Abstract
The effects of short-time fasting on appetite, growth, and nutrient were studied in Atlantic salmon (Salmo salar) smolts. Feed deprivation did change the energy metabolism with reduced plasma protein and muscle indispensible amino acid levels. Plasma levels of ghrelin were significantly higher in starved salmon compared with fed fish after 2 days, but no differences in circulating ghrelin were found between treatments after 14 days. Two mRNA sequences for ghrelin-1 and ghrelin-2, 430 and 533 bp long, respectively, were detected. In addition, the growth hormone secretagogues-receptor like receptor (GHSR-LR) 1a and 1b were identified. Ghrelin-1 but not ghrelin-2 mRNA levels were affected by starvation in the stomach. Lower ghrelin-1 mRNA levels were detected at day 2 in starved fish compared with fed fish. The mRNA levels of GHSR-LR1a were not affected by starvation. Fasting reduced the phenotypic growth and the transcription of insulin-like growth factor (IGF)-II together with IGF-IIR, but IGF-I mRNA were not regulated in fasted salmon after 14 days. Three IGF-binding proteins (IGFBP) at 23, 32, and 43 kDa were found in salmon, and circulating 23 kDa was significantly increased after 14 days of starvation compared with fed fish, indicating increased catabolism. The levels of IGFBP-1 mRNA were significantly higher in fed and starved fish after 14 days compared to those at the start of the experiment, but no significant difference was observed between the treatments. In conclusion, we have shown that circulating ghrelin and ghrelin-1 mRNA is related to changes in energy metabolism in Atlantic salmon.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For salmonids and many other species of fish, periods of reduced food intake is a normal phase of their lives. Endocrine regulation of food deprivation and growth in fish is relative complex. Ghrelin is one of the many neuropeptide hormones that have been documented to be involved in appetite stimulatory (orexigenic) responses in rodents (Kojima et al. 1999). Ghrelin is a growth hormone secretagogue and functions primarily as a GH-releasing hormone and as an orexigen, but has also been documented to be involved in the immune system, stress, energy metabolism, and growth in fish (Kaiya et al. 2008). Ghrelin in fish is as in mammals predominantly produced in endocrine cells of the stomach (Kaiya et al. 2003; Sakata et al. 2004).
In fish, circulating ghrelin directly or indirectly acts as an endocrine peptide to stimulate the release of some hormones, but the regulative mechanism is not yet clear (Kaiya et al. 2008). In shorter timescale, Unniappan et al. (2004) have observed a postprandial decrease in ghrelin mRNA expression in the hypothalamus and gut, and a postprandial decrease in serum ghrelin levels in goldfish. Pankhurst et al. (2008a, b) showed elevated plasma ghrelin levels 24 h after the last meal in rainbow trout. These results suggest that ghrelin is a possible orexigen. In longer-term scale, Jönsson et al. (2007) reported that plasma levels of ghrelin decreased during fasting over a period of 1–3 weeks in a study with rainbow trout. On the other hand, Murashita et al. (2009) reported increased mRNA levels on ghrelin-1, but not ghrelin-2 in the stomach of salmon (Salmo salar) starved for 6 days.
The ghrelin receptor sequences (growth hormone secretagogues-receptor like receptor, GHSR-LR) are recently published in rainbow trout (Kaiya et al. 2009a). Gene structure and characterization of the protein sequence identified it to be closely similar to GHS-R1a in pufferfish Arothron meleagis (Palyha et al. 2000), black seabream Acanthopagrus schlegeli (Chan and Cheng 2004), and tilapia (Kaiya et al. 2009b), although further study is required to confirm receptor activations by ghrelin or GHS (Kaiya et al. 2009a). Exogenous (human) ghrelin intraperitoneally (i.p.) and intracerebroventricularly administered stimulates feeding in goldfish (Unniappan et al. 2004) and reduces food intake in rainbow trout (Jönsson et al. 2010), but i.p. administered trout ghrelin did not affect food intake during 12-h postinjection in rainbow trout (Jönsson et al. 2007). Pre-smolts and smolts of Atlantic salmon following stress showed elevated levels of plasma levels of cortisol and glucose and a suppression of plasma ghrelin levels (Pankhurst et al. 2008b).
Insulin-like growth factor I (IGF-I) is the major anabolic agent responsible for tissue growth (Duan 1998; Thissen et al. 1999), but there is clear evidence that insulin-like growth factor II (IGF-II) is related to local paracrine/autocrine regulation of muscle tissue growth in teleost fishes (Vong et al. 2003; Hevrøy et al. 2007). We have earlier examined the possible nutritional regulation of the somatotropic system by a single indispensable amino acids (IAA) lysine, methionine, quality of dietary protein, and dietary protein level in Atlantic salmon (Hevrøy et al. 2007; Espe et al. 2008; Hevrøy et al. 2008; El-Mowafi et al. 2010). All these studies have shown prominent responses on the GH-IGF system in Atlantic salmon in relation to dietary protein deficiency resulting in reduced feed intake and/or growth reductions. The availability of IGF-I to bind to receptors is regulated by a family of high-affinity IGF-binding proteins (IGFBPs). Shimizu et al. (2005) suggested that regulation of free IGF levels gives the IGFBP-1 a crucial role in regulating the metabolic action of IGF-I in chinook salmon (Oncorhynchus tshawytscha).
During starvation periods, it is necessary to allocate energy reserves away from growth and into support of vital processes (Sumpter et al. 1991). Both protein and fat were important sources of energy during starvation of adult Atlantic salmon (Einen et al. 1998). Changes in energy stores have not been studied in detail regarding free IAAs in the muscle, where limitation of specific amino acids is suggested to limit protein synthesis and growth of salmon (Carter et al. 2000). Smolts are probably vulnerable to food deprivation during the early postsmolt phase, and little is known about how this affects free amino acids in the muscle growth phase. In this study, we used newly adapted 1+ postsmolt for a short experimental study to examine the relationship between nutrients, IGFs, IGF-binding proteins, and the responses of the hormone ghrelin during feed deprivation.
