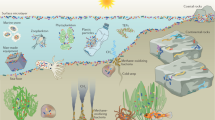
Abstract
In the marine environment, biofilms on submerged surfaces can promote or discourage the settlement of invertebrate larvae and macroalgal spores. The settlement-mediating effects of biofilms are believed to involve a variety of biofilm attributes including surface chemistry, micro-topography, and a wide range of microbial products from small-molecule metabolites to high-molecular weight extracellular polymers. The settled organisms in turn can modify microbial species composition of biofilms and thus change the biofilm properties and dynamics. A better understanding of biofilm dynamics and chemical signals released and/or stored by biofilms will facilitate the development of antifouling and mariculture technologies. This review provides a brief account of 1) existing knowledge of marine biofilms that are relevant to settlement mediation, 2) biotechnological application of biofilms with respect to developing non-toxic antifouling technologies and improving the operation of aquaculture facilities, and 3) challenges and future directions for advancing our understanding of settlement-mediating functions of biofilms and for applying this knowledge to real-life situations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most sessile marine organisms produce planktonic propagules referred to as larvae for invertebrates and spores for algae. The planktonic phase lasts for minutes, hours, days, weeks, or even months while the propagules grow, develop, and drift for various distances in the water column before they settle in new habitats (Qian 1999; Levin 2006). The settlement process is characterized by habitat exploration, which is often followed by a cascade of ontogenetic events and terminates planktonic existence when propagules finally attach to substrates. The establishment of a new community of invertebrates or algae on solid surfaces is commonly referred to as colonization.
Sessile marine organisms can be either a nuisance or a valuable resource to maritime activities, depending on the species of concern and the location of the settlement. For instance, the colonization of man-made structures by invertebrates and algae poses serious threats to the safe and efficient operation of vessels and equipment and consequently leads to enormous economic losses for maritime industries (Yebra et al. 2004). The nuisance phenomenon is commonly referred to as “biofouling” and the associated mitigation measures are called “antifouling” techniques (Clare 1998). There is a substantial industrial and commercial interest in controlling the biofouling process. In contrast, a big challenge for the operation of aquaculture farms (i.e., shellfish and algae) and the rehabilitation of marine habitats is how to effectively and/or purposely induce the settlement of larvae and spores to replenish the broodstocks of commercially valuable or ecologically important sessile organisms. In addition, aquaculture farm operations are also challenged by the settlement of biofouling species that blocks nets and cages, and outcompete the farmed species for space and food (Braithwaite and McEvou 2005). Although the purpose of antifouling technologies is completely opposite to that of aquaculture farming and marine habitat conservation, the operational principle is the same; we can manipulate the behavior and physiology of invertebrate larvae and algal spores during their habitat exploration and settlement.
In 1935, Zobell and Allen first observed that the settlement of invertebrate larvae was enhanced by a thin layer of microbial aggregates present on artificial substrates. These thin microbial aggregates, referred to as biofilms, are ubiquitous on all aquatic surfaces (Cooksey and Wiggleworth-Cooksey 1995; Allison 2003) and are composed of bacteria, diatoms, fungi, unicellular algae, and protozoa (Marshall and Bowden 2000; Maki 2002). During habitat exploration and settlement planktonic propagules typically interface with biofilms. In recent years, there has been a resurgence of research interest on interaction between settling propagules and biofilms, fueled by new experimental evidence that marine biofilms are instrumental to habitat selection and the onset of settlement events for many sessile marine organisms (Maki 1999; Huang and Hadfield 2003; Railkin 2004; Qian et al. 2003). For example, recent laboratory studies suggest that invertebrate larvae can distinguish between biofilms having different microbial community structure and can “choose” to settle on or to reject biofilms of different origins and physiological qualities (Lau et al. 2005; Hung et al. 2005a, b; Dobretsov and Qian 2006). The properties of biofilms responsible for the mediation of settlement include surface chemistry, micro-topography, and a wide variety of microbial products ranging from small-molecule metabolites to high-molecular weight extracellular polymers (Harder et al. 2002a; Lau et al. 2003a; Lam et al. 2005a, b).
There has also been increased evidence that propagule settlement can be induced or inhibited by manipulating settlement-relevant attributes of biofilms or by presenting biofilm mimetic of those attributes to invertebrate larvae and algal spores (Maki 1999; Callow and Callow 2000; Maki et al. 2000; Qian et al. 2003). This novel approach to controlling the settlement process of sessile marine organisms constitutes a promising direction in the development of novel biotechnologies for mitigating biofouling in a nontoxic manner, increasing the yield of aquaculture, and facilitating conservation management of benthic marine habitats.
The aims of this review are to discuss (1) existing knowledge of marine biofilms that are relevant to settlement mediation; (2) biotechnological implications of biofilms with respect to developing nontoxic antifouling technologies, improving the operation of aquaculture facilities, and facilitating the rehabilitation of benthic marine habitats; and (3) current challenges and future directions for advancing our understanding of settlement-mediating functions of biofilms and for applying this knowledge to real-life situations.
Biofilm Formation and Dynamics
Much of the early biofilm work focused on the biofilm development process. It became clear that within seconds following the initial exposure of a surface to seawater, dissolved organic matter adheres to the surface and forms a thin (<100 nm) conditioning film referred to as molecular fouling (Little and Zsolnay 1985). Consequently, the surface receives new physiochemical properties (Charaklis and Cooksey 1983), which subsequently affect the type and kinetics of primary colonizers to be recruited to the surface (Schneider and Marshall 1994). Among the primary colonizers bacteria are usually dominant components owing to their high abundance in seawater (∼105 × 106cells ml−1) (Dang and Lovell 2000). Bacterial cells establish the first surface contact and interact with the conditioning films. It is thus possible to alter primary colonization by manipulating surface physiochemical properties, as demonstrated by Ista et al. (1996). At any time, other microbes that displace or cooperate effectively with existing members will join the biofilm community and go through the same life cycle (Marshall and Bowden 2000).
Meanwhile, over the past decade, quorum sensing signaling has become an emerging field of biofilm research. It is now generally accepted that bacteria produce and respond to chemical signals and that cell-to-cell communications leads to the coordination and reorganization of microbial activities (Parsek and Greenberg 1999). Quorum sensing also controls how biofilms develop, what kind of microbial structure the biofilms will have, and how individual microbes disperse in biofilms (Parsek and Greenberg 1999; Greenberg 2003 and references cited therein). The link between quorum sensing and biofilm development has stimulated recent work in bacterial community dynamics.
The succession of a biofilm community can be influenced by a number of physiological and biological events initiated by primary colonizers, as well as by the modification of physical and biochemical properties of the surface, which determine the types/species of bacteria to be recruited as secondary colonizers (Lappin-Scott and Costerton 1989). Synergistic and/or competitive interactions among colonizers, together with the arrival of new recruits and/or loss of previous colonists, continuously shape the biofilm community (Wimpenny 1996). Biofilm succession is also influenced by physical and chemical conditions of the external environment. For example, microbes capable of rapid adhesion have a distinct selective advantage in turbulent flows. Moreover, nutrient loads in the aqueous phase determine the number, type, and metabolic state of planktonic microbes available to surfaces, as well as the tendency of the microbes to adhere to surfaces and become part of the biofilm community. On the other hand, microbes also have the ability to deliberately detach from biofilms when the local conditions become unfavorable (Maki 1999).
Diatoms will attach to the surface at any time during biofilm development and grow when light is sufficient (Cooksey et al. 1984). The proliferation of diatoms and bacteria in biofilm results in a patchy distribution of microcolonies. As the thickness of the biofilm increases, sharp gradients of pH and dissolved oxygen develop within the biofilms. The metabolic byproducts of microbes living in the deeper zones have important implications in metal corrosion (Acuňa et al. 2006).
The structure and composition of biofilms as well as their mediatory effects on microbial and larval propagules are very dynamic owing to changes in environmental variables. Since even a mature biofilm is never static, the relationship between habitat and biofilm community is tight and reciprocal so that the structure, composition and/or physiology of a biofilm community effectively reflect key environmental factors of a substratum.
Response of Larvae and Algal Spores to Biofilms
Natural biofilms can either enhance (Kirchman et al. 1982; Mitchell and Maki 1988; Callow and Callow 2000; Qian et al. 2003; Huang and Hadfield 2003; Hung et al. 2005a, b; Lau et al. 2005), or inhibit settlement of marine invertebrate larvae and macroalgal spores (Maki et al. 1988; Holmstrøm et al. 1992; Egan et al. 2001; Dobretsov and Qian 2002; Dobretsov and Qian 2004), or have no observable effect (Todd and Keough 1994; Lau et al. 2003a). The ability of larvae and spores to differentiate between biofilms of varying composition (e.g., Patel et al. 2003; Lau et al. 2005), density (e.g., Maki et al. 1988; Joint et al. 2002a), metabolic activity (e.g., Holmstrøm and Kjelleberg 1999), age (e.g., Szewzyk et al. 1991; Keough and Raimondi 1996), or origin (Qian et al. 2003; Thiyagarajan et al. 2005; Dobretsov and Qian 2006) is remarkable, indicating a finely tuned response by dispersal stages to mediatory cues present in biofilms. In general, bacterial biofilms of different species differ significantly with respect to their mediatory effect on larval and spore settlement (Holmstrøm et al. 1996; Lau et al. 2002; Huang and Hadfield 2003; Patel et al. 2003). The relative proportion of inhibitive and inductive strains in biofilms varies with environmental conditions (Dobretsov et al. 2006). However, there is no predictive relationship between the phylogenetic affiliation of bacteria and their effects on larval or spore settlement (Lau et al. 2002; Patel et al. 2003; Dobretsov and Qian 2004).
While some invertebrate species only settle on biofilms with viable bacterial cells, (Lau et al. 2003b), others can settle on biofilms of nonviable cells (Hung et al. 2005a). In an effort to identify the settlement signal of the common fouling tubeworm Hydroides elegans, it has been suggested that bacterial extracellular polymeric substance (EPS) facilitates larval settlement by scavenging waterborne cues produced by bacteria (Harder et al. 2002a; Lau et al. 2003a). Similar to the studies on monospecies bacterial films, monospecies benthic diatom films are categorized in regard to their mediatory effect on larval settlement (Harder et al. 2002b). Contrary to bacterially induced larval settlement by waterborne bacterial metabolites, the mediatory effect of diatoms is evoked by diatom EPS in two studies by Lam et al. (2003, 2005a). Differences (i.e., molecular weight and monomer composition) between EPS obtained from diatoms grown under different environmental conditions are clearly reflected in the larval settlement response (Lam et al. 2005b), confirming the ability of larvae to distinguish between biofilms of varying composition, physiological condition, and growth phase (Wieczorek and Todd, 1998f).
As mentioned earlier, in recent years numerous studies have shown that quorum sensing signals are involved in cell-to-cell communication within prokaryotes and between prokaryotes and eukaryotes (Parsek and Greenberg 1999; Joint et al. 2002b; Yan et al. 2003; Tait et al. 2005). At least one class of quorum sensing signals, acetylated homoserine lactones (AHLs), is recognized as a mediatory cue for algal spore settlement (Joint et al. 2002a). For instance, zoospores of the marine green alga Ulva intestinalis respond to quorum sensing signal molecules on bacterial films for the selection of surface sites for permanent attachment (Joint et al. 2002a) and spore settlement is positively correlated with the density of wild-type V. anguillarum biofilms, but not to the density of bacterial strains unable to produce AHLs (Tait et al. 2005). In fact, U. intestinalis zoospores appear to be able to sense a range of different AHL molecules, which is indicated by the fact that spores often settle directly on bacterial cells and on the sites of concentrated AHL production. More recently, Wheeler et al. (2006) shows that U. intestinalis zoospores rapidly reduce swimming speed when they are exposed to specific AHLs. The effect on swimming speed becomes more pronounced with proximity to a point source of AHL, resulting in a massive accumulation of zoospores around the point source. The authors argue that the decreased swimming speed may contribute to increased settlement on bacterial biofilms by both concentrating zoospore numbers and increasing the likelihood that settlement will occur. These studies clearly demonstrate the recognition of quorum sensing signals between prokaryotes and eukaryotes, suggesting that quorum sensing signals released by marine bacteria in biofilms are important not only for biofilm formations but also for settlement of propagules of marine organisms.
Since cells in natural biofilms are conglutinated in a blend of EPS, a combined or synergistic effect of EPS of the resident cells and metabolites on larvae and spores may be a likely mode of action (Harder et al. 2002a). A wide range of hitherto nearly unknown “infochemicals” affecting biofouling processes are encrypted in marine biofilms (Harder et al. 2002b; Dahms and Qian 2005; Dahms et al. 2004). In summary, biofilm components, such as bacteria and diatoms, can be regarded as small-scale proxies, indicating the suitability of substrata for larvae of sessile invertebrates and algal spores seeking a permanent settlement site.
Technological Implications of Biofilms
Antifouling technologies
The recruitment and growth of sessile organisms on man-made structures causes serious problems to maritime industries and navies around the world (Yebra et al. 2004, Whelan and Regan 2006). Micro- and macrofouling reduces the hydrodynamic efficiency of ships and propellers, leads to pipeline blockage, and sensor malfunction, and increases the weight of appliances deployed in seawater. Since biofilms are important cues for larval settlement, effective control and/or manipulation of biofilm formation on the surfaces of marine structures should be a primary target of biofouling control (Evans and Hoagland 1986). One well-known example of such defense in nature is the red alga Delisea pulchra, which produces halogenated furanones that interfere with bacterial quorum sensing signals, inhibit bacterial growth, biofilm formation (Rice et al. 1999), and larval settlement (de Nys et al. 1995) and algal spore attachment (Dworjanin et al. 2006).
Marine microbes are a potent source of antifouling compounds. Yet so far, only a small number of marine bacteria have been screened and only several anti-settlement compounds have been isolated and identified (Fusetani 2002; Dobretsov et al. 2006; Paul et al. 2006). Inhibitor of the barnacle Balanus amphitrite larval settlement ubiquinone-8 (1) was produced by the marine bacterium Alteromonas sp. isolated for the marine sponge Halichondria okadai (Kon-ya et al. 1995). Similarly, the bacterium Acinetobacter sp., isolated from the surface of the ascidians Stomosoa murrayi produces inhibiting compounds for larval settlement of B. amphitrite (Olguin-Uribe et al. 1997). Recently, five antifouling diketopiperazines were isolated from the deep sea bacterium Streptomyces fungicidicus (Li et al. 2006). Fifty percent effective concentrations (EC50) of these compounds against larval settlement were quite low compare to the other antifouling agents and above mentioned bacterial antifouling compound. These examples showed that bacterial compounds may be used for biofouling control.
Antifouling compounds have also been isolated from the tissues of macroorganisms (Fusetani 2004; Blunt et al. 2006; Paul et al. 2006), but using microbes as sources of antifouling compounds has clear advantages. First of all, compound supply has always been a bottleneck of marine natural product chemistry; it is almost impossible to harvest sufficient macroorganisms from the natural environment or from mariculture farms to meet the needs of commercialization of bioactive compounds. In contrast, it is possible to develop large-scale bacterial cultures for compound productions on a commercial scale. Secondly, microorganisms that produce the compounds can be genetically modified to increase the yield (Demain and Davies 1999). Thirdly, bacterial strains of the same species can produce different compounds under different culture conditions, thereby increasing the potential number of bioactive compounds (Armstrong et al. 2001). Since only a small proportion of microorganisms have been screened for antifouling compounds more effort should be made to isolate and culture microbes, especially from extreme environments, since microbes from such environments have rarely been studied and may produce novel chemical compounds. Furthermore, paint matrices that are compatible with the chemistry of the novel antifouling compounds may need to be developed. This process will require an intense co-operation between academic institutions and industry (Rittschof 2000).
Chemical compounds produced by microorganisms can not only modify biofilms (i.e. disrupt or enhance the growth of existing biofilms) but also induce or inhibit larval settlement (Dobretsov et al. 2006). For example, antifungal, antibacterial, anti-algal, and antilarval compounds were isolated from the epibiotic bacterium Pseudoalteromonas tunicate (Holmstrøm et al. 1996; Holmstrøm and Kjelleberg 1999, Egan et al. 2001). Therefore, microorganisms, which not only directly inhibit the formation of biofilms and the growth of microfoulers but also indirectly affect the settlement of invertebrate larvae and algal spores, will be promising source for future antifouling applications.
Along these lines of reasoning, Wahl (1997) proposed that industrial appliances can be protected by bacteria growing on their surfaces. Possibility of surface protection by “living bacterial paints” was demonstrated by Holmstrøm et al. (2002). The question of whether bacterial biofilms can persist for months or years and successfully protect man-made structures from biofouling deserves further investigation, particularly in the following respects: 1) whether there is a technically feasible matrix to keep microbes alive on ship hulls or other man-made surfaces, 2) provided there is a suitable matrix, whether the “live paint” can provide service time similar to the conventional organometal-based paints, both in terms of the longevity of the microbial cells and the durability of the paint matrix, 3) since microbial physiology varies with the external environment, whether microbial synthesis in “live paint” will work independently of environmental factors, 4) whether or not the antifouling effects of the “live paint” are affected by other biofilms that will inevitably form on the paint surface, and 5) what will be environmental impact from introduction of biologically active bacterial species into new environments? In summary marine microbes may be considered as important source of both antimicrobial and antifouling compounds. Isolation and identification of antifouling compounds from marine microbes deserves further studies.
Aquaculture
Biofouling is a nuisance for fish farms because of the dense layers of macrofoulers covering facilities like net cages and rafts (Braithwaite and McEvou 2005). On the other hand, larval settlement can be fostered for the farmed species. In fact, considerable efforts have been made to enhance the larval settlement of trade shellfishes (Morse and Morse 1984; Kang et al. 2004). Previous studies have shown that many species of benthic diatoms in biofilms can enhance larval settlement of commercially valuable invertebrate species, such as abalone (Slattery 1992; Daume et al. 1999) and that settlement is often correlated with diatom cell density in biofilms (Daume et al. 1999). In addition, Zhao et al. (2003) show that biofilms induce larval settlement of the pearl oyster Pinctada maxima. Biofilms can also induce larval settlement of the oyster Crassostrea virginica although it is not clear whether inductive cues are from diatoms or bacteria, or both (Weiner et al. 1993). In fact, oyster larvae can respond similarly to waterborne substances released by either adult oysters or bacterial biofilms (Weiner et al. 1993; Tamburri et al. 1996). However, up to this stage, no settlement cues from natural biofilms have been isolated or identified. Identification of inductive cues for commercial aquaculture species will certainly be of interest to the mariculture industry in the years to come. In particular, identification of waterborne inductive cues that are largely ignored nowadays (Fusetani 2004; Paul et al. 2006) should be a focal points of future studies.
On the other hand, biofilms can not only enhance larval settlement but also provide a suitable food source to newly metamorphosed juveniles of some shellfish. This is important to abalone hatcheries that are currently suffering from excessively high juvenile mortality, which is largely due to unsuitable biofilms as a food source for the spats. To achieve breakthroughs in shellfish hatchery industries, we need to gain a better understanding of 1) what kind of inductive chemical signals are produced by biofilms, 2) which microbial species produce the inductive chemical cues, 3) under what conditions microbes release the cues, 4) what kind of microbial composition in biofilms is necessary for high juvenile growth and survival, and 5) how to maintain desirable biofilms in aquaculture systems.
Additionally, biofilms can benefit aquaculture by serving as biofilters to absorb and biodegrade excessive nutrients. For example, some bacteria can be involved in oxygen- or nitrate-dependent sulfide oxidation in the biofilters of a marine circulation aquaculture system (Golz et al. 1999). Bacteria and diatoms immobilized in biofilms within biofilters that remove ammonia and nutrients in seawater supply systems are worthy of further exploration.
Microbial biofilms may transmit infectious diseases to cultivated organisms (Zhao et al. 2003). As yet, this phenomenon has hardly been pursued in the context of epibiotic marine biofilms. A “take-over” in a given biofilm community by strains that are pathogenic for certain organisms may take place. This holds true particularly for epibiotic microbial communities on animate substrata where diseases and tissue necroses commonly occur (Mitchell and Chet 1975).
In addition, microbes from marine biofilms may suppress microbial infection and may be useful in preventing diseases. In most cases, such biofilms cover invertebrates and algae as well as their dispersal stages. For example, the surface of estuarine shrimp eggs (Palaemon macrodactylus) is often colonized by a monospecies biofilm of the bacterium Alteromonas sp. (Gil-Turnes et al. 1989). When the bacteria are suppressed by antibiotic treatment, the eggs become susceptible to infection by crustacean pathogens that subsequently kill eggs (Gil-Turnes et al. 1989). Similarly, a Gram-negative bacterium that lives on the surface of embryos of the American lobster Homarus americanus produces the antifungal compound tyrosol, which inhibits the growth of pathogenic fungi in vitro (Gil-Turnes and Fenical 1992). It is also suggested that compounds produced by a bacterium associated with the aquaculture of bivalve Agropecten purpuratus control the growth of pathogenic strains, such as Vibrio alginolyticus and V. anguillarum (Riquelme et al. 1996). Overall, using microbes as pathogen control agents may be an alternative in disease control of marine fisheries and aquaculture industries.
Looking Ahead
Genetic Diversity of Biofilms
Up to this stage, several major components of biofilms have not been studied with respect to their contribution to the bioactivity of entire biofilms. This holds for cyanobacteria that are dominating components in biofilms of certain euphotic environments such as the upper intertidal and the spray zones. Similarly, a possible effect of fungi on the settlement of marine organisms has rarely been investigated (Dobretsov et al. 2006). Furthermore, in almost all the documented analysis of marine biofilms, focus has been limited to particular microbial taxa; the combined effects of microbes from different taxa have yet to be explored, owing to the following technical challenges. First, although DNA fingerprinting techniques such as denaturing gradient gel electrophoresis (DGGE), terminal restriction fragment length polymorphism (TRFLP), single-strand conformational polymorphism (SSCP), clone libraries, and fluorescein in situ hybridization (FISH) are powerful tools used to reveal microbial diversity and composition in biofilms, each of these techniques has its own limitations in accuracy, sensitivity, and specificity. Second, none of these fingerprinting techniques is substantially quantitative. Thirdly, it is even more difficult to analyze bacteria, fungi, diatoms, and other microbes in biofilms in a single study using molecular techniques because we need to first identify effective specific primers (genes) for each major taxon and ensure that those primers will be amplified under the same polymerase chain reaction (PCR) settings in order to provide any relative quantitative estimation. Real-time PCR is quantitative in nature but is unrealistically labor intensive if one wants to analyze all the different phyla in biofilms. This is due to the fact that standard curves for each pair of primers need to be established although real-time PCR does not require the same or similar PCR conditions for quantitative estimation. The microarray technique may overcome some of these technical difficulties in the future but again it is not quantitative in nature. Finally, the greatest challenge is how to correlate settlement effect with the presence, absence, or quantity of certain microbes so as to deduce which microbes might be responsible for the observed effects. Although coupling larval settlement assays with DNA fingerprinting can show two varying factors (larval settlement and microbial composition) in the assay dishes, it is never possible to tell “which organisms” are responsible for the observed effects, even with an ultra-high density microarray printed with a half-million probes, to reveal microbial diversity in the finest resolution current technology allows. We will be nowhere closer to knowing “which organism” is actually producing a signaling effect for larval settlement induction or inhibition. Therefore, a more promising direction is to first determine the chemical cues from natural biofilms that really facilitate the communication between the larvae and the microbes. Once we know the cues, we will be able to trace them down stream along the larval receptor/signal transduction pathways and to search upstream to the microbial producers (see later). However, direct evidence for the in situ role of chemical cues for larval settlement induction or inhibition remains scarce.
Chemical Diversity of Biofilms
The complex relationships between microbial community composition, chemical compound production, and settlement-mediating effects of biofilms remain highly speculative because of the lack of proper analytical methods. First of all, the identification of cues derived from natural biofilms made of many microbial species has not been a common practice in laboratory bioassays although it is ecologically more relevant, perhaps because it has proven difficult. So far, the isolation and identification of larval settlement inducers and inhibitors are primarily based on monospecies biofilm developed under laboratory conditions. As indicated by several studies, cultivable bacteria are often functionally inactive minorities in the natural environment but can adapt well to laboratory culture conditions (Eilers et al. 2000). In other words, cultivable bacteria involved in previous colonization studies possibly represent only a minor and functionally insignificant subset of microbes present in natural marine environments (Lau et al. 2003b).
Second, chemical signal profiles released from multispecies biofilms may be complex, posing serious analytical challenges to laboratory bioassays and chemical analysis. To overcome these challenges, it is desirable to analyze natural biofilms with contrasting bioactivity for larval settlement to narrow down the search for bioactive compounds. For instance, in our previous study, we found that mid intertidal biofilms induced cyprid settlement of Balanus amphitrite while subtidal biofilms from the same site induced cyprid settlement of Balanus trigonus (Thiyagarajan et al. 2005). Thus, intertidal biofilms contain inductive cues for Balanus amphitrite while the subtidal biofilms do not. It may be feasible to identify unique compounds in the extracts if we can compare chemical profiles of extracts of the natural biofilms from both tidal heights using liquid chromatography-nuclear magnetic resonance (LC-NMR) and LC-NMR-MS, which allow separation of extracts, trapping of compounds and their identification. The challenge of this method is that these cues may be extremely waterborne and nonstable, which may seriously affect their isolation and identification.
Lastly, the synthesis of bioactive metabolites by bacteria changes with environmental conditions (Kjelleberg et al. 1993). Changes in the type and amount of compound production in entire multispecies biofilms are likely to occur (Cooksey and Wiggleworth-Cooksey 1995), but have not been characterized either under laboratory or field conditions. Therefore, it is necessary not only to isolate bioactive compounds from microbes under reproducible laboratory conditions but also to monitor their production and release rates under natural conditions where the cells occur in heterogeneous consortia in biofilms (Paerl and Pinckney 1996). In fact, microbes may exhibit additive or synergistic effects in biofilm. Further, a wide range of hitherto unknown “infochemicals” that facilitate maritime fouling processes may be encrypted in marine biofilms. However, no “infochemical” from any bacterial source with antifouling properties has been fully identified so far. More knowledge about such “infochemicals” will facilitate the development of novel, environmentally benign means for the avoidance or enhancement of fouling and will improve our understanding of the interaction between biofilm constituents and colonization.
Receptor Studies
Taking the settlement of invertebrate larvae and algal spores as a whole, there has been substantial effort in identifying structure, nature, and origin of natural larval settlement cues (Morse and Morse 1984; Morse et al. 1984; Tamburri et al. 1992, 1996; Zimmer-Faust and Tamburri 1994; Turner et al. 1994; Decho et al. 1998; Eri et al. 1999; see review by Hadfield and Paul 2001; Dreanno et al. 2006, in press), larval receptors (Trapido-Rosenthal and Morse 1986; Hadfield et al. 2000), possible signal transduction pathways and regulatory mechanisms (Degnan and Morse 1995; Carpizo-Ituarte and Hadfield 2003), and potential genes involved in larval settlement and metamorphosis (Seaver et al. 2005; Seaver and Kaneshige 2006). However, we find there is still a long way to go before we can draw conclusions on any of the above-mentioned aspects. Although a variety of chemical compounds have been proposed as larval settlement cues (see review by Qian 1999; Hadfield and Paul 2001; Steinberg et al. 2002), few cues have been isolated from natural habitats or natural biofilms and tested in the field. There is considerable evidence suggesting that the process of perceiving natural or artificial inductive substances is ascribed to an external epidermal sensory cell (Baloun and Morse 1984; Trapido-Rosenthal and Morse 1986; Baxter and Morse 1992) or to the cephalic sensory organ (Chia and Koss 1984), rhinophores (Chia and Koss 1982), and the anterior portion of the propodium (Chia and Koss 1989, Chia et al. 1992), there is little direct evidence to suggest that these structures are predisposed to perceived settlement or metamorphic cues. Therefore, whether the receptors responsible for larval settlement response to chemical cues are located on larval surfaces or on internal tissues remains an open question. The conclusion of this issue will certainly affect our interpretation of observable larval behavior in laboratory bioassays, owing to potential differences in duration required for larvae to detect and then respond to the cues. Although G protein-coupled receptors and cAMP signal transduction pathways are involved in morphogenetic signal transduction pathways in red abalone (Trapido-Rosenthal and Morse 1986), it is also suggested that G protein-coupled receptors are not involved in larval settlement of the polychaete Hydroides elegans (Carpizo-Ituarte and Hadfield 2003). There is evidence that two distinct developmental pathways governing gene expression xand metamorphosis in Haliotis and the morphogenetic pathways are directly linked to the processes of settlement and metamorphosis (Degnan and Morse 1995). But it is unclear whether this will apply to other marine invertebrate larvae.
To identify and locate the larval receptors triggered by chemical cues, we must know if a particular cue acts directly or indirectly on larvae since there is strong evidence that chemical cues can either directly act on larvae or alternatively affect larval settlement by modifying biofilms developed in the assay vessels (Beckmann et al. 1999; Jin and Qian 2004, 2005), which complicates the relationship between settling larvae and chemical cues. In situ hybridization with whole-mounted larvae may help us to localize the receptor(s) that are affected by chemical cues and successful expression of segmentation genes during larval development in the polychaete Capitella sp. I and Hydroides elegans. Work by Seaver and her collaborators (Seaver et al. 2005; Seaver and Kaneshige 2006) sheds some light on this aspect of research as well as providing detailed methodology for future studies to follow. Genes involved in larval segmentation are most likely involved in larval settlement and metamorphosis of those invertebrates and may provide hints for receptor studies. Further, to narrow down the targeted receptor(s) we also need to know the mode of action of the chemical cues on settling larvae.
To overcome these challenges, one possible practical approach is to first identify the genes that are affected by specific chemical cues, examine the expression level of those genes in larvae treated versus those untreated with chemical cues, and then try to understand the potential role of those genes in signal transduction pathways. For instance, if one can identify effective inductive and inhibitive compounds derived from natural biofilms for larval settlement of a particular species, one can then expose the settling larvae of this species to those compounds. Then both the up-regulated and down-regulated genes of the settled larvae versus competent larvae can be cloned and identified, using differential display PCR (DD-PCR) techniques. The expression level of those genes can then be determined using real-time PCR. Through these methods, the genes actively involved in larval settlement and metamorphosis may be identified.
In conclusion, biofilms are promising tools for marine technological applications. However, a better understanding of the genetic basis as well as signal transduction pathways underlying settlement processes will be essential to the development of new antifoulants or attractants, which will allow us to develop nontoxic biotechnological approaches where targeted species are either prevented from colonization or promoted to settle. To gain an understanding of biofilm derived signal recognition we need to (1) identify both effective inducers and inhibitors that are released from biofilms for the settlement of larvae and spores (see above), (2) to identify and locate larval chemo-receptors being activated by cues, (3) to elucidate the signal transduction pathways involved in the settlement of dispersal stages, and 4) to identify potential genes regulating signal transduction pathways. Once genetic regulation of the colonization process is better understood, chemicals can be identified and applied that will directly interfere with genetic transcription itself or inhibit or foster any step along the signal transduction pathway. Without a thorough understanding of these issues, it will be difficult to develop either a nontoxic antifouling technology or a biotechnology in which surface colonization is enhanced.
References
Acuňa N, Ortega-Morales BO, Valadez-Gonzalez A (2006) Biofilm colonization dynamics and its influence on the corrosion resistance of austenitic UNS S31603 stainless steel exposed to Gulf of Mexico seawater Mar Biotech (in press)
Allison DG (2003) The biofilm matrix Biofouling 19, 139–150
Armstrong E, Yan LM, Boyd, KG, Wright PC, Burgess JG (2001) The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461, 37–40
Baloun AJ, Morse DE (1984) Ionic control of metamorphosis in larval Haliotis rufescens (Gastropoda). Biol Bull 167, 124–138
Baxter GT, Morse DE (1992) Cilia from abalone larvae contain a receptor-dependent G protein transduction system similar to that in mammals. Biol Bull 183, 147–154
Beckmann M, Harder T, Qian PY (1999) Induction of larval attachment and metamorphosis in the serpulid polychaete Hydroides elegans by dissolved free amino acids: mode of action in laboratory bioassays. Mar Ecol Prog Ser 190, 167–178
Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR (2006) Marine natural products. Natural Prod Rep 23, 26–78
Borenstein SB (1994) Microbiologically Influenced Corrosion Handbook (New York: Industrial Press)
Braithwaite RA, McEvou LA (2005) Marine biofouling on fish farms and its remediation. Adv Mar Biol 47, 15–52
Callow ME, Callow JA (2000) Substratum location and zoospore behavior in the fouling alga Enteromorpha. Biofouling 15, 49–56
Carpizo-Ituarte EJ, Hadfield MG (2003) Transcription and translation inhibitors permit metamorphosis up to radiole formation in the serpulid polychaete Hydroides elegans Haswell. Biol Bull 204, 114–125
Charaklis WG, Cooksey B (1983) Biofilm and microbial fouling. Adv Appl Microbiol 29, 93–148
Chia F-S, Koss R (1982) Fine structure of the larval rhinophores of the nudibranch, Rostanga pulchra, with emphasis on the sensory receptor cells. Cell Tissue Res 225, 235–248
Chia F-S, Koss R (1984) Fine structure of the cephalic sensory organ in the larva of the nudibranch Rostanga pukhra (Mollusca, Opisthobranchia, Nudibranchia). Zoomorphology 104, 131–139
Chia F-S, Koss R (1989) The fine structure of the newly discovered propodial ganglia of the veliger larva of the nudibranch, Onchidoris bilamellate. Cell Tissue Res 256, 17–26
Chia F-S, Koss R, Stevens S, Goldberg JI (1992) Isolation of neurons of a nudibranch veliger. Biol Bull 182, 66–76
Clare AS (1998) Towards non-toxic antifouling. J Mar Biotechnol 6, 3–6
Cooksey B, Cooksey KE, Miller CA, Paul JP, Webster D, Rubin RW (1984) The attachment of microfouling diatoms. In: Marine Biodeterioration: An Interdisciplinary Study, Costlow JD, Tipper RD, eds. (Annapolis, MD: US Naval Institute Press)
Cooksey KE, Wiggleworth-Cooksey B (1995) Adhesion of bacteria and diatoms to surfaces in the sea-a review. Aquat Microb Ecol 9, 87–96
Dahms H-U, Dobretsov S, Qian P-Y (2004) The effect of bacterial and diatom biofilms on the settlement of the bryozoan Bugula neritina. J Exp Mar Biol Ecol 313, 191–209
Dahms H-U, Qian P-Y (2005) Exposure of biofilms to meiofaunal copepods affects the larval settlement of Hydroides elegans (Polychaeta). Mar Ecol Prog Ser 297, 203–214
Dang H, Lovell CR (2000) Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl Environ Microbiol 66, 467–475
Daume S, Brand-Gardner S, Woelkerling WJ (1999) Preferential settlement of abalone larvae: diatom films vs non-geniculate coralline red algae. Aquaculture 174, 243–254
Decho AW, Browne KA, Zimmer-Faust RK (1998) Chemical cues: Why basic peptides are signal molecules in marine environments. Limnol Oceanogr 43, 1410–1417
Degnan BM, Morse DE (1995) Developmental and morphogenetic gene regulation in Haliotis rufescens larvae at metamorphosis. Am Zool 35, 391–398
Demain AL, Davies JE, eds. (1999) Manual of Industrial Microbiology and Biotechnology. (Washington, DC: ASM Press)
De Nys R, Steinberg PD, Willemson P, Dworjanyn SA, Gabelish CL, King RJ (1995) Broad spectrum effects of secondary metabolites from the red alga Delisea pulchra in antifouling assays. Biofouling 8, 259–271
Dobretsov S, Qian P -Y (2002) Effect of bacteria associated with the green alga Ulva reticulata on marine micro- and macrofouling. Biofouling 18(3), 217–228
Dobretsov S, Qian P-Y (2004) The role of epibotic bacteria from the surface of the soft coral Dendronephthya sp in the inhibition of larval settlement. J Exp Mar Biol Ecol 299, 35–50
Dobretsov S, Qian P-Y (2006) Facilitation and inhibition of larval attachment of the bryozoan Bugula neritina in association with mono-species and multi-species biofilms. J Exp Mar Biol Ecol 333, 263–274
Dobretsov S, Dahms H-U, Qian P -Y (2006) Inhibition of biofouling by marine microorganisms and their metabolites. Biofouling 22, 43–54
Dreanno C, Kirby RR, Clare AS (In press) Locating the barnacle settlement pheromone: spatial and ontogenetic expression of the settlement-inducing protein complex of Balanus amphitrite. Proc R Soc B (Doi: 101098/rspb20063649online)
Dreanno C, Matsumura K, Dohmae N, Takio K, Hirota H, Kirby RR, Clare AS (2006) An a2-macroglobulin-like protein is the cue to gregarious settlement of the barnacle Balanus Amphitrite. PNAS 103, 14396–14401
Dworjanin SA, De Nys R, Steinber P (2006) Chemically mediated antifouling in the red alga Delisea pulchra. Mar Ecol Prog Ser 318, 153–163
Egan S, James S, Holmstrøm C, Kjelleberg S (2001) Inhibition of algal spore germination by the marine bacterium Pseudoalteromonas tunicata. FEMS Microbiol Ecol 5, 67–73
Eilers H, Pernthaler J, Gloeckner FO, Amann R (2000) Culturability and in situ abundance of pelagic bacteria in the North Sea. Appl Environ Microbiol 66, 3044–3051
Eri R, Arnold JM, Hinman VF, Green KM, Jones MK, Degan BM, Lavin MF (1999) Hemps, a novel EGF-like protein, plays a central role in ascidian metamorphosis. Development 126, 5809–5818
Evans LN, Hoagland KD (1986) Algal Biofouling. (Amsterdam: Elsevier)
Fusetani N (2004) Biofouling and antifouling. Nat Prod Rep 21, 94–104
Gil-Turnes MS, Fenical W (1992) Embryos of Homarus americanus are protected by epibiotic bacteria. Biol Bull 182, 105–108
Gil-Turnes MS, Hay ME, Fenical W (1989) Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science 246, 116–118
Golz WJ, Rusch KA, Malone RF (1999) Modeling the major limitations on nitrification in floating-bead filters. Aquacult Eng 20, 43–62
Greenberg EP (2003) Bacterial communication and group behavior. J Clin Invest 112, 1288–1290
Hadfield MG, Paul VJ (2001) Natural chemical cues for settlement and metamorphosis of marine invertebrate larvae. In: Marine Chemical Ecology, McClintock JB, Baker W, eds. (Boca Raton, FL: CRC Press) pp 431–461
Hadfield MG, Meleshkevitch E, Boudko D (2000) The apical sensory organ of a gastropod veliger is a receptor for settlement cues. Biol Bull 198, 67–76
Harder T, Lam C, Qian, P-Y (2002a) Induction of larval settlement in the polychaete Hydroides elegans by marine biofilms: an investigation of monospecific diatom films as settlement cues. Mar Ecol Prog Ser 229, 105–112
Harder T, Lau SCK, Dahms H-U, Qian P-Y (2002b) Isolation of bacterial metabolites as natural inducers for larval settlement in the marine polychaete Hydroides elegans (Haswell). J Chem Ecol 28(10), 2029–2043
Holmstrøm C, Kjelleberg S (1999) Marine Pseudoalteromonas species are associated with higher organisms and produce active extracellular compounds. FEMS Microbiol Ecol 30, 285–293
Holmstrøm C, Rittschof D, Kjelleberg S (1992) Inhibition of settlement by larvae of Balanus amphitrite and Cliona intestinalis by a surface-colonizing marine bacterium. Appl Environ Microb 58, 2111–2115
Holmstrøm C, James S, Egan S, Kjelleberg S (1996) Inhibition of common fouling organisms by marine bacterial isolates with special reference to the role of pigmented bacteria. Biofouling 10, 251–259
Holmstrøm C, Egan S, Francks A, McCloy S, Kjelleberg S (2002) Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol Ecol 41, 47–58
Huang S, Hadfield MG (2003) Composition and density of bacterial biofilms affect metamorphosis of the polychaete Hydroides elegans. Mar Ecol Prog Ser 260, 161–172
Hung OS, Thiyagarajan V, Wu RSS, Qian P-Y (2005a) Effects of ultraviolet radiation on films and subsequent settlement of Hydroides elegans. Mar Ecol Prog Ser 304, 155–166
Hung OS, Gosselin LA, Thiyagarajan V, Wu RSS, Qian P-Y (2005b) Do effects of ultraviolet radiation on microbial films have indirect effects on larval attachment of the barnacle Balanus amphitrite. J Exp Mar Biol Ecol 323, 16–26
Ista LK, Fan HY, Baca O, Lopez GP (1996) Attachment of bacteria to model solid surfaces: oligo(ethylene glycol) surfaces inhibit bacterial attachment. FEMS Microbiol Lett 142, 59–63
Jin T, Qian PY (2004) Effect of mono-amino acids on larval metamorphosis of the polychaete Hydroides elegans. Mar Ecol Prog Ser 267, 223–232
Jin T, Qian PY (2005) Amino acid exposure modulates the bioactivity of biofilms for larval settlement of Hydroides elegans by altering bacterial community components. Mar Ecol Prog Ser 297, 169–179
Joint I, Callow ME, Callow JA, Clarke KR (2002a) The attachment of Enteromorpha zoospores to a bacterial biofilm assemblage. Biofouling 16, 151–158
Joint I, Tait K, Callow ME, Callow JA, Milton D, Williams P, Camara M (2002b) Cell-to-cell communication across the prokaryote-eukaryote boundary. Science 298, 1207–1207
Kang KH, Kim BH, Kim JM (2004) Induction of larval settlement and metamorphosis of the abalone, Haliotis discus hannai larvae using bromomethane and potassium chloride. Aquaculture 230, 249–259
Keough MJ, Raimondi PT (1996) Responses of settling invertebrate larvae to bioorganic films: effects of large-scale variation in films. J Exp Mar Biol Ecol 207(1–2), 59–78
Kirchman D, Graham D, Reish D, Mitchell R (1982) Lectins may mediate in the settlement and metamorphosis of Janua (Dexiospira) brasiliensis Grube (Polychaeta: Spirorbidae). Mar Biol Lett 3, 201–222
Kjelleberg S, Albertson N, Flärdh K, Holmquist L, Jouper-Jaan A, Marouga R, Östling J, Svenblad B, Weichart D (1993) How do non-differentiating bacteria adapt to starvation? Anton v Leeuwen 63, 333–341
Kon-ya K, Shimidzu N, Otaki N, Yokoyama A, Adachi K, Miki W (1995) Inhibitory effect of bacterial ubiquinones on the settling of barnacle, Balanus amphitrite. Experientia 51, 153–155
Lam C, Harder T, Qian P-Y (2003) Induction of larval settlement in the polychaete Hydroides elegans by surface-associated settlement cues of marine benthic diatoms. Mar Ecol Prog Ser 263, 83–92
Lam C, Harder T, Qian P -Y (2005a) Induction of larval settlement in the polychaete Hydroides elegans by extracellular polymers of benthic diatoms. Mar Ecol Prog Ser 286, 145–154
Lam C, Harder T, Qian P-Y (2005b) Different growth conditions of benthic diatoms affect quality and quantity of extracellular polymeric larval settlement cues. Mar Ecol Prog Ser 294, 109–116
Lappin-Scott HM, Costerton JM (1989) Bacterial biofilms and surface fouling. Biofouling 1, 323–342
Lau SCK, Mak KK, Chen F, Qian P-Y (2002) Bioactivity of bacterial strains from marine biofilms in Hong Kong waters for the induction of larval settlement in the marine polychaete Hydroides elegans. Mar Ecol Prog Ser 226, 301–310
Lau SCK, Harder T, Qian P-Y (2003a) Induction of larval settlement in the serpulid polychaete Hydroides elegans (Haswell): role of bacterial extracellular polymers. Biofouling 19, 197–204
Lau SCK, Thiyagarajan V, Qian P-Y (2003b) The bioactivity of bacterial isolates in Hong Kong waters for the inhibition of barnacle (Balanus amphitrite Darwin) settlement. J Exp Mar Biol Ecol 282, 43–60
Lau SCK, Thiyagarajan V, Cheung SCK, Qian P-Y (2005) Roles of bacterial community composition in biofilms as a mediator for larval settlement of three marine invertebrates. Aquat Microb Ecol 38, 41–51
Levin LA (2006) Recent progress in understanding larval dispersal: new directions and digressions. Integr Comp Biol 46, 282–297
Li X, Dobretsov S, Xu Y, Xiao X, Qian P-Y (2006) Antifouling diketopiperazines produced by a deep-sea bacterium Streptomyces fungicidicus. Biofouling (in press)
Little BJ, Zsolnay ZA (1985) Chemical fingerprinting of adsorbed organic materials on metal surfaces. J Colloid Interface Sci 104, 79–86
Maki JS (1999) The influence of Marine microbes on biofouling. In: Recent Advances in Marine Biotechnology, Vol. 3: Biofilms, Bioadhesion, Corrosion, and Biofouling. Fingerman M, Nagabhushanam R, Thompson F, eds. (Enfield, NH: Science Publishers) pp 147–171
Maki JS (2002) Biofouling in the marine environment. In: Encyclopedia of Environmental Microbiology. Bitton G, ed. (New York: John Wiley & Sons) pp 610–619
Maki JS, Rittschof D, Costlow JD, Mitchell R (1988) Inhibition of attachment of larval barnacles, Balanus amphitrite, by bacterial surface films. Mar Biol 97, 199–206
Maki JS, Ding L, Stokes J, Kavouras JH, Rittschof D (2000) Substratum/ bacterial interactions and larval attachment: films and exopolysaccharides of Halomonas marina (ATCC 25374) and their effect on barnacle cyprid larvae, Balanus amphitrite Darwin. Biofouling 16, 159–170
Marshall PD, Bowden GHW (2000) Microbial community interaction in biofilms. In Community Structure and Cooperation in Biofilms. Society for General Microbiology Symposia 59, Allison DG, Gilbert P, Lappin-Scott HM, Wilson M, eds. (Cambridge) pp 167–198
Mitchell R, Chet I (1975) Bacterial attack of corals in polluted seawater. Microb Ecol 2, 227–233
Mitchell R, Maki JS (1988) Microbial surface films and their influence on larval settlement and metamorphosis in the marine environment. In: Marine Biodeterioration: Advanced Techniques Applicable to the Indian Ocean. Thompson M-F, Sarojini R, Nagabushanam R, eds. (New Delhi: Oxford & IBH) pp 489–497
Morse ANC, Morse DE (1984) Recruitment and metamorphosis of Haliotis larvae induced by molecules uniquely available at the surfaces of crustose red algae. J Exp Mar Biol Ecol 74, 191–215
Morse ANC, Froyd CA, Morse DE (1984) Molecules from cyanobacteria and red algae that induce larval settlement and metamorphosis in the mollusk Haliotis rufescens. Mar Biol 81, 293–298
Olguin-Uribe G, Abou-Mansour E, Boulander A, Debard H, Francisco C, Combaut G (1997) 6-Bromoindole-3-carbaldehyde from an Acinetobacter sp bacterium associated with the ascidian Stomoza murrayi. J Chem Ecol 23, 2507–2521
Paerl HW, Pinckney JL (1996) A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microb Ecol 31, 225–247
Parsek MR, Greenberg EP (1999) Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Methods Enzymol 310, 43–55
Patel P, Callow ME, Joint I, Callow JA (2003) Specificity in larval settlement modifying response of bacterial biofilms towards zoospores of the marine alga. Enteromorpha Environ Microb 5, 338–349
Paul VJ, Puglisi MP, Ritson-Williams R (2006) Marine chemical ecology. Nat Prod Rep 23, 153–180
Qian P-Y (1999) Larval settlement of polychaetes. Hydrobiologia 402, 239–253
Qian P-Y, Thiyagarajan V, Lau SCK, Cheung SCK (2003) Bacterial community profile in biofilm and attachment of the acorn barnacle Balanus amphitrite. Aquat Microb Ecol 33, 225–237
Railkin AI (2004) Marine Biofouling: Colonization Processes and Defenses. CRC Press
Rice SA, Givskov M, Steinberg P, Kjelleberg S (1999) Bacterial signals and antagonists: the interaction between bacteria and higher organisms. J Mol Microbiol Biotechnol 1, 23–31
Riquelme C, Hayashida G, Araya R, Uchida A, Satomi M, Ishida Y (1996) Isolation of a native bacterial strain from the scallop Agropecten purpuratus with inhibitory effects against pathogenic Vibrios. J Shellfish Res 15, 369–374
Rittschof D (2000) Natural product antifoulants: one perspective on the challenges related to coatings development. Biofouling 15, 119–125
Schneider RP, Marshall KC (1994) Retention of the Gram negative marine bacterium SW8 on surfaces—effects of microbial physiology, substratum nature and conditioning films. Colloids Surf B Biointerfaces 2, 387–396
Seaver EC, Kaneshige LM (2006) Expression of ‘segmentation’; genes during larval and juvenile development in the polychaetes Capitella sp I and H elegans. Dev Biol 289, 179–194
Seaver EC, Thamm K, Hill S (2005) Growth patterns during segment formation in the two annelids Capitella sp I and Hydroides elegans: comparisons at distinct life history stages. Evol Dev 7, 312–326
Slattery M (1992) Larval settlement and juvenile survival in the red abalone (Haliotis rufescens): an examination of inductive cues and substrate selection. Aquaculture 102, 143–153
Steinberg PD, De Nys R, Kjelleberg S (2002) Chemical cues for surface colonization. J Chem Ecol 28, 1935–1951
Szewzyk U, Holmstrøm C, Wrangstadh M, Samuelsson MO, Maki JS, Kjelleberg S (1991) Relevance of the exopolysaccharide of marine Pseudomonas sp strain S9 for the attachment of Ciona intestinalis larvae. Mar Ecol Prog Ser 75, 259–265
Tait K, Joint I, Daykin M, Milton DL, Williams P, Camara M (2005) Disruption of quorum sensing in seawater abolishes attraction of zoospores of the green alga Ulva to bacterial biofilms. Environ Microbiol 7, 229–240
Tamburri MN, Zimmer-Faust RK, Tamplin LL (1992) Natural sources and properties of chemical inducers mediating settlement of oyster larvae: a re-examination. Biol Bull 183, 327–338
Tamburri MN, Finelli CM, Wethey DS, Zimmer-Faust RK (1996) Chemical induction of larval settlement. Biol Bull 191, 367–373
Thiyagarajan V, Hung OS, Chiu JMY, Wu RSS, