). Specifically, we determined if the differences in gene expression profiles between large and small offspring of a XX male differ from those detected between large and small offspring of a XY male. We also compared the gene expression profiles of the large XX male offspring to large XY male offspring and the small XX male offspring to the small XY male offspring. Our objective was to determine whether the progeny of XX males have altered gene expression profiles and altered growth compared to the progeny of XY males.
Method
Fish Husbandry and Tissue Collection
Rainbow trout from the Lyndon strain (Lyndon Fish Hatcheries Inc., New Dundee, ON) were reared at the Alma Aquaculture Research Station, using a protocol approved by the Animal Care Committee at the University of Guelph. Five maternal half-sibling (half-sib) families were created on September 18, 2008 (Fig. 1). Five female rainbow trout were crossed with a XX male (neomale) and the offspring were reared communally under a natural photoperiod. These fish are referred to as the XX offspring, as they are all female. They were fed a commercial rainbow trout ration, containing 44–55 % protein and 15–22 % fat, at a rate corresponding to the thermal growth coefficients devised for rainbow trout (2–3 % of body weight daily) (Dumas et al. 2007). Details of production, rearing, and sampling of families are the same as those used to rear the progeny produced by mating the same five females to an XY male (Kocmarek et al. 2014). The offspring of the normal male are referred to as XX/XY offspring as this lot contains both males and females. The common husbandry practices facilitate the comparison of the two lots.
Parental crosses used to produce the families used for the microarrays. A single XX male (♂) was crossed with five females (♀); a single cross is depicted. The two largest (L) and two smallest (S) fish were selected from each female’s offspring pool. The largest fish (L1) from each female parent was pooled and compared to a pool of the second smallest fish (S2). A pool of the second largest fish from each female was compared a pool of the smallest fish from each female. This process was repeated using a XY male (♂) and the same five female fish. 254 × 190 mm (96 × 96 DPI)
Tank densities were adjusted periodically according to optimal rearing densities and feeding schedules as regulated by the bulk biomass of the fish (Fig. 2). Bulk weights were monitored biweekly during the initial growth period until April 14, 2009, and then every 28 days until the completion of the experiment. Feed rations were adjusted according to fish average weights and water temperature. Water came from deep underground wells, with temperatures that varied from 8 to 10 °C (averaging 8.5 °C), throughout the experimental period. Mortalities per interval, thermal growth co-efficients (TGC) (Dumas et al. 2007), tank densities, and weight-specific growth rates (SGR) were calculated throughout the experimental period (Fig. 2).
Growth profiles of the XX and XX/XY offspring. Bulk tank weights were recorded biweekly until April 14, 2008, and then every 4 weeks (28 days) until June 23, 2009, when the fish were combined in a single tank. Measurements are shown from the first interval following the first bulk weighing. Thermal growth coefficient (TGC), specific growth rate (SGR), tank density, and average weight based on bulk measurements are depicted above the mortalities plot. The asterisk indicates when the fish were moved to a larger tank to reduce density. 374 × 245 mm (96 × 96 DPI)
No disease outbreaks occurred, and mortalities were minimal throughout the experimental period with the largest percentage occurring during the early rearing stages. On June 23, 2009, the two lots of half-sib families were combined in a single tank and keep together until the experiment’s completion.
On December 2, 2009 (15 months of age from fertilization), weight and length measurements were collected from 53–55 fish per half-sib family. As premature maturation occurs in male rainbow trout at two years of age, fish were sampled at an early age to ensure that gene expression profiles were not confounded due to the onset of precocious maturation. All fish were actively growing juveniles at the time of sampling as indicated by the presence of thin gonadal strands. The body weight of the XX offspring and XX/XY offspring and across the female half-sib families were compared using a two-factor (male parent and female parent) ANOVA with interaction. An F test for equality of two variances was used to determine if the XX and XX/XY offspring showed significant differences in variance for either weight or length. The male and female offspring within the XX/XY offspring group were also tested for significant differences in variance. A t test was used to compare the average weight and length of the male and female XX/XY offspring.
Tissue Collection
Samples of liver and white muscle were collected for DNA and RNA extraction from 53 XX offspring and 55 XX/XY offspring. The fish were purposefully size-selected to represent the fastest and slowest growing fish in each family. White muscle was excised immediately below the dorsal fin and well above the lateral line to avoid contamination with red muscle. A small square plug, ≤1cm2, was sampled and the surface skin removed. Liver and muscle myofibril samples were placed in the RNA-preserving solution (495 g L−1 (NH4)2SO4, 40 mL L−1 EDTA, 25 mL L−1 1 M Na3C6H5O7, adjusted to pH 5.2) and stored at −80 °C.
Parentage and Sex Determination
Using a standard phenol chloroform protocol (Taggart et al. 1992), DNA was extracted from the liver samples of parents and progeny. The purity and concentration of the DNA was quantified using a Nanodrop 8000 spectrophotometer (Thermo Scientific, Waltham, MA). Samples were stored at −20 °C. Microsatellite markers BX887563, Omm1220, Omm5147, and Omy1212UW (Danzmann et al. 2008) were used to determine parentage (methods of Haidle et al. 2008). Polymerase chain reaction (PCR) mixtures were made in 7 μL volumes (2.6 ng genomic DNA μL−1, 1× PCR buffer, 0.125 mM dNTP, 1.5 mM MgCl2, 0.1 mg ml−1 BSA, 0.3 μM of each forward or reverse marker primers labeled with tetrachloro-6-carboxy-flourescent, 0.021 U μL−1 Taq DNA polymerase). PCR conditions began with initial denaturation (95 °C for 10 min), followed by 35 cycles of denaturation (95 °C for 1 min), annealing (30 s), and extension (72 °C for 30 s) and concluded with final extension (72 °C for 5 min). An annealing temperature of 58 °C was used for all primers except BX887563, which used a temperature of 54 °C. Polyacrylamide gel electrophoresis (6 % polyacrylamide gel, 19:1 ratio of acrylamide to bisacrylamide, 8 M urea, 0.5× TBE buffer) was used to detect the PCR product. Gels were scanned using an FMBIO III Fluorescence scanner (MiraiBio Inc., Alameda, CA, USA).
The sex of the offspring was confirmed using the Y-specific marker OmyY1 (Brunelli et al. 2008). Polymerase chain reaction (PCR) mixtures were made in 20 μL volumes (3 ng genomic DNA μL−1, 1× PCR buffer, 0.2 mM dNTP, 1.75 mM MgCl2, 0.2 μM of each forward or reverse marker primers, 0.01 U μL−1 Taq DNA polymerase). PCR conditions began with initial denaturation (95 °C for 5 min), followed by 30 cycles of denaturation (95 °C for 30 s), annealing (58 °C for 1 min), and extension (72 °C for 1 min), and concluded with final extension (72 °C for 10 min). The OmyY1 PCR products were run on 1.5 % agarose gels containing ethidium bromide. The gels were scanned on an AlphaImager 3400 (ProteinSimple, San Jose, CA, USA).
RNA Isolation
To compare the gene expression levels of progeny from XX and XY sires across the common female (dam) genetic backgrounds total RNA was isolated from liver and muscle from the two largest and two smallest fish by weight in each of the ten families (Fig. 1). TRIzol (Invitrogen, Carlsbad, CA) extractions were performed according to the manufacturer’s instructions and a Nanodrop 8000 spectrophotometer was used to determine the RNA concentrations. Samples were stored at −80 °C. To confirm the RNA was not degraded distinct 18S and 28S rRNA bands were detected after agarose gel electrophoresis. The extraction process was repeated until a minimum of 40 μg RNA was collected from each sample. Since large and small fish can potentially contribute proportionally different amounts of mRNA to a total RNA pool, mRNA was used instead of total RNA to create cDNA, eliminating this potential bias. mRNA was purified from the total RNA using μMACS mRNA Isolation Kits – Small Scale (Miltenyi Biotec Inc., Auburn, CA) according to the manufacturer’s instructions.
Gene Expression
mRNA pools (0.5 μg total) composed of 0.1 μg of mRNA per individual were created for both liver and white muscle (Fig. 1). The size differences between the full-sib families meant we could not compare the largest fish to the smallest fish within each lot as this would have resulted in the majority of large fish coming from one or two families. The same would be true of the small fish. The mRNA from the largest individual from each of the five families within the XX offspring families was pooled (L1) and then the process was repeated for the second largest individuals (L2), smallest individuals (S1), and next smallest (S2) individuals. Gene expression of the L1 fish was compared to that of the S2 fish and the expression of the L2 fish were compared to those of the S1 fish. This resulted in approximately equal weight differences between the largest and smallest fish (Table 1). This process was repeated for the XX/XY offspring.
Gene expression was quantified using the Agilent microarray (catalog number - G2519F-020938) which contains DNA probes from 43 663 genes selected from Atlantic salmon expressed sequence tag databases (Aglient Technologies, Mississauga, ON). Atlantic salmon microarrays are suitable for use in gene expression studies of other salmonids (Rise et al. 2006; von Schalburg et al. 2005). This chip contains genes related to muscle growth including muscle component, metabolic, apoptotic, and stress response genes etc. Approximately 0.2–0.3 μg of cDNA was prepared from the mRNA using a SuperScript® Plus Direct cDNA Labeling System with Alexa Fluor® aha-dUTPs (Invitrogen, Burlington, ON) and labeled with either Alexa Fluor 555 or Alexa Fluor 647. Briefly, following the manufacturer’s specifications, the mRNA was reverse transcribed using an anchored oligod(T)20 primer and random hexamers in cDNA synthesis reactions that incorporated Alexa Fluor-labeled nucleotides. A Nanodrop 8000 spectrophotometer was used to calculate the concentration and dye incorporation rates of the cDNA. Samples were stored at −20 °C until hybridization on the same or the following day.
The cDNA was hybridized overnight to the microarrays according to the hybridization protocol described in two-color microarray-based gene expression analysis (Agilent Technologies). Dye flips were included to compensate for any dye effects. The “preventing ozone-related problems” version of the hybridization protocol was used as air ozone levels were above the level that could affect the Alexa Fluor 555 signal. A GenePix 4200A scanner (Axon Instruments, Weatherford Texas) was used to scan the microarrays, and Genepix Pro 6.1 software was used to process images, align the spots, integrate the Genepix Array List file with the microarray images, and quantify the spots. The arrays were subject to manual review and all unacceptable spots were flagged and excluded.
The data were analyzed by Genespring GX 7.3.1 software. To normalize the spot intensity from various replicates, Lowess (locally weighted scatter plot smoothing) normalization was applied. Genes that did not have a value for all of the replicates within a lot were removed from the analysis. The Genespring software was used to calculate t statistic, p value (probability), Gaussian p value, and the ratio between the two dyes for each spot. Spots with ≥1.2-fold change or more were considered differentially expressed (p value ≤ 0.05), while spots showing ≥2.4-fold change were designated as highly upregulated. Microarray data were submitted under the submission number GSE42584 to the NCBI Gene Expression Omnibus (GEO).
Blast2go (www.blast2go.de/b2ghome) assigned GO terms to the differently expressed microarray sequences. All parameters were set to the default with the exception of the Blast expectation value threshold which was set to 1.0E-6. If Blast2go was unable to assign GO terms to a sequence, manual evaluations were considered using AmiGO (http://amigo.geneontology.org/cgi-bin/amigo/go.cgi) assignments. A heterogeneity G test (www.uoguelph.ca/~rdanzman/ with links to the software module) was used to compare the number of genes differently expressed in the GO categories at GO biological process levels 2 and 3 (GOslim generic categories) between groups using a backwards elimination procedure. The category with the greatest heterogeneity was reported if significance was detected. This category was removed from the analysis and the remaining categories were reanalyzed in a stepwise fashion until no significant differences remained. The paired groupings evaluated by the heterogeneity G tests were as follows: large vs. small XX offspring, large XX vs. large XX/XY offspring, and small XX vs. small XX/XY offspring. GO category paired comparisons were considered as being significant if their relative gene counts differed by ≥5 % from one another.
The comparison between the large and small XX/XY offspring and the validation of the microarray results with real-time PCR have been reported previously (Kocmarek et al. 2014).
Results
Sex and Body Weight
All of the XX offspring were female based on the sex-linked marker. The XX and XX/XY offspring did not differ significantly in body weight or length and showed similar mortality rates, average bulk weights, SGR, and TGC throughout the experimental period (Fig. 2). However, the XX offspring showed less variance in weight and length (Fig. 3). Male and female offspring within the XX/XY offspring family did not differ significantly in average weight or length or in the variance of those traits. When the XX and XX/XY offspring were combined, the offspring of different females did differ significantly, with the offspring of females 1 and 2 being significantly heavier than the offspring of females 3 and 5 (Fig. 4).
Large and Small XX Offspring
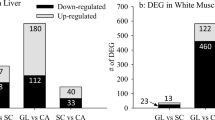
The XX offspring showed upregulation of 341 sequences in the liver (p ≤ 0.05, fold change ≥ 1.2; Online Resources 1–4), with 302 upregulated in the small fish (Online Resource 2). Blast2Go and Agilent were unable to identify 183 sequences in the XX male offspring (Online Resources 3 and 4). A list of genes showing ≥2.4-fold upregulation (i.e., classified as highly upregulated genes) for both the large and small XX offspring is shown in Table 2.
In the muscle, 404 sequences were upregulated in the XX offspring (Online Resources 5–8). As in the liver, the majority of genes showed upregulation in the small fish (260 gene probes) (Online Resource 6). One hundred and thirteen sequences were unidentified in the XX offspring (Online Resource 7 and 8). Highly upregulated sequences in the muscle of the large and small XX offspring are shown in Table 3.
The large and small XX offspring showed significant differences at the biological process GO level 2 in the cellular process (GO:0009987), metabolic process (GO:0008152), cellular component organization (GO:0016043), and response to stimulus (GO:0050896) categories in the muscle (Fig. 5a). Large XX fish had a higher proportion of genes upregulated in the “response to stimulus” and “cellular component organization” categories than did small XX fish, while the reverse was true for the “metabolic process” and “cellular process” categories. In addition, small XX fish had a higher proportion of genes differentially expressed in these latter two categories than small XX/XY fish. A list of the specific genes upregulated within the large and small fish in these categories are presented in Online Resources 9A, 10A, 11A, and 12, respectively. For a list of all sequences falling within all GO categories, in both the liver and the muscle, see Online Resources 13–16. In the small XX offspring 25.6 % of genes were recorded in the metabolic process category while 15.9 % of genes were found in this category in the large XX offspring (Online Resource 17). Genes in the cellular process category made up 33.7 % of the total genes in the small XX offspring and 18.3 % of the genes in the large XX offspring. Genes in the cellular component organization category made up 0 and 6.1 % of the total genes in the small and large XX offspring, respectively. Genes in the response to stimulus category made up 6.0 and 11.0 % of the total genes in the small and large XX offspring, respectively.
Gene ontology term distributions in the muscle. For biological process a levels 2 and b 3, significant deviation from the expected number of sequences (total sequences in a category related to total upregulated sequences) between the two data sets are marked with asterisks. Only terms differing by at least 5% are considered significantly different. (*Large XX offspring compared to small XX offspring; +small XX offspring compared to small XX/XY offspring, p ≤ 0.05). 254 × 190 mm (96 × 96 DPI)
At biological process level 3, significant differences between the large and small XX offspring in the macromolecule metabolic process (GO:0043170), small molecule metabolic process (GO:0044281), and the cellular metabolic process (GO:0044237) categories were detected (Fig. 5b). Lists of the genes within these categories are presented in Online Resources 18, 19, and 20A, respectively. Genes in the macromolecule metabolic process, small molecule metabolic process, and the cellular metabolic process made up 10.8, 4.9, and 15.3 % of the genes in the small XX offspring and 2.6, 0, and 6.8 % of the genes in the large XX offspring, respectively (Online Resource 21). No significant differences were seen between the GO categories of the large and small XX offspring in the liver (Online Resources 22 and 23).
XX and XX/XY offspring
Liver
Many of the sequences showed similar patterns of upregulation in the XX and XX/XY offspring. For example, complement C1q-like protein 4 precursor (A_05_P332842) was highly upregulated in the large offspring from both male parents (Table 2). Other genes showed a consistent pattern of upregulation at the ≥1.2-fold level including apolipoprotein l3-like (A_05_P451048) in the large fish and glutathione peroxidase 3 precursor (A_05_P388822) and ictacalcin (A_05_P488907) in the small fish (Online Resource 2). 39S ribosomal protein mitochondrial-like, ras-related protein rab-37-like, and group xiia secretory phospholipase a2 precursor were also upregulated in the liver of small XX/XY offspring and small XX offspring albeit for different sequences which may represent duplicated or orthologous copies of the genes. A few genes showed opposite patterns of expression between the families (i.e., upregulated in the large XX offspring and the small XX/XY male offspring or vice versa) and these included nicotinamide phosphoribosyltransferase (A_05_P355253), lymphocyte-specific protein tyrosine kinase (A_05_P322837), and kunitz bovine pancreatic trypsin inhibitor domain protein (A_05_P433482).
Neither the large XX and XX/XY offspring nor the small XX and XX/XY offspring showed significant differences in the observed number of genes assigned to either the level 2 or level 3 GO categories (Online Resources 22 and 23). The biological process GO level 2 category with the largest number of upregulated sequences in large and small XX/XY offspring and XX offspring was cellular process (GO:0009987) (Online Resource 22). A list of the Agilent probe IDs that correspond to the individual GO level 2 and level 3 categories is provided in Online Resources 13 and 14. At level 3, the biological process GO category cellular metabolic process (GO:0044237) had the most upregulated sequences in the large and small fish from both family lots, although in the large XX offspring the primary metabolic process category (GO:0044238) had the same number of sequences (Online Resource 23).
Muscle
Many of the genes showed a consistent pattern of upregulation within a size category for both the XX offspring and XX/XY offspring. DNA-damage-inducible transcript 4-like protein (A_05_P249334), max protein (A_05_P478412), and ADP/ATP translocase 2 (A_05_P276614) were highly upregulated in the large fish from both offspring groups and calcium-binding and coiled-coil domain-containing protein 1 isoform 2 (A_05_P276634) was highly upregulated in the small fish from both offspring groups (Table 3). Other genes were highly upregulated in one offspring group and upregulated ≥1.2-fold in the other offspring group. In the large fish, genes following this pattern included beta-enolase-like isoform 1 (A_05_P482637) and mid1-interacting protein 1 (A_05_P364592) (Online Resource 5). Highly upregulated genes in the small XX/XY offspring including parvalbumin 2 (A_05_P423182), parvalbumin beta-2 (A_05_P429727), glyceraldehyde-3-phosphate dehydrogenase (A_05_P419277), thrombospondin 2 (A_05_P322687), and alpha actin (A_05_P449267) were also upregulated ≥1.2-fold in the small XX offspring (Online Resource 6). Ten other sequences showed ≥1.2-fold upregulation in the large fish from both offspring groups as did 17 additional sequences in the small fish from both offspring groups (Online Resources 5 and 6). Small ubiquitin-related modifier 3 precursor (A_05_P418747) and genes identified by different sequences in the different offspring groups, including alpha actin, calmodulin, and member ras oncogene family, were upregulated in the large XX offspring and downregulated in the small XX/XY offspring.
The gene expression patterns of the large XX/XY offspring did not differ significantly from the large XX offspring using the G test for heterogeneity (Online Resources 17 and 21). In contrast, the small XX offspring differed from the small XX/XY offspring in the proportion of genes falling within the GO level 2 categories of cellular process, metabolic process, and cellular component organization (Fig. 5a). Lists of the individual genes falling within these categories are presented in the Online Resources 9–11, respectively. Genes in the cellular component organization category composed 7.7 % of the genes in the small XX/XY offspring while no genes from this category were upregulated in the small XX offspring (Online Resource 17). The metabolic process category contained 25.6 % of the genes in the XX offspring and 15.9 % of genes in the small XX/XY offspring. In the small XX and XX/XY offspring, the cellular process category had 33.8 and 17.5 % of the upregulated genes, respectively. At GO level 3, significant differences were found in the cellular metabolic process category and the small molecule metabolic process category (Fig. 5b). These categories contained 15.3 and 4.9 % of the genes in the small XX offspring, respectively, and 9.0 and 0 % of the genes in the small XX/XY offspring, respectively (Online Resource 21). Lists of the genes falling within these categories are present in the Online Resources 19 and 20.
In the large XX/XY offspring, the GO level 2 category containing the most differentially regulated sequences was metabolic process (GO:0008152), followed by cellular process (GO:0009987) (Online Resource 17). In all other fish, the category containing the most sequences was cellular process, followed by metabolic process. The GO level 3 categories, primary metabolic process (GO:0044238) and regulation of biological process (GO:0050789) contained the first and second greatest number of sequences in the large and small XX/XY offspring (Online Resource 21). The categories cellular metabolic process (GO:0044237) and regulation of biological process contained the most sequences in the small and large XX offspring, respectively. The category containing the second greatest number of sequences was “primary metabolic process” in both the large and small XX offspring.
Discussion
The comparison of growth-related traits and gene expression profiles between the offspring of the XX and XY males detected few differences in their relative performance. Although the offspring of XX and XY males showed no differences in the average values of growth-related traits and mortality, XX offspring had smaller variance for body length and weight. This supports the suggestions that the use of all-female populations of fish should decrease size variation as this would eliminate rapidly growing male juveniles which are likely to undergo early maturation (Luo et al. 2011). In addition, lower variation in body size might lead to reduced crowding stress because of lower propensity for undesirable behavioral interactions (Hopkins and Todd 1991). Overall the gene expression patterns of XX and XX/XY offspring were also very similar in that a large number of upregulated genes were shared within size classes and no GO category differences were seen in the liver. In the muscle, significant GO category differences were seen in the small XX and small XX/XY offspring but not between the large XX and large XX/XY offspring. The small XX offspring contained a greater percentage of genes in the “small molecule metabolic process” and “cellular metabolic process” categories compared to the small XX/XY offspring, although several other GO categories also differed between the small fish at lower levels of differentiation.
Although no significant differences were detected in liver, large and small XX offspring and small XX/XY and XX offspring did differ significantly for the same level 2 GO categories in white muscle. Small XX fish expressed higher levels of “cellular process” and “metabolic process” genes than either large XX fish or small XX/XY fish, while the reciprocal was true for genes within the “cellular component organization” category. Similarly, at GO level 3 a higher proportion of genes related to “cellular metabolic process” and “small molecule metabolic process” were upregulated in small XX fish than they were in either large XX fish or small XX/XY fish. This suggests that genes within these classes may be signatures of both “female-based” and size-related metabolic differences given that small XX/XY fish would also carry signatures of male-specific metabolic differences. Child terms within “cellular metabolic process” include pathways by which individual cells transform chemical substances. “Small molecule metabolic process” involves any pathway or chemical reaction related to a small molecule such as a monomeric unit, small molecular weight substance, or a non-encoded molecule.
Comparison of Large and Small XX Offspring
Although the major differences in gene expression between large and small XX fish were related to metabolic process functions, a number of significant level 3 GO categories related to “response to stress,” “multicellular organism development,” “cell death,” “catabolic process,” “response to external stimulus,” “anatomical structure development,” and “establishment of localization” were more upregulated in large XX fish compared to small XX fish, albeit at 3–5 % levels of differentiation. Similarly, “nitrogen compound metabolic process” and “primary metabolic process” were categories with significantly higher numbers of GO terms associated within small fish at the 3–5 % level of differentiation.
Several genes related to carbohydrate and lipid metabolism were significantly upregulated in the white muscle of large XX offspring highlighting their importance in the metabolic processes leading to growth enhancement. The carbohydrate metabolism genes included enolase 3–2, lactate dehydrogenase-a, mannose-6-phosphate isomerase, phosphoglucose isomerase, transketolase, pyruvate kinase, utp–glucose-1-phosphate uridylyltransferase, fructose-bisphosphatase isozyme 2, phosphoglycerate kinase 1 and glycogen genes. In addition, lipid metabolism genes, including lipoprotein lipase genes, heart and intestinal fatty acid-binding protein genes, adiponectin receptor protein 1, and delta-6 fatty acyl desaturase were upregulated in the large XX fish. Other studies have shown a link between genes related to carbohydrate and lipid metabolism and energy production and growth in fish (Tymchuk et al. 2009; Devlin et al. 2013; White et al. 2013). Paradoxically, genes within the GO level 2 categories “metabolic process” and “cellular process,” which are parent terms of the carbohydrate and lipid metabolizing genes were more upregulated in the small XX offspring compared to large XX fish. Within the small XX fish, genes related to transcription, translation, translational processing, cell cycle, and signaling were more common within the “metabolic process” category which suggests that metabolic pathways that differ from carbohydrate and lipid metabolism are of greater importance in regulating the growth patterns of small fish. The same pattern was observed in the GO level 3 category “cellular metabolic process,” which is a child term of both “metabolic process” and “cellular process,” and in the categories, “macromolecule metabolic process,” “primary metabolic process,” and “small molecule metabolic process,” which are child terms of the “metabolic process” category.
In the small XX offspring, a large number of differentially expressed genes in the macromolecule metabolic process and cellular metabolic process categories were related to transcription and translation with functions that can be involved in suppressing growth. These genes included cellular repressor of e1a-stimulated genes 1 (CREG1), cold shock domain-containing protein e1 (CSED1), eukaryotic translation initiation factor (EIF2b4), and transcription factor E2f4. EIF2b4 is known to inhibit protein synthesis in response to stress (GO term: 0032057). E2f4 in conjunction with lysine (K)-specific demethylase 5A causes transcriptional repression during cell differentiation (Beshiri et al. 2012). CSED1 acts as a positive or negative regulator of cell death, depending on the cell type in mammals (Dormoy-Raclet et al. 2007), and CREG1 inhibits cellular proliferation by inhibiting cyclins A and B expression (Moolmuang and Tainsky 2011). Conversely, the large XX offspring showed an upregulation of only a few of genes in these process categories: transcription elongation factor 1 homolog, vacuolar protein sorting 52, and t-complex protein 1 subunit epsilon.
In contrast to the functions listed above, genes stimulating growth were also upregulated in the small XX fish. This list includes genes such as insulin growth factor I receptor (IGF1R), inhibitor of nuclear factor kappa-b kinase subunit beta (IKBKB), paired box protein (Pax1), and calcium calmodulin-dependent protein kinase iv (CAMK4). IGF1R prevents apoptosis and protein breakdown and promotes myogenesis, glucose uptake, glycogen synthesis, and lipid storage. The activation of CAMK4 as a result of elevated Ca2+ levels may also inhibit further cellular degradation, as would the activation of NF-κB via IKBKB (Mora et al. 2005). Pax1 is a developmental regulatory gene involved in early developmental processes and osteogenesis. Additionally, other protein subcomponents of the translation initiation complex (i.e., EIF1b and EIF3a) were upregulated in the small XX fish. EIF1b expression has been reported to be upregulated during proteasome degradation suppression in humans (Kinyamu et al. 2008), suggesting that cellular sparing is enhanced in small XX fish. This is also supported by the observation that the level 3 term “cell death” is elevated in large XX fish which suggests greater cellular turnover rates.
Myofibrillar and cytoskeletal genes were upregulated in the muscle of the large XX offspring, as evidenced by the fact that “anatomical structure development” was one of the level 3 GO categories that was significantly elevated in large XX compared to small XX fish. Many of these sequences, including 3 myosin sequences, actinin alpha 2, alpha actin, collagen type i alpha 3 chain, and keratin 19 were found within the level 2 GO categories “cellular component organization” and “response to stimulus.” These GO categories also contained retinoid x receptor alpha and retinol binding protein cellular, which are related to the myogenesis activator retinoic acid (Hamade et al. 2006), and tripartite motif-containing protein 55, which is involved in muscle cell turnover (Perera et al. 2012), in the large fish. This pattern is similar to that reported in previous work where myofibrillar and muscle regulatory proteins were upregulated in growing female rainbow trout compared to females showing atrophying muscle (Salem et al. 2006b) and in sex-unspecified GH-transgenic and domestic rainbow trout compared to wild-type controls (Tymchuk et al. 2009). This is in contrast to the results reported for the XX/XY offspring which showed upregulation of genes related to muscle components in small fish and upregulation of genes related to the connective tissue in large fish (Kocmarek et al. 2014).
The liver of large and small XX offspring showed no differences in the proportion of upregulated genes in GO categories demonstrating that gene expression in this organ is a poor indicator of growth differences relative to muscle. This is similar to the results reported for the large and small XX/XY offspring (Kocmarek et al. 2014). IGFBP1 was one of the few genes identified as upregulated in the liver in both small XX/XY and XX offspring. IGF-binding proteins have been linked to increased protein catabolism in Atlantic salmon (Hevrøy et al. 2011) and with heightened cessation of bone growth and repair in aging mammals (Parker et al. 2007). Given that IGF-I is known to stimulate muscle growth (Fuentes et al. 2013), the binding of IGFs by IGFBP-1 may contribute to decreased growth in the small fish. IGFBP1a and IGFBP1b, which are both expressed in zebrafish muscle, showed increased expression during fasting (Amaral and Johnston 2011). Overexpression of IGFBP in grass carp, Ctenopharyngodon idella, embryos causes phenotypes that resemble hypoxia-induced developmental delay and growth retardation (Sun et al. 2011). Additionally, IGFBP1a/b was also reported to be upregulated in small embryonic zebrafish compared to large zebrafish (Amaral and Johnston 2012). These fish were derived from lines selected for either enhanced or decreased growth over four generations, and the findings indicate that elevated IGFBP expression is linked to decreased growth capacity.
Several genes related to calcium metabolism that were upregulated in both large and small fish are of interest in relation to their association with muscle function. Calcium signaling can mediate both positive and negative growth events in vertebrates and is important in regulating apoptotic events (Orrenius et al. 2003; Harr and Distelhorst 2010). Parvalbumin 2 (PVALB) and parvalbumin beta-2, genes involved in calcium binding, were upregulated in the muscle of the small fish. PVALB expression has been coupled to protein phosphatase 1 regulatory subunit 9a (PPP1R9A) expression (http://string-db.org), a multimeric gene that is involved with numerous cellular functions. Our finding that protein phosphatase 1 regulatory subunit 12c (PPP1R12C) is also upregulated in small fish indicates a coupling of functions related to cytoskeletal modifications to actin and myosin that appear to be related to the action of the PPP1R12C subunit (Tan et al. 2001; Cohen 2002). Parvalbumins, when over-expressed, lead to shorter quick-twitch muscle relaxation times, but paradoxically have been coupled to lower mitochondrial densities in slow-twitch muscle groups (Racay et al. 2006; Seebacher and Walter 2012). Individuals with lower parvalbumin levels have enhanced mitochondrial volumes and metabolic flux capabilities, as evidenced by inverse associations between SIRT1/PGG-1α activities and parvalbumins levels (Ducreux et al. 2012). Calmodulin was observed to be upregulated in large XX fish and a variant isoform of the gene was also upregulated in small XX/XY offspring. Calmodulin is a major mediator of intracellular calcium signaling, and its actions are tied to the regulation of translation and cell cycle events as well as the regulation of calcineurin (PPP3) and several protein kinases. Genes integrated with calmodulin expression and related to scaffolding (http://string-db.org) such as actinin alpha 2 were upregulated in large XX offspring, while genes such as filamin-c, lim-domain binding 3 were more upregulated in small XX offspring. As mentioned previously, CAMK4 was expressed at a higher level in small XX offspring, and this gene as well as CAMK2d is also upregulated in small XX/XY fish (Kocmarek et al. 2014). Calcium signaling produces a complex array of differential gene expression patterns in large and small fish and perhaps most revealing is the fact that calmodulin is upregulated in both size classes of fish indicating the importance of this 2nd messenger pathway in regulating growth overall.
Comparison of XX/XY and XX Offspring
Differences in patterns of gene expression between XX and XX/XY fish were minimal. As previously mentioned possible sex-specific gene expression patterns resulting from the comparison of all-female and mixed sex offspring might explain any expression differences. We did, however, examine the fish as juveniles prior to the onset of the sexual maturation process. Even though this should have minimized any sex-specific differences in gene expression, early maturation cannot be entirely discounted as males often mature precociously at 2 years of age with demonstrable increases in 11-ketotestosterone(11-KT) levels several months preceding the onset of maturation (Campbell et al. 2003). It could be hypothesized, therefore, that elevated levels of 11-KT in any male fish we sampled that were destined to precociously mature could have caused associated changes in other metabolic pathways thus confounding expression pattern differences. According to this hypothesis, we would have predicted that the greatest differences in expression patterns would occur between large XX and large XX/XY fish as precociously maturing males are also known to have elevated growth rates compared to males that have delayed maturation rates (Thrope 1977; Naevdal et al. 1979). We also might have predicted larger expression differences between large XX and XX/XY offspring given that the RNA pool generated for the large XX/XY offspring was produced from six males and four females, while the reverse numbers were used to produce the small XX/XY fish RNA (Kocmarek et al. 2014). This sampling effect resulted from the need to match large and small fish of approximately the same size differential for the paired replicate pools that were produced for the microarray analysis. Our data do not support the hypothesis that the expression patterns are the result of precocious sexual maturation of XX/XY offspring because the greatest differences were seen between the small XX and small XX/XY offspring. However, these differences may be related to other sex-specific differences as sexually dimorphic gene expression has been reported in zebrafish, which are not known to undergo precocious male maturation (discussed below). Due to the fact that RNA samples were pooled in this experiment we cannot examine this possibility, however, this is an issue that could be explored in the future.
Although both XX and XX/XY progeny lots had similar expression patterns overall, they differed in the counts of several genes contributing to the GO categories. For example, a different set of transcription and translation genes were upregulated in the small XX/XY offspring and only the small XX/XY offspring showed upregulation of a large number of protein synthesis genes, including among others, mitochondrial ribosomal protein s16, peptidase (mitochondrial processing) alpha, t-complex protein 1 subunit gamma, and ras-related protein rab-2a. Protein degradation genes, including proteasome- and ubiquitin-related sequences, as well as DNA damage-binding protein 1, and selenoprotein s, which is involved in ER-associated degradation (Christensen et al. 2012) were only upregulated in the small XX/XY offspring. This suggests more protein turnover is occurring in the small XX/XY offspring than in the small XX offspring. Also, a significantly greater percentage of the small XX/XY offspring with differentially expressed genes compared to small XX offspring fell in the GO level 3 categories “cell death” and “catabolic process” at the 3–5 % level of differentiation (Online Resource 21). If small XX/XY offspring express higher protein degradation rates than XX offspring this may be coupled to greater metabolic variability in this group leading to greater variation in growth rates.
The differences we observed in metabolic process and protein turn-over genes may also be due to the mixed sex and all-female offspring groups that were used. Several studies in zebrafish have found sexually dimorphic gene expression in somatic tissues (Zheng et al. 2013; Small et al. 2009; Sreenivasan et al. 2008), including differential expression of candidate genes for muscle growth (Ulloa et al. 2013). This latter study indicated that males showed greater differentiation in certain muscle growth regulating genes than did females when fed a variable plant-based dietary regime. If supported by additional research, this would suggest that male-specific differences in gene expression may elicit a broader range of phenotypic outcomes compared to females. Differences in apoptotic rates between the sexes have also been reported but with no clear consensus as to whether levels are elevated in either sex. Elevated gene expression levels in the brain tissue of female Medaka fish following exposure to environmental estrogens have been reported (Stokes et al. 2004). Studies in mammals indicate significantly higher male-specific spontaneous (Molloy et al. 2003) and disease-stressed (Hofmann-Lehmann et al. 1998) apoptotic rates in white blood cells and in the spleen in response to low-dose radiation (Cassie et al. 2006). The lack of consensus across studies on sex-specific patterns in gene expression indicates that we cannot discount such sex-specific differences as an explanation for the observed heterogeneity in gene expression profiles between small XX/XY versus small XX fish.
Comparisons between large and small fish indicated that both large XX and XX/XY fish had greater proportions of genes expressed in the “response to stress,” “response to external stimulus,” and “establishment of localization” categories than did small XX and XX/XY fish. While these similarities in the classifications of both large XX and XX/XY fish indicate overall congruence in the physiological profiles of these two groups of fish, some differences in the assemblages of genes assigned to these categories exist. For example, in the “response to stress” ontology, transcriptional regulatory genes such as pyruvate kinase, max protein, and beta-enolase were common to both groups, as was the oxidative stress gene glutathione peroxidase, and the protein-processing gene ap-1 complex subunit gamma-1. Additional genes related to blood components and immune function were also shared by large fish in both categories. Conversely, components related to cytoskeletal structure (e.g., telethonin and fibronectin) were only evident in the large XX/XY fish, while a greater diversity of cell signaling components (e.g., retinoid x receptor alpha, phosphatidylinositol 3-kinase regulatory subunit) was detected in large XX fish. “Response to external stimulus” category genes largely overlapped with “response to stress” genes as they are related categories in GO classifications. In the “establishment of localization” category, large XX/XY fish had a greater proportion of blood component genes (e.g., hemoglobin alpha and beta subunits) expressed that were lacking in large XX fish. In contrast to this, large XX fish expressed a couple of collagen alpha chain types, and troponin subunits not found in the large XX/XY fish, along with vitellogenin aa. The detection of this latter gene may not be surprising given that XX fish are all female. However, the differential distribution of varying cytoskeletal and myofibrillar components to the GO categories identified is unclear at present. While higher levels of cytoskeletal component genes were previously observed in larger XX/XY fish, the converse was true for myofibrillar component genes (Kocmarek et al. 2014). The finding that myofibrillar component genes were more upregulated in small fish was unexpected, and our observation that troponin components are more upregulated in the large XX fish is more consistent with expectations. Whether the myofibrillar component genes show greater cyclic variation in their expression levels to account for these differences or whether sex-specific differences (i.e., lower expression of myofibrillar component genes in male fish) may account for the observed differences require further studies.
Conclusions
The values of growth-related traits and mortality rates in the XX and XX/XY offspring suggest that the two offspring groups have similar growth trajectories and could be expected to have comparable husbandry costs from an aquaculture perspective. The similar gene expression profiles of the XX/XY offspring and XX offspring indicate that candidate genes identified in one group are likely representative of those in the other group. Further characterization of what could be considered female-specific growth profiles is warranted, however, as current research identified GO signatures of large fish growth that may be confounded by male versus female metabolic differences. For example, different transcription and translation genes were upregulated in the small XX and small XX/XY offspring with suppression of metabolic and cellular processes occurring in the small XX offspring and increased protein degradation suggested in the small XX/XY offspring. To what degree are these differences related to sex-specific influences on gene expression? Juvenile sex-specific gene expression patterns may relate to the decreased size variation seen in XX fish and should be investigated in future studies.
References
Amaral IP, Johnston IA (2011) Insulin-like growth factor (IGF) signalling and genome-wide transcriptional regulation in fast muscle of zebrafish following a single-satiating meal. J Exp Biol 214:2125–2139
Amaral IPG, Johnston IA (2012) Experimental selection for body size at age modifies early life-history traits and muscle gene expression in adult zebrafish. J Exp Biol 215:3895–3904
Berejikian BA, Van Doornik DM, Endicott RC, Hoffnagle TL, Tezak EP, Moore ME, Atkins J (2010) Mating success of alternative male phenotypes and evidence for frequency-dependent selection in Chinook salmon, Oncorhynchus tshawytscha. Can J Fish Aquat Sci 67:1933–1941
Beshiri ML, Holmes KB, Richter WF, Hess S, Islam AB, Yan Q, Plante L, Litovchick L, Gévry N, Lopez-Bigas N, Kaelin WG Jr, Benevolenskaya EV (2012) Coordinated repression of cell cycle genes by KDM5A and E2F4 during differentiation. Proc Natl Acad Sci U S A 109:18499–18504
Brunelli JP, Wertzler KJ, Sundin K, Thorgaard GH (2008) Y-specific sequences and polymorphisms in rainbow trout and Chinook salmon. Genome 51:739–748
Campbell B, Dickey JT, Swanson P (2003) Endocrine changes during onset of puberty in male spring Chinook salmon, Oncorhynchus tshawytscha. Biol Reprod 69:2109–2117
Carpio Y, Acosta J, Morales R, Santisteban Y, Sanchéz A, Estrada MP (2009) Regulation of body mass growth through activin type IIB receptor in teleost fish. Gen Comp Endocrinol 160:158–167
Cassie S, Koturbash I, Hudson D, Baker M, Ilnytskyy Y, Rodriguez-Juarez R, Weber E, Kovalchuk O (2006) Novel retinoblastoma binding protein RBBP9 modulates sex-specific radiation responses in vivo. Carcinogenesis 27:465–474
Christensen LC, Jensen NW, Vala A, Kamarauskaite J, Johansson L, Winther JR, Hofmann K, Teilum K, Ellgaard L (2012) The human selenoprotein VCP-interacting membrane protein (VIMP) is non-globular and harbors a reductase function in an intrinsically disordered region. J Biol Chem 287:26388–26399
Cohen PT (2002) Protein phosphatase 1—targeted in many directions. J Cell Sci 115:241–256
Danzmann RG, Davidson EA, Ferguson MM, Gharbi K, Koop BF, Hoyheim B, Lien S, Lubieniecki KP, Moghadam HK, Park J, Phillips RB, Davidson WS (2008) Distribution of ancestral proto-Actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon). BMC Genomics. doi:10.1186/1471-2164-9-557
Devlin RH, Sakhrani D, White S, Overturf K (2013) Effects of domestication and growth hormone transgenesis on mRNA profiles in rainbow trout (Oncorhynchus mykiss). J Anim Sci. doi:10.2527/jas. 2013-6612
Dormoy-Raclet V, Markovits J, Malato Y, Huet S, Lagarde P, Montaudon D, Jacquemin-Sablon A, Jacquemin-Sablon H (2007) Unr, a cytoplasmic RNA-binding protein with cold-shock domains, is involved in control of apoptosis in ES and HuH7 cells. Oncogene 26:2595–2605
Ducreux S, Gregory P, Schwaller B (2012) Inverse regulation of the cytosolic Ca2+ buffer parvalbumin and mitochondrial volume in muscle cells via SIRT1/PGC-1α axis. PLoS One. doi:10.1371/journal.pone.0044837
Dumas A, France J, Bureau DP (2007) Evidence of three growth stanzas in rainbow trout (Oncorhynchus mykiss) across life stages and adaptation of the thermal-unit growth coefficient. Aquaculture 267:139–146
Espinosa E, Josa A, Gil L, González N (2011) Sex steroid levels in XY males and sex-reversed XX males, of rainbow trout (Oncorhynchus mykiss), during the reproductive cycle. Reprod Domest Anim 46:8–14
Fuentes EN, Valdés JA, Molina A, Björnsson BT (2013) Regulation of skeletal muscle growth in fish by the growth hormone–insulin-like growth factor system. Gen Comp Endocrinol 192:136–148
Guiry A, Flynn D, Hubert S, O’Keeffe AM, LeProvost O, White SL, Forde PF, Davoren P, Houeix B, Smith TJ, Cotter D, Wilkins NP, Cairns MT (2010) Testes and brain gene expression in precocious male and adult maturing Atlantic salmon (Salmo salar). BMC Genomics. doi:10.1186/1471-2164-11-211
Haidle L, Janssen JE, Gharbi K, Moghadam HK, Ferguson MM, Danzmann RG (2008) Determination of quantitative trait loci (QTL) for early maturation in rainbow trout (Oncorhynchus mykiss). Mar Biotechnol 10:579–592
Hamade A, Deries M, Begemann G, Bally-Cuif L, Genêt C, Sabatier F, Bonnieu A, Cousin X (2006) Retinoic acid activates myogenesis in vivo through Fgf8 signalling. Dev Biol 289:127–140
Harr MW, Distelhorst CW (2010) Apoptosis and autophagy: decoding calcium signals that mediate life or death. Cold Spring Harb Perspect Biol. doi:10.1101/cshperspect.a005579
Hevrøy EM, Azpeleta C, Shimizu M, Lanzén A, Kaiya H, Espe M, Olsvik PA (2011) Effects of short-term starvation on ghrelin, GH-IGF system, and IGF-binding proteins in Atlantic salmon. Fish Physiol Biochem 37:217–232
Hofmann-Lehmann R, Holznagel E, Lutz H (1998) Female cats have lower rates of apoptosis in peripheral blood lymphocytes than male cats: correlation with estradiol-17 beta, but not with progesterone blood levels. Vet Immunol Immunopathol 65:151–160
Hopkins CL, Todd PR (1991) Production of monosex and triploid salmon stocks. New Zealand Freshwater Fisheries Report No. 128. MAF Fisheries, Christchurch
Ji XS, Liu HW, Chen SL, Jiang YL, Tian YS (2011) Growth differences and dimorphic expression of growth hormone (GH) in female and male Cynoglossus semilaevis after male sexual maturation. Mar Genomics 4:9–16
Kinyamu HK, Collins JB, Grissom SF, Hebbar PB, Archer TK (2008) Genome wide transcriptional profiling in breast cancer cells reveals distinct changes in hormone receptor target genes and chromatin modifying enzymes after proteasome inhibition. Mol Carcinog 47:845–885
Kocmarek AL, Ferguson MM, Danzmann RG (2014) Differential gene expression in small and large rainbow trout derived from two seasonal spawning groups. BMC Genomics 15:57. doi:10.1186/1471-2164-15-57
Kocour M, Linhart O, Gela D, Rodina M (2005) Growth performance of all-female and mixed-sex common carp Cyprinus carpio L. populations in the central Europe climatic conditions. J World Aquacult Soc 36:103–113
Liu H, Guan B, Xu J, Changchun H, Tian H, Chen H (2012) Genetic manipulation of sex ratio for the large-scale breeding of YY super-male and XY all-male yellow catfish (Pelteobagrus fulvidraco (Richardson)) Mar. Biotechnologyl 15:321–328
Luo K, Xiao J, Liu S, Wang J, He W, Hu J, Qin Q, Zhang C, Tao M, Liu Y (2011) Massive production of all-female diploids and triploids in the crucian carp. Int J Biol Sci 7:487–495
Molloy EJ, O’Neill AJ, Grantham JJ, Sheridan-Pereira M, Fitzpatrick JM, Webb DW, Watson RW (2003) Sex-specific alterations in neutrophil apoptosis: the role of estradiol and progesterone. Blood 102:2653–2659
Moolmuang B, Tainsky MA (2011) CREG1 enhances p16(INK4a) -induced cellular senescence. Cell Cycle 10:518–530
Mora AL, LaVoy J, McKean M, Stecenko A, Brigham KL, Parker R, Rojas M (2005) Prevention of NF-kappaB activation in vivo by a cell-permeable NF-kappaB inhibitor peptide. Am J Physiol Lung Cell Mol Physiol 289:L536–L544
Naevdal G, Holm M, Leroy R, Moller D (1979) Individual growth rate and age at sexual maturity in rainbow trout. FiskDir Skr Ser HavUnders 17:1–10
Nynca J, Kuźmiński H, Dietrich GJ, Hliwa P, Dobosz S, Liszewska E, Karol H, Ciereszko A (2012a) Biochemical and physiological characteristics of semen of sex-reversed female rainbow trout (Oncorhynchus mykiss, Walbaum). Theriogenology 77:174–183
Nynca J, Kuźmiński H, Dietrich GJ, Hliwa P, Dobosz S, Liszewska E, Karol H, Ciereszko A (2012b) Changes in sperm parameters of sex-reversed female rainbow trout during spawning season in relation to sperm parameters of normal males. Theriogenology 77:1381–1389
Orrenius S, Nicotera P, Zhivotovsky B (2003) Cell death mechanisms and their implications in toxicology. Toxicol Sci 119:3–19
Parker EA, Hegde A, Buckley M, Barnes KM, Baron J, Nilsson O (2007) Spatial and temporal regulation of GH-IGF-related gene expression in growth plate cartilage. J Endocrinol 194:31–40
Perera S, Mankoo B, Gautel M (2012) Developmental regulation of MURF E3 ubiquitin ligases in skeletal muscle. J Muscle Res Cell Motil 33:107–122
Phumyu N, Boonanuntanasarn S, Jangprai A, Yoshizaki G, Na-Nakorn U (2012) Pubertal effects of 17α-methyltestosterone on GH-IGF-related genes of the hypothalamic-pituitary-liver-gonadal axis and other biological parameters in male, female and sex-reversed Nile tilapia. Gen Comp Endocrinol 177:287–292
Racay P, Gregory P, Schwaller B (2006) Parvalbumin deficiency in fast-twitch muscles leads to increased 'slow-twitch type' mitochondria, but does not affect the expression of fiber specific proteins. FEBS J 273:96–108
Rescan P-Y, Montfort J, Rallière C, Le Cam A, Esquerré D, Hugot K (2007) Dynamic gene expression in fish muscle during recovery growth induced by a fasting-refeeding schedule. BMC Genomics 8:438–455
Rise ML, Douglas SE, Sakhrani D, Williams J, Ewart KV, Rise M, Davidson WS, Koop BF, Devlin RH (2006) Multiple microarray platforms utilized for hepatic gene expression profiling of GH transgenic coho salmon with and without ration restriction. J Mol Endocrinol 37:259–282
Rougeot C, Nicayenzi F, Mandiki SN, Rurangwa E, Kestemont P, Mélard C (2004) Comparative study of the reproductive characteristics of XY male and hormonally sex-reversed XX male Eurasian perch, Perca fluviatilis. Theriogenology 62:790–800
Salem M, Kenney PB, Rexroad CE 3rd, Yao J (2006a) Microarray gene expression analysis in atrophying rainbow trout muscle: a unique nonmammalian muscle degradation model. Physiol Genomics 28:33–45
Salem M, Kenney PB, Rexroad CE 3rd, Yao J (2006b) Molecular characterization of muscle atrophy and proteolysis associated with spawning in rainbow trout. Comp Biochem Physiol D Genomics Proteomics 1:227–237
Seebacher F, Walter I (2012) Differences in locomotor performance between individuals: importance of parvalbumin, calcium handling and metabolism. J Exp Biol 215:663–670
Small CM, Carney GE, Mo Q, Vannucci M, Jones AG (2009) A microarray analysis of sex- and gonad-biased gene expression in the zebrafish: evidence for masculinization of the transcriptome. BMC Genomics. doi:10.1186/1471-2164-10-579
Sreenivasan R, Cai M, Bartfai R, Wang X, Christoffels A, Orban L (2008) Transcriptomic analyses reveal novel genes with sexually dimorphic expression in the zebrafish gonad and brain. PLoS One. doi:10.1371/journal.pone.0001791
Stejskal V, Kouřil J, Musil J, Hamáčková J, Policar J (2009) Growth pattern of all-female perch (Perca fluviatilis L.) juveniles—is monosex perch culture beneficial? J Appl Ichthyol 25:432–437
Stokes EA, Lonergan W, Weber LP, Janz DM, Poznanski AA, Balch GC, Metcalfe CD, Grober MS (2004) Decreased apoptosis in the forebrain of adult male Medaka (Oryzias latipes) after aqueous exposure to ethinylestradiol. Comp Biochem Physiol C Toxicol Pharmacol 138:163–167
Sun CF, Tao Y, Jiang XY, Zou SM (2011) IGF binding protein 1 is correlated with hypoxia-induced growth reduce and developmental defects in grass carp (Ctenopharyngodon idellus) embryos. Gen Comp Endocrinol 172:409–415
Taggart JB, Hynes RA, Prodöuhl PA, Ferguson A (1992) A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol 40:963–965
Tan I, Ng CH, Lim L, Leung T (2001) Phosphorylation of a novel myosin binding subunit of protein phosphatase 1 reveals a conserved mechanism in the regulation of actin cytoskeleton. J Biol Chem 276:21209–21216
Thrope JE (1977) Bimodal distribution of length of juvenile Atlantic salmon (Salmo salar L.) under artificial rearing conditions. J Fish Biol 11:175–184
Tymchuk W, Sakhrani D, Devlin R (2009) Domestication causes large-scale effects on gene expression in rainbow trout: analysis of muscle, liver and brain transcriptomes. Gen Comp Endocrinol 164:175–183
Ulloa PE, Peña AA, Lizama CD, Araneda C, Iturra P, Neira R, Medrano JF (2013) Growth response and expression of muscle growth-related candidate genes in adult zebrafish fed plant and fishmeal protein-based diets. Zebrafish. doi:10.1089/zeb.2012.0823
von Schalburg KR, Rise ML, Cooper GA, Brown GD, Gibbs AR, Nelson CC, Davidson WS, Koop BF (2005) Fish and chips: various methodologies demonstrate utility of a 16,006-gene salmonid microarray. BMC Genomics 15:126–133
White SL, Sakhrani D, Danzmann RG, Devlin RH (2013) Influence of developmental stage and genotype on liver mRNA levels among wild, domesticated, and hybrid rainbow trout (Oncorhynchus mykiss). BMC Genomics. doi:10.1186/1471-2164-14-673
Zheng W, Xu H, Lam SH, Luo H, Karuturi RK, Gong Z (2013) Transcriptomic analyses of sexual dimorphism of the zebrafish liver and the effect of sex hormones. PLoS One. doi:10.1371/journal.pone.0053562
Acknowledgments
This investigation was supported by a Natural Sciences and Engineering Research Council of Canada Strategic Grant. For providing the experimental fish used in this study, the authors wish to thank Sean Pressey and Lynn and Clarke Rieck from Lyndon Fish Hatcheries Inc. We also wish to thank Michael Burke and the staff of the Alma Aquaculture Research Station for the care and maintenance of the experimental fish. Additionally, Xia Yue and Anne Easton helped with the collection of the phenotypic data and tissue samples.
Conflict of Interest
The authors state this research is free of conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Online Resources 1–8
Tables displaying up-regulated genes in the large and small XX and XX/XY offspring for the liver and the white muscle. Tables contain gene names (when known), the Agilent identification numbers, the fold-change between the large and small fish, and the p-value. (DOCX 141 kb)
Online Resource 9
Displays the genes, with their Agilent identification numbers, fold-change between the large and small fish, and the p-values, up-regulated in the white muscle falling within the GO ‘cellular process category’ for the large and small XX offspring and the small XX/XY offspring in the white muscle. (DOCX 33 kb)
Online Resource 10
Displays the genes, with their Agilent identification numbers, fold-change between the large and small fish, and the p-values, up-regulated in the white muscle falling within the GO ‘metabolic process’ category for the large and small XX offspring and the small XX/XY offspring in the white muscle. (DOCX 29 kb)
Online Resource 11
Displays the genes, with their Agilent identification numbers, fold-change between the large and small fish, and the p-values, up-regulated in the white muscle falling within the GO ‘cellular component organization’ category for the large XX offspring and the small XX/XY offspring in the white muscle. (DOCX 22 kb)
Online Resource 12
Displays the genes, with their Agilent identification numbers, fold-change between the large and small fish, and the p-values, up-regulated in the white muscle falling within the GO ‘response to stimulus’ category for the large and small XX offspring in the white muscle. (DOCX 18 kb)
Online Resource 13–16
Contains the list of the sequences falling within each GO category in the liver and the white muscle for the XX/XY offspring and the XX offspring. (XLSX 52 kb)
Online Resource 17
Contains the G-test of heterogeneity results for the white muscle at GO level 2 (XLSX 13 kb)
Online Resource 18
Displays the genes, with their Agilent identification numbers, fold-change between the large and small fish, and the p-values, up-regulated in the white muscle falling within the GO ‘macromolecule metabolic process’ category for the large and small XX offspring in the white muscle. (DOCX 17 kb)
Online Resource 19
Displays the genes, with their Agilent identification numbers, fold-change between the large and small fish, and the p-values, up-regulated in the white muscle falling within the GO ‘small molecule metabolic process’ category for the small XX offspring in the white muscle. (DOCX 16 kb)
Online Resource 20
Displays the genes, with their Agilent identification numbers, fold-change between the large and small fish, and the p-values, up-regulated in the white muscle falling within the GO ‘cellular metabolic process’ category for the large and small XX offspring and the small XX/XY offspring in the white muscle. (DOCX 25 kb)
Online Resource 21
Contains the G-test of heterogeneity results for the white muscle at GO level 3. (XLSX 16 kb)
Online Resources 22 and 23
Contains the G-test of heterogeneity results for the liver at GO levels 2 and 3. (XLSX 22 kb)
Rights and permissions
About this article
Cite this article
Kocmarek, A.L., Ferguson, M.M. & Danzmann, R.G. Comparison of Growth-Related Traits and Gene Expression Profiles Between the Offspring of Neomale (XX) and Normal Male (XY) Rainbow Trout. Mar Biotechnol 17, 229–243 (2015). https://doi.org/10.1007/s10126-015-9612-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-015-9612-5