Materials and methods
Fish and experimental condition
The starvation experiment was conducted at Nofima Ingredients, Austevoll (60º05′N, 05º16′E), Norway. Salmon postsmolt (Salmo salar L., 1+) of the NLA strain was used in the experiment. After an acclimation period to seawater for 34 days in 2 × 2 0.8-m3 tanks, the fish with an average body weight of 128 ± 19 g (N = 280, n = 70) were randomly distributed into four similar tanks all with feed collectors (Hølland Teknologi AS, Sandnes, Norway). Experimental tanks were kept indoor under continuous light, and all tanks were supplied with 75 l min−1 seawater pumped from a depth of 50 m. The salmon in two tanks were starved, while the fish in two other tanks were fed a commercial diet (pellet size of 3.5–4.5 mm) consisting of mainly marine protein and marine lipids (analysis dry matter 950 g kg−1, protein 426 g kg−1, lipid 278 g kg−1, and gross energy 23.8 MJ kg−1). During the experiment salinity, ~31.5 gl−1 and temperature 10.1 ± 0.1°C were stable, and the levels of oxygen in the outlet water were never lower than 7.0 mg l−1 (saturation 80%). The fish were fed by automatic feeders that were adjusted every day to maintain 10% feed in excess. Feeding was offered every day in two main periods (5 a.m. to 8 a.m. and 2 p.m. to 3 a.m.). Bulk weight of all fish was obtained at the start of the experiment 10th of June and at the end of experiment 24th of June 2008. All samplings were performed 5 h after the fed fish was fed to ab libitum in the morning meal, and the starved fish was sampled at the same time. This was executed to secure comparable sampling regime of plasma nutrients and hormones. The fish were sedated with 12 mg l−1 AQUI-S in tanks before netting to reduce stress (Olsvik et al. 2007). Individually sampled fish (n = 6 per tank) were killed by a blow to the head, and weights and fork lengths were measured to the nearest g and nearest 0.5 cm at the start, after 2 days and after 14 days of the experiment. The bulk weights were quantified for the remaining fish in each tank at the start and the end of experiment. Samples of brain, liver, stomach and white muscle from the medial part of the fillet were collected and flash-frozen in liquid nitrogen and stored at −80°C until analyzed. Blood was withdrawn from the caudal veins using sterile needles (Vacuette, Greiner bio-one) with vacutainer and put on 10-ml EDTA −2Na tubes on ice before centrifugation (1,250×g for 10 min). The study was performed in accordance with the guidelines drawn by the Norwegian State Commission for Laboratory Animals.
Chemical analyses
Crude chemical analyses of the diets were conducted as briefly described. Nitrogen was determined after total combustion using a Nitrogen–Analyser. Dry weight and ash content was determined gravimetrically after freeze-drying the samples and dried to final weight in an oven at 550°C. Amino acids in de-proteinized muscle tissues and plasma were determined on a Biochrom 20 plus amino acid analyzer (Amersham Pharmacia Biotech, Sweden) equipped with a lithium column using postcolumn derivatization with ninhydrin as described (Espe et al. 2006). Plasma samples were analyzed for the enzymes alanine aminotransferase (ALAT), aspartate aminotransferase (ASAT) and for the nutrient glucose, total protein, cholesterol and triacylglycerol (TAG) on a Maxmat Biomedical Analyser (SM1167, Maxmat S.A., France). For the analyses, Maxmat reagents and the appropriated calibrators and controls for the different methods were used.
RNA extraction
Total RNA was extracted using Trizol reagent according to the manufacturer’s recommendation (Invitrogen). In order to eliminate genomic DNA, the total RNA samples were subjected to DNase treatment (DNeasy, Ambion), using the manufacturer’s protocol. Quantity and quality of RNA were assessed with the NanoDrop® ND-1000 UV–Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA), where a 260/280 nm absorbance ratio of 1.8–2.0 indicates a pure RNA sample. To evaluate the RNA integrity, the 6000 Nano LabChip® kit (Agilent Technologies, Palo Alto, CA, USA) was used, with average RNA Integrity Numbers (RIN) of 9.7 (SD ± 0.6) in brain, 10 (SD ± 0.0) in liver, 9.7 (SD ± 0.2) in stomach, and 10 (SD ± 0.0) in white muscle tissue.
Cloning and bioinformatic analysis of ghrelin mRNA
PCR primers for the cloning of the ghrelin transcripts in Atlantic salmon were designed based on homologous ghrelin mRNA sequences in rainbow trout (accession NM_001124588). These are listed in Table 2. PCR was carried out using a mix of the primers on an Atlantic salmon stomach cDNA sample run for 45 cycles. The obtained products were run on a 2% agarose DNA gel, and individual products cut out from the gel and sequenced. These sequences were compared to known ghrelin sequences in GenBank Protein, using BLASTX. Based on this comparison, it was clear that pure products encoding ghrelin-1 (ghr1) as well as ghrelin-2 (ghr2) mRNA had been obtained. The sequences were assembled using the Phred/Phrap package (Ewing and Green, 1998), and based on the assembled sequences, RACE-PCR primers specific for each transcript were designed (Table 1). The 5′ and 3′ ends of the sequences were obtained by rapid amplification of the cDNA ends (RACE)-PCR (GeneRacer, Invitrogen) according to the manufacturer’s protocol, including the use of nested PCR. PCR products were ligated into a pCR4-TOPO plasmid vector and transferred to a chemically competent TOP10 Escherichia coli cells (TOPO TA cloning kit, Invitrogen). After selection on agar, plasmids of transformed cells were isolated (miniprep; Bio-Rad). Sequencing was performed by the dideoxynucleotide procedure using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) on the ABI Prism 310 automated sequencer (Applied Biosystems).
Resulting sequence data from each gene was combined with earlier sequences and re-assembled using Phred/Phrap. Resulting contigs for each transcript were manually inspected and compared to publicly available ghrelin sequences using BLASTN. The preproghrelin-1 was aligned to available homologous sequences from a selection of fish species as well as the sequence from chicken (Gallus gallus) and mouse (Mus musculus), using PRANK default parameters. The first 24 residues of the predicted protein sequence in chum salmon Oncorhynchus keta (A8R5N8) were not included in the alignment due to probable incorrect annotation of the translation start site in this protein, which does not start with a valid start codon. Using PHYML, the resulting multiple alignments was used to find a maximum likelihood evolutionary tree based on this protein sequence.
Real-time RT-PCR
PCR primers for the targets genes IGF-I, IGF-II, GH-R, IGF-R, and the reference gene acidic ribosomal phosphoprotein (ARP) in S. salar were designed with the Primer Express software and Biosoft software as previously described (Hevrøy et al. 2007). PCR primers for S. salar IGF-IIR have been described by Bower et al. (2008). GHSR-LR1a and GHSR-LR1b were designed with the Primer Express software based on sequences of O. mykiss (Kaiya et al. 2009a) (Table 1). Amplified PCR products of all GHSR-LR1a and GHSR-LR1b cDNAs were sequenced to ensure that the correct mRNA sequences were quantified. The fragments were sequenced with BigDye version 3.1 fluorescent chemistry (Applied Biosystems) and run on an ABI PRISM® 377DNA apparatus at the University of Bergen Sequencing Facility. The identified gene of S. Salar GHSR-LR1a and GHSR-LR1b were given the GenBank accession numbers GQ373171 and GU181419, respectively. All the designed primers were BLAST searched against any salmon sequence in the NCBI database to verify specificity. As reference genes, the ARP (Hevrøy et al. 2007), β-actin, and elongation factor 1αβ (EF1-αβ) (Olsvik et al. 2005) were used (Table 2). For verification, PCR products were run on a 2% agarose DNA gel with ethidium bromide staining and analyzed with the Bio-Rad Gel Doc 2000.
Reverse transcription (RT) was performed using a modified Multiscribe reverse transcription protocol (GeneAmp PCR 9700, Applied Biosystems, AB) as previously described (Hevrøy et al. 2006). Brain, liver, stomach, and white muscle samples from each fish were run in duplicate (250 ng, ±5%) on 96-well plates for the RT and real-time PCR. Real-time PCR amplification and analysis were performed on a LightCycler 480 Real-time PCR system (Roche Applied Science, Basel, Switzerland). The real-time PCR SYBR Green Master Mix (LightCycler 480 SYBR Green master mix kit, Roche) contained FastStart DNA polymerase and gene specific primers at final concentration of 500 nM. Two microliters of cDNA from each well was transferred to a new plate, and 10 μl of real-time master mix was added. The following real-time PCR protocol was employed; (stage 1) heating to 50°C for 2 min, (stage 2) heating to 95°C for 10 min, and (stage 3) 50 cycles of 95°C for 15 s and 60°C for 60 s. Efficiency of real-time PCR was monitored using twofold dilution curves of total RNA using 5 points (range 500–31.25 ng/μl).
Radioimmunoassay
Ghrelin levels in plasma were measured with a heterologous assay using a 125I-radioimmunoassay kit for human ghrelin (Linco Research Inc, St Charles, Missouri), which is specific for biologically active (octanoylated) ghrelin. One milliliter of plasma was immediately acidified (HCl to a final concentration of 0.1 M) for measurements of active ghrelin (Hosoda et al. 2000). The assay has been validated in use for Atlantic salmon by Pankhurst et al. (2008b).
Western ligand blotting using digoxigenin-labeled IGF-I
Recombinant human IGF-I (hIGF-I; Novozymes GroPep Ltd., Adelaide, SA, Australia) was labeled with digoxigenin-3-o-metylcarbonyl-e-aminocaproic acid-N-hydroxysuccimimide ester (DIG; Roche, Basel, Switzerland) according to the method of Shimizu et al. (2000). DIG was stored at –20°C until use. Western ligand blotting was carried out according to the method of Hossenlopp et al. (1986) with modifications. One microliter of normal human serum (Sigma), and 2 μl of salmon plasma, was separated by discontinuous 12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) under nonreducing condition. Proteins were transferred to a nitrocellulose membrane after SDS–PAGE. The nitrocellulosemembrane was incubated with 60 ng/ml of DIG-hIGF-I in Superblock (Pierce, Rockford, IL) at 4°C overnight. After washing, the membrane was incubated with antibody against DIG (Roche) at a dilution of 1:2500 for 2 h at room temperature. IGFBP was visualized by the use of the enhanced chemiluminescence (ECL) Western blotting reagents (Amersham Life Science Inc., Arlington Heights, IL) and the use of autoradiography film (Amersham Life Science Inc.). Molecular masses of Atlantic salmon IGFBPs were estimated by the Precision Plus Protein Standard Kaleidoscope™ (Biorad).
Calculation and statistical analysis
CT values were calculated using the E-Method from Roche Applied Science. The E-Method analyzes the amplification efficiency of target and reference genes by using relative standards. The different reference genes were evaluated by the geNorm VBA applet for Microsoft Excel (Vandesompele et al. 2003) where the rank of the best-fitted reference gene in brain tissue was as follows: β-actin (M-value 0.483) = EF1-αβ (0.483), for liver tissue; EF1-αβ (0.378) < ARP (0.389) < β-actin (0.441), for stomach tissue; EF1-αβ (0.247) = ARP (0.247) and for white muscle; EF1-αβ (0.405) < ARP (0.418) < β-actin (0.524). All of the reference genes were used for normalization of the target genes as neither of them were affected by treatment (evaluated by geNorm and REST 2005). Data for other statistical treatments were tested for normal distribution using the Kolomogorov–Smirnov test (Zar, 1984). A one-way ANOVA was conducted to test the effects of the treatments on individual length, weight, condition factor, and effects of treatments in muscle and liver gene expression. Two-way ANOVA was conducted to test the effects of plasma nutrients, amino acids, ghrelin concentrations, and on the treatments on gene expression in brain and stomach tissue, excluding the start levels from the statistical analysis. Levenes test was used to test the homogeneity of the ANOVA data. Significant differences between groups were tested by Student–Newman–Keuls test. The level of statistical significance was set at P < 0.05.
Results
Phenotypic responses and feed intake
Starved salmon had lower growth (TGC), weight gain, and condition factor compared with fed salmon after 14 days (Table 3). Compared to the start of the experiment, individual lengths were significantly increased in fed fish after 14 days (one-way ANOVA, P = 0.014, n = 12, Table 3). Starved fish had significantly reduced condition factor after 14 days (one-way ANOVA, P = 0.017, n = 12, Table 3). No significant effects of individual body weight (BW) were detected between the two treatments (one-way ANOVA, P = 0.063, n = 12, data not shown). Mean feed intake was 0.7% of BW during the 14 days (Table 3).
Plasma ghrelin and IGF-binding proteins
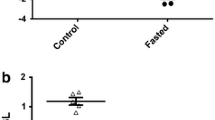
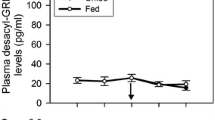
Plasma concentrations of ghrelin were significantly higher in starved salmon compared with fed salmon (two-way ANOVA, P = 0.035, Fig. 1). There were differences in plasma concentrations of ghrelin between starved and fed salmon after 2 days, but no differences after 14 days (Newman–Keuls, P > 0.05, Fig. 1). Three IGFBPs at 23 kDa, 32 kDa, and 43 kDa were indentified in the plasma of Atlantic salmon (Fig. 2a). There were significant effects of treatment on 23 kDa after 14 days of starvation compared with fed fish (two-way ANOVA, P = 0.025, Fig. 2b), with higher plasma levels of 23 kDa in starved fish compared with fed fish (Newman–Keuls, P < 0.05, Fig. 2b). The levels of 32 and 43 kDa were unaltered after 14 days of treatment.
a Western ligand blotting with digoxigenin-labeled human IGF-I of plasma from Atlantic salmon (pooled samples of six fish, n = 2). Fed or starved fish were sampled after 14 days. Normal human serum (NHS; 1 μl) was used as internal standards. Arrows indicate migration position of newly identified Atlantic salmon IGFBPs with molecular mass (kDa) or nonspecific band (NS). b Relative intensity of 23, 32, and 43 kDa insulin-like binding protein in fed (filled) and starved (blank) groups of Atlantic salmon after 14 days (pooled samples of six fish, n = 2, SE). Western blots were quantified by using Image-J, and values are expressed as relative intensity according to normal human serum sample. Superscript letters indicate significant difference (Newman–Keuls test, P < 0.05)
Cloning and analysis of ghrelin mRNA sequences
Quality clipped mRNA sequences for preproghrelin-1 and -2 are 430 bp and 533 bp long, respectively, and these have been deposited in the GenBank with accession numbers EU513378 and EU513379. Both contain the full coding sequence for their respective protein products. Of the 111 residues in the predicted protein sequence of Atlantic salmon preproghrelin-1, only two residues differ from the homologous protein sequence in rainbow trout. Neither of these two sites belongs to the three residues unique for preproghrelin-1, and the same difference is found between the predicted protein sequence for S. salar preoproghrelin-2 and its O. mykiss homolog, supporting their authenticity.
Phylogenetic analysis
The topology of the evolutionary tree agrees with the taxonomic relationships of the included organisms (Fig. 3).
PHYML maximum likelihood evolutionary tree of Atlantic salmon (Salmo salar, EU513378) Ghr 1 and ghrelin-1 in rainbow trout (Oncorhynchus mykiss, NM_001124588), ghrelin-1 in chum salmon (Oncorhynchus keta, AB36005), ghrelin in Japanese eel (Anguilla japonica, AB062427), ghrelin-1 in zebrafish (Danio rerio, NM_001083872), ghrelin in common carp (Cyprinius carpio, AB332394), chicken (Gallus gallus, Q8AV73), and ghrelin in mouse (Mus musculus, A2RSS9)
Gene expression
In brain, the mRNA levels of GHS-R1a were significantly up-regulated in both starved and fed fish compared to at 14 days of treatment (two-way ANOVA, P < 0.001, Fig. 4), but no significant differences were detected between starved and fed fish. GHS-R1b was not expressed in the brain of Atlantic salmon as detected by real-time RT-PCR. In liver, the mRNA levels of IGFBP-1 were significantly up-regulated by both treatments (one-way ANOVA, P < 0.001, Fig. 5). The levels of IGFBP-1 mRNA were significantly lower at the start of the experiment compared to fed and starved fish after 14 days (Newman–Keuls, P < 0.05), but no significant difference was observed between the treatments. The mRNA levels of ghrelin-1 in the stomach were significantly higher in fed compared with starved fish (two-way ANOVA, 0.031, Fig. 6), and there was a trend that mRNA levels of ghrelin-2 (two-way ANOVA, P = 0.052) were higher in fed fish compared with starved fish (Fig. 6). Levels of ghrelin-1 were significantly higher in fed fish compared with starved fish at day 2, but not at day 14. Levels of IGF-II and IGF-IIR mRNA in white muscle were significantly down-regulated by treatment (one-way ANOVA, P = 0.001 and P = 0.018, Fig. 7). Levels of IGF-II and IGF-IIR mRNA were significantly higher at the start of the experiment compared to fed and starved fish after 14 days (Newman–Keuls, P < 0.05, Fig. 7), but no significant difference was detected between fed and starved fish after 14 days.
Mean normalized expression (MNE, mean ± SE) of growth hormone secretagogues-receptor like receptor (GHSR-LR1a) in the brain of Atlantic salmon (n = 12) at the start of the experiment and after fed and starved for 2 days and 14 days. The target genen GHS-R1a was normalized with ARP, EF-1αβ, and β-actin using geNorm. Superscript letters indicate significant difference (Newman–Keuls test, P < 0.05)
Mean normalized expression (MNE, mean ± SE) of insulin-like growth factor I (IGF-I), insulin-like growth factor II (IGF-II), and IGF-binding protein 1 in liver of Atlantic salmon (n = 12) at the start of the experiment and fed and starved for 2 days and 14 days. The target genes were normalized with ARP, EF-1αβ, and β-actin using geNorm. Superscript letters indicate significant difference (Newman–Keuls test, P < 0.05)
Mean normalized expression (MNE, mean ± SE) of ghrelin-1 (Ghr1) and ghrelin-2 (Ghr2) in stomach of Atlantic salmon (n = 12) at the start of the experiment and fed and starved for 2 and 14 days. The target genes were normalized with ARP, EF-1αβ, and β-actin using geNorm. Superscript letters indicate significant difference (Newman–Keuls test, P < 0.05)
Mean normalized expression (NME, mean ± SE) of insulin-like growth factor I (IGF-I), insulin-like growth factor II (IGF-II), insulin-like growth factor I receptor (IGF-IR), and IGF II receptor (IGF-IIR) in white muscle of Atlantic salmon (n = 12) at the start of the experiment and fed and starved for 14 days. The target genes were normalized with ARP, EF-1αβ, and β-actin using geNorm. Superscript letters indicate significant difference (Newman–Keuls test, P < 0.05)
Plasma nutrient and clinical parameters
Plasma glucose concentrations were higher in starved compared with fed salmon (two-way ANOVA, P = 0.029, Table 4), but there were no significant differences within treatments on day 2 or after 14 days (Newman–Keuls, P > 0.05, Table 4). The plasma protein concentrations were affected by treatment (two-way ANOVA, P = 0.020, Table 4), and there were lower protein levels in starved fish after 2 and 14 days compared with fed fish (Newman–Keuls, P > 0.05, Table 4). Aspartate amino transferase (ASAT) was significantly affected by time (two-way ANOVA, P = 0.020, Table 4), but there were no differences between starved and fed fish (Newman–Keuls, P > 0.05). Plasma triacylglycerol, cholesterol, and alanine amino transferase (ALAT) concentrations were equal in starved and fed fish.
Free amino acids
The plasma histidine concentration was significantly reduced by time (two-way ANOVA, P = 0.001, Table 5), and there was a significant interaction effect between treatment and time (two-way ANOVA, P = 0.001), with lower histidine concentration when starved at 14 days compared with higher concentration at starved fish compared to fed fish at 2 days (Newman–Keuls, P < 0.05). There was a significant reduction in lysine concentration by time (two-way ANOVA, P = 0.049), but no treatment-mediated effects were observed (Newman–Keuls, P > 0.05, Table 5). No major effects were observed on the concentrations of other plasma-free amino acids after 2 days of starvation, but lower concentration of the sum of the indispensable amino acids (IAAs) and total free amino acids were observed after 14 days of starvation (Table 5). Free IAA concentrations in white muscle were significantly affected by treatment (two-way ANOVA, P = 0.010, Table 6), with reduced concentrations in starved fish compared to fed salmon (Newman–Keuls, P < 0.05, Table 6). Muscle lysine concentration was significantly affected by treatment (two-way ANOVA, P = 0.009, Table 6), and significantly lower concentrations were detected in starved fish compared with fed fish (Newman–Keuls, P < 0.05). No difference was seen in the sum of dispensable amino acids in fed or starved fish (data not shown), and in the total pool of muscle amino acids between starved and fed fish (Tables 5 and 6).
Discussion
Short-time food deprivation reduced both body weight and condition factor, in line with weight loss data reported after 15 days of starvation in Atlantic salmon and rainbow trout of similar size (Wilkinson et al. 2006; Cleveland et al. 2009).
Starved salmon showed higher plasma concentrations of ghrelin after 2 days, but no effects were seen on ghrelin plasma level in starved vs. fed fish after 14 days of treatment, supporting the results of Pankhurst et al. (2008a). In the current study, we can see large alternation of the energy metabolism with particular lower levels of circulating protein and free muscle amino acids lysine in starved fish, but we cannot exclude effects of possible changes in other nutrients, such as triacylglycerol and cholesterol, indicating changes in the energy metabolism. Plasma amino acid concentrations were clearly reduced after 14 day of food deprivation, suggesting a need for additional amino acids for energetic purposes. Plasma glucose levels were unaltered during food deprivation in our study, and similar effects have been shown in rainbow trout after food deprivation for 9 days, and in Atlantic salmon fed restricted satiation ration (Sundby et al. 1991). Our results indicate that plasma ghrelin is related to changes in energy metabolism in Atlantic salmon, and after 14 days on starvation, the salmon has slowed down the metabolism to reduce energy spending, and therefore there is no response in ghrelin circulation levels of starved fish. Fasting for 24 h also increased ghrelin plasma levels in Japanese quail (Shousha et al. 2005), and all these observations are indicating ghrelin to be an orexigen or a short-time regulator of energy homeostasis as shown in goldfish (Unniappan et al. 2004). This is the opposite result compared with what was reported in starved rainbow trout by Jönsson et al. (2007), which found increased plasma level of ghrelin in fed fish compared with starved fish after 7, 14, and 21 days. Recent findings may suggest ghrelin to be a short-time regulator of energy metabolism in salmonids and that starvation for longer periods does not alter the plasma ghrelin level as shown by Jönsson et al. (2007). In a recent study by Fox et al. (2009), it was shown that ghrelin is acting as a long-term indicator of negative energy balance in tilapia Oreochromis mossambicus, and not as a meal-initiated signal. They detected significantly higher plasma ghrelin levels in starved fish after 2 and 4 weeks of starvation that correlated with reduced glucose level. Atlantic salmon is a carnivore fish highly adapted for protein and not so much carbohydrate metabolism as omnivorous species such as tilapia (Hemre et al. 2002), and these differences in dietary requirements between species possibly influence when changes in energy metabolism occur during starvation and hence plasma ghrelin levels.
Independent of this work, Murashita et al. (2009) submitted full-length mRNA sequences for preproghrelin-1 and -2 in S. salar to the GenBank. These sequences are almost identical to our sequences (EU513378 and EU613379), with the exception of a single base difference in preprogrehlin-1 that would cause a synonymous substitution. In the current study, we observed significant lower levels of ghrelin-1 mRNA in starved fish compared with fed fish after 2 days, but no altered levels after 14 days of treatment. Increased mRNA levels on ghrelin-1 but not ghrelin-2 in stomach of salmon starved for 6 days were observed by Murashita et al. (2009). The results of Murashita et al. (2009) regarding ghrelin mRNA regulation are thus contradicting to our results, most likely due to the timing of sampling. Ghrelin mRNA levels and circulating plasma ghrelin have been shown to rise significantly prior to a meal and decrease following feeding in mammals (Bagnasco et al. 2002), and similar findings have been reported in the goldfish (Unniappan et al. 2004). Recently, Peddu et al. (2009) documented stomach ghrelin mRNA levels in tilapia to rise preprandially and remain unchanged following a meal. In fish from the same study that missed the scheduled feeding, ghrelin mRNA levels in the stomach dropped significantly 1 h after the meal. Therefore, orexigenic signals are likely to be rapidly induced and relative short lived. Our result showing down-regulation of ghrelin mRNA in starved fish after 2 days may be due to that starved fish missed the schedule feeding as suggested by Peddu et al. (2009).
The significant up-regulation of GHSR-LR1a mRNA levels in brain of starved and fed salmon after 2 days may be linked to the sampling of whole brain, where GHSR-LR1a are differently expressed in different areas (Kaiya et al. 2009a), but this needs further investigations. Since we do not have different expression of GHSR-LR1a between treatments, we cannot relate our responses to changes in energy homeostasis or orexigenic signal. For future studies of ghrelin, plasma and tissue samples for gene expression should be sampled both before and after a meal for detailed study of the rapid alternation of circulating plasma levels and mRNA transcription of ghrelin and GHSR-LR (Pankhurst et al. 2008a; Peddu et al. 2009). We have developed a protocol where we sample fish 5 h postprandial, in order to secure high group homogeneity of plasma amino acids and hormones (Espe et al. 1999; Hevrøy et al. 2007). In the current examination, the mRNA levels of ghrelin may have been higher prior to the anticipated meal, but this needs to be addressed in future studies. Jönsson et al. (2007) used a protocol where they did not feed the fish the last 24 h before sampling, and this may have affected the reported plasma ghrelin levels of starved fish in their study. In future examinations, standardized sampling regimes should be used in order to fully understand regulation of circulating and tissue mRNA levels of ghrelin.
No observed significant regulation of IGF-I in muscle and liver of starved or fed Atlantic salmon indicates that IGF-I was not regulated at the transcriptional level in the current study. Earlier studies have shown up-regulation of muscle IGF-I in Atlantic salmon experiencing a restricted feeding regime (Bower et al. 2008). A similar result has been shown in a study with rainbow trout where re-fed animals showed up-regulation of muscle IGF-I and IGF-II compared to starved fish, but IGF-I more prominent (Chauvigné et al. 2003). A longer experimental period in our study would probably have been better for the assessment of the anabolic muscle endocrine response. IGF-II in muscle tissue was significantly down-regulated in starved salmon, which may be caused by the lower level of free indispensable amino acids in the muscle tissue. We have documented a sevenfold up-regulation of IGF-II mRNA level in white muscle of Atlantic salmon when fed a diet with high level of dietary lysine compared with low lysine (Hevrøy et al. 2007). The structural protein compounds of the muscle are rich in the amino acid lysine. In the current study, lower levels of free lysine in muscle may be a cue for the autocrine or paracrine down regulation of IGF-II in starved salmon. Amino acids are potent regulators of hormone secretion from endocrine cells (Newsholme et al. 2005). In mammals, there is evidence that suggests that amino acids (leucine, glycine, tyrosine, phenylalanine, proline, methionine, tryptophan and histidine in liver tissue, and leucine in the skeletal muscle) together with insulin may regulate signaling directly and regulate cell growth through protein kinase cascades, such as mTor, Erk, elF2α. (Kadowaki and Kanazawa 2003). Among amino acids, branched-chain amino acid (BCAAs) and especially leucine seem to be major regulating factors of muscle protein synthesis (Dardevet et al. 2000). In our study, all the indispensable amino acids with the exception of histidine were reduced in starved fish compared with fed fish after 14 days, and this reduction may have been the main factors responsible for IGF-II down-regulation of the paracrine or autocrine regulation of muscle growth. Further study is needed to distinguish which of the indispensible amino acids, or which amino acid combination, that are responsible for reduced anabolic signal, but we would like to speculate that lysine may be involved. Feed restriction in Atlantic salmon mediated significantly down-regulation of IGF-II mRNA muscle level in the early phase of re-feeding before showing increasing levels after 30 days of appetite feeding (Bower et al. 2008). The same response has been showed in muscle of channel catfish (Ictalurus punctatus), with higher IGF-II levels in fed animals compared with restricted animals after 30 days of feeding (Peterson and Waldbieser 2009). The IGF-IR isoform 1b in the muscle of Atlantic salmon examined in the current study was not affected by starvation as documented earlier in salmon and trout in food restriction studies (Chauvigné et al. 2003; Bower et al. 2008). Function of IGF-IIR in fish is not yet clear, but this protein has been reported to target ligands for the lysosomal degradation pathway (Bower et al. 2008). The significant down-regulation of IGF-IIR in muscle of starved fish in the current study correlated well with a significant IGF-II down-regulation, and the same response was shown by Bower et al. (2008) when salmon were in a catabolic or anabolic state.
Shimizu et al. (2006) showed that physiological regulation of the chinook salmon (O. tshawytscha) 22-kDa IGFBP, which is an orthologue of mammalian IGFBP-1, increased in catabolic states such as fasting and stress. A six-week period of fasting increased the circulated 22-kDa IGFBP level and IGFBP-1 gene expression in liver of chinook salmon (Shimizu et al. 2006). In the current study, we have detected the circulating 23-, 32-, and 43-kDa IGFBPs for the first time in Atlantic salmon. This is in good agreement with the patterns observed in other fish species, exhibiting three major IGFBP bands at 20–25, 29–32, and 40–50 kDa (Kelley et al. 2006). In chinook salmon, 22-kDa IGFBP has been identified as an ortholog of mammalian IGFBP-1 (Shimizu et al. 2005). The Atlantic salmon 23-kDa IGFBP should correspond to chinook salmon IGFBP-1 based on the almost identical size and physiological response. Although the identities of two other salmon IGFBPs are still under debate, we have recently identified chinook salmon 41-kDa IGFBP as one of two paralogs of IGFBP-2 (Shimizu et al., unpublished data) and proposed chinook salmon 28-kDa IGFBP to be either a paralog of salmon IGFBP-1 or IGFBP-4 (Shimizu et al. 2005), which may be applicable for the classification of the two Atlantic salmon IGFBPs. Our results showed higher levels of circulating 23-kDa IGFBP and a significant higher level of IGFBP-1 mRNA after 14 days of starvation, suggesting higher catabolic activity in starved fish. However, we also found high IGFBP-1 mRNA levels in fed fish compared with the mRNA levels at the start of the experiment, indicating a catabolic and not proper anabolic phase in the fed salmon. Results from a study with channel catfish (Peterson and Waldbieser 2009) showed a significant up-regulation of IGFBP-1 mRNA in liver of fish starved for 30 days compared to fed fish, but interestingly 15 days of re-feeding did not restore the concentration of IGFBP-1. In conclusion, IGFBP-1 is a good molecular marker of catabolism in feed restriction or starvation studies.
Reduced plasma protein concentrations were detected after 2 days and 14 days of starvation, probably related to a metabolism favoring protein deposition in seawater-adapted fish compared with lipid deposition in freshwater-adapted postsmolts as suggested by Usher et al. (1991). Rapid protein mobilization in Atlantic salmon during starvation has been documented by others (Krogdahl and Bakke-McKellep 2005), who reported a rapid reduction in tissue mass and protein levels of gastrointestinal tissue after only 2 days of starvation. The reduction of plasma and muscle-free indispensable amino acids in salmon starved for 14 days compared with fed fish does support that salmon need protein and indispensable amino acids to maintain high metabolism and to secure an optimum protein growth. All the indispensable amino acids were reduced after 2 and 14 days except histidine, which was not altered during starvation. In addition, the histidine-containing component anserine (beta-alanyl-N-methylhistidine) was maintained at a stable concentration. Stable concentration of free imidazole compounds (histidine and anserine) in muscle may support a priority for the salmon in keeping a high buffer and stable cell osmotic status (Breck, 2004). Anserine is likely to function as an important cellular buffer and as an antioxidant (Kohen et al. 1988), and the observed reduced plasma histidine and increased concentration of anserine and 1-methyl-L-histidine (1-mhis) in starved salmon compared with fed salmon support that histidine and anserine in muscle of salmon may have antioxidant functions, as suggested by Mommsen (2001). Major responses on white muscle-free amino acids status related to nutritional status have been documented during maturation (Mommsen et al. 1980) and feed restrictions (Carter and Houlihan, 2001). The current study supports that food deprivation effects energy metabolism of starved fish because of the reduced plasma protein and muscle indispensable amino acids concentrations and that these large effects are related to the high amino acid demand of the Atlantic salmon (El-Mowafi et al. 2010).
In conclusion, we have showed that circulating ghrelin is related to changes in energy metabolism in Atlantic salmon and that circulating ghrelin together with ghrelin-1 transcription may be regulated relative rapidly during starvation. The circulating indispensable amino acids concentrations and the pool of indispensable amino acids in white muscle are reduced with feed deprivation, and there may be a link to the down-regulation of IGF-II mRNA levels in muscle of fasted salmon after 14 days of starvation. The significantly increased IGFBP-1 mRNA levels in starved fish seem to be the most prominent molecular marker of catabolism in feed restriction or starvation studies.
References
Bagnasco M, Kalra PS, Kalra SP (2002) Ghrelin and leptin pulse discharge in fed and fasted rats. Endocrinol 143:726–729
Bower NI, Li X, Taylor R, Johnston IA (2008) Switching to fast growth: the insulin-like growth factor (IGF) system in skeletal muscle of Atlantic salmon. J Exp Biol 211:3859–3870
Breck O (2004) Histidine nutrition and cataract development in Atlantic salmon, Salmo salar L. Ph.D. thesis, University of Bergen, Bergen, 83
Carter CG, Houlihan DF (2001) Protein synthesis. In: Wright PA, Anderson PM (eds) Nitrogen extraction, fish physiology, vol 20. Academic Press, San Diego, pp 31–75
Carter CG, Houlihan DF, He ZY (2000) Changes in tissue free amino acid concentrations in Atlantic salmon, Salmo salar L., after consumption of a low ration. Fish Physiol Biochem 23:295–306
Chan CB, Cheng CH (2004) Identification and functional characterization of two alternatively spliced growth hormone secretagogue receptor transcripts from the pituitary of black seabream, Acanthopagrus schlegeli. Mol Cell Endocrinol 214:81–95
Chauvigné F, Gabillard JC, Weil C, Rescan PY (2003) Effect of refeeding on IGFI, IGFII, IGF receptors, FGF2, FGF6, and myostatin mRNA expression in rainbow trout myotomal muscle. Gen Comp Endocrinol 132:209–215
Cleveland BM, Weber GM, Blemings KP, Silverstein JT (2009) Insulin-like growth factor-I and genetic effects on indexes of protein degradation in response to feed deprivation in rainbow trout (Oncorhynchus myskiss). Am J Physiol-Reg I 297:R1332–R1342
Dardevet D, Sornet C, Balage M, Grizard J (2000) Stimulation of in vitro rat muscle protein synthesis by leucine decreases with age. J Nutr 130:2630–2635
Duan CM (1998) Nutritional and developmental regulation of insulin-like growth factors in fish. J Nutr 128:306S–314S
Einen O, Waagan B, Thomassen MS (1998) Starvation prior to slaughter in Atlantic salmon (Salmo salar) I. Effects on weight loss, body shape, slaughter-and fillet-yield, proximate and fatty acid composition. Aquaculture 166:85–104
El-Mowafi A, Ruohonen K, Hevrøy EM, Espe M (2010) Impact of digestible energy levels at three different dietary amino acid levels on growth performance and protein accretion in Atlantic salmon. Aquac Res 41:373–384
Espe M, Sveier H, Høgøy I, Lied E (1999) Nutrient absorption and growth of Atlantic salmon (Salmo salar L.) fed fish protein concentrate. Aquaculture 174:119–137
Espe M, Lemme A, Petri A, El-Mowafi A (2006) Can Atlantic salmon grow on diets devoid of fish meal? Aquaculture 255:255–262
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Fox BK, Breves JP, Hirano T, Grau EG (2009) Effects of short- and long-term fasting on plasma and stomach ghrelin, and the growth hormone/insulin-like growth factor I axis in the tilapia, Oreochromis mossambicus. Domest Anim Endocrinol 37:1–11
Hemre G-I, Mommsen TP, Krogdahl Å (2002) Carbohydrates in fish nutrition: effects on growth, glucose metabolism and hepatic enzymes. Aquacult Nutr 8:175–194
Hevrøy EM, Jordal AE, Hordvik I, Espe M, Hemre G, Olsvik PA (2006) Myosin heavy chain mRNA expression correlates higher with muscle protein accretion than growth in Atlantic salmon, Salmo salar. Aquaculture 252:453–461
Hevrøy EM, El-Mowafi A, Taylor R, Olsvik PA, Norberg B, Espe M (2007) Lysine intake affects gene expression of anabolic hormones in Atlantic salmon (Salmo salar). Gen Comp Endocrinol 152:39–46
Hosoda H, Kojima M, Matsuo H, Kengawa K (2000) Ghrelin and Des-acyl Ghrelin: Two major forms of rat ghrelin peptide in gastrointestinal tissue. Biochem Biophys Res Commun 279:909–913
Hossenlopp P, Seurin D, Segovia-Quinson B, Hardouin S, Binoux M (1986) Analysis of serum insulin-like growth factor binding proteins using Western blotting: use of the method for titration of the binding proteins and competitive binding studies. Anal Biochem 154:138–143
Jönsson E, Forsman A, Einarsdottir IE, Kaiya H, Rouhonen K, Björnsson BT (2007) Plasma ghrelin levels in rainbow trout in response to fasting, feeding and food composition, and effects of ghrelin on voluntary food intake. Comp Biochem Phys A 147:1116–1124
Jönsson E, Kaiya H, Björnsson BT (2010) Ghrelin decrease food intake in juvenile rainbow trout (oncorhynchus mykiss) through the central anorexigenic corticotrophin-releasing factor system. Gen Comp Endochrinol 166:39–46
Kadowaki M, Kanazawa T (2003) Amino acids as regulators of proteolysis. J Nutr 133:2052S–2056S
Kaiya H, Kojima M, Hosoda H, Morriyama S, Takahashi A, Kawauchi H, Kangawa K (2003) Peptide purification, complementary deoxyribonucleic acid (DNA) and genomic DNA cloning, and functional characterization of ghrelin in rainbow trout. Endocrinology 144:5215–5226
Kaiya H, Miyazato M, Kangawa K, Richard EP, Unniappan S (2008) Ghrelin: a multifunctional hormone in none-mammalian vertebrates. Comp Biochem Phys A 149:109–128
Kaiya H, Mori T, Miyazato M, Kangawa K (2009a) Ghrelin receptor (GHS-R)-like receptor and its genomic organization in rainbow trout, Oncorhynchus mykiss. Comp Biochem Phys A 153:438–450
Kaiya H, Riley LG, Janzen W, Hirano T, Grau EG, Miyazato M, Kangawa K (2009b) Identification and genomic sequence of a ghrelin receptor (GHS-R)-like receptor in the Mozambique Tilapia, Oreochromis mossambicus. Zool Sci 26:330–337
Kelley KM, Price TD, Galima MM, Sak K, Reyes JA, Zepeda O, Hagstrom R, Truong TA, and Lowe CG (2006) Fish endocrinology. In: Reinecke M, Zaccone G, Kapoor BG (eds) Science Publishers, Enfield, pp 167–195
Kohen R, Yamamoto Y, Cundy KC, Ames BN (1988) Antioxidant activity of carnosine, homocarnosine, and anserine present in muscle and brain. Proc Natl Acad Sci USA 85:3175–3179
Kojima M, Hosoda H, Date Y, Nakazao M, Matuso H, Kangawa K (1999) Ghrelin is a growth-hormone-releasing acylated peptide. Nature 402:656–660
Krogdahl A, Bakke-McKellep AM (2005) Fasting and refeeding cause rapid changes in intestinal mass and digestion enzyme capacities of Atlantic salmon (Salmo salar L.). Comp Biochem Phys A 141:450–460
Mommsen TP (2001) Paradigms of growth in fish. Comp Biochem Phys B 129:207–219
Mommsen TP, French CJ, Hochachka PW (1980) Sites and pattern of protein and amino acid utilization during the spawning migration of salmon. Can J Zool 58:1785–1799
Murashita K, Kurokawa T, Nilsen TO, Rønnestad I (2009) Ghrelin, cholecystokinin, and peptide YY in Atlantic salmon (Salmo salar): molecular cloning and tissue expression. Gen Comp Endochrinol 160:223–235
Newsholme P, Brennan L, Rubi B, Maechler P (2005) New insights into amino acid metabolism, beta-cell function and diabetes. Clin Sci 108:185–194
Olsvik PA, Lie KK, Jordal OA-E, Nilsen TO, Hordvik I (2005) Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Molecul Biol 6:21
Olsvik PA, Lie KK, Hevrøy EM (2007) Does anaesthetics and sampling strategies affect transcription analysis of fish tissues? BMC Molecular Biol 8:48
Palyha OC, Feighner SD, Tan CP, McKee KK, Hreniuk DL, Gao YD, Schleim KD, Yang L, Morriello GJ, Nargund R, Patchett AA, Howard AD, Smith RG (2000) Ligand activation domain of human orphan growth hormone (GH) secretagogue receptor (GHS-R) conserved from pufferfish to human. Mol Endocrinol 14:160–169
Pankhurst NW, King HR, Ludke SL (2008a) Relationship between stress, feeding and plasma ghrelin levels in rainbow trout, Oncorhynchus mykiss. Mar Freshw Behav Phy 41:53–64
Pankhurst NW, Ludke SL, King HR, Peter RE (2008b) The relationship between acute stress, food intake, endocrine status and life history stage in juvenile farmed Atlantic salmon, Salmo salar. Aquaculture 275:311–318
Peddu SC, Breves JP, Kaiya H, Grau GE, Riley LG (2009) Pre-and postprandial effects on ghrelin signalling in the brain and on the GH/IGF-I axis in the Mozambique tilapia (Oreochris mossambibicus). Gen Comp Endocrinol 161:412–418
Peterson BC, Waldbieser GC (2009) Effects of fasting on IGF-I, IGF-II, and IGF-binding protein mRNA concentrations in channel catfish (Ictalurus punctatus). Domest Anim Endocrinol 37:74–83
Sakata I, Mori T, Kaiya H, Yamazaki M, Kangawa K, Inoue K, Sakai T (2004) Localization of ghrelin-producing cells in the stomach of the rainbow trout (Oncorhynchus mykiss). Zool Sci 21:757–762
Shimizu M, Swanson P, Fukada H, Hara A, Dickhoff WW (2000) Comparison of extraction methods and assay validation for salmon insulin-like growth factor-I using commercially available components. Gen Comp Endocrinol 119:26–36
Shimizu M, Dickey JT, Fukada H, Dickhoff WW (2005) Salmon serum 22 kDa insulin-like growth factor-binding protein (IGFBP) is IGFBP-1. J Endocrinol 184:267–276
Shimizu M, Beckman BR, Hara A, Dickhoff WW (2006) Measurement of circulating salmon IGF binding protein-1: assay development, response to feeding ration and temperature, and relation to growth parameters. J Endocrinol 188:101–110
Shousha S, Nakahara K, Kojima M, Miyazato M, Hosoda H, Kangawa K, Murakami N (2005) Different effects of peripheral and central ghrelin on regulation of food intake in the Japanese quail. Gen Comp Endocrinol 141:178–183
Sumpter JP, Le Bail PY, Pickering AD, Pottinger TG, Carragher JF (1991) The effect of starvation on growth and plasma growth hormone concentration of Rainbow trout, Oncorhynchus mykiss. Gen Comp Endocrinol 83:94–102
Sundby A, Eliassen KA, Blom AK, Åsgård T (1991) Plasma insulin, glucagon, glucagon-like peptide and glucose levels in response to feeding, starvation and life long restricted feed ration in salmonids. Fish Physiol Biochem 3:253–259
Thissen JP, Underwood LE, Ketelslegers JM (1999) Regulation of insulin-like growth factor-I in starvation and injury. Nutr Rev 57:167–176
Unniappan S, Canosa LF, Peter RE (2004) Orexigenic action of ghrelin in goldfish: Feeding-induced changes in brain and gut mRNA expression and serum levels, and responses to central and peripheral injections. Neuroendocrinology 79:100–108
Vandesompele J, de Preter K, Pattyn F, Poppe B, van Roy N, de Paepe A, Speleman F (2003) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:1465–1476
Vong QP, Chan KM, Cheng CH (2003) Quantification of common carp (Cyprinus carpio) IGF-I and IGF-II mRNA by real-time PCR: differential regulation of expression by GH. J Endocrinol 178:513–521
Wilkinson RJ, Porter M, Woolcott H, Longland R, Carragher JF (2006) Effects of aquaculture related stressors and nutritional restriction on circulating growth factors (GH, IGF-I and IGF-II) in Atlantic salmon and rainbow trout. Comp Biochem Phys A145:214–224
Zar JH (1984) Biostatistical analysis. Prentice-Hall, Englewood Cliffs, pp 253–260
Acknowledgments
The authors thank Eva Mykkeltvedt, Hui-shan Tung, Jacob Wessels, and Christine Hunskår for great technical help in the molecular laboratory at NIFES. This work has been carried out with financial support from Ministry of Fisheries and Costal Affairs and Research Council of Norway project no. 199683/S40.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hevrøy, E.M., Azpeleta, C., Shimizu, M. et al. Effects of short-term starvation on ghrelin, GH-IGF system, and IGF-binding proteins in Atlantic salmon. Fish Physiol Biochem 37, 217–232 (2011). https://doi.org/10.1007/s10695-010-9434-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-010-9434-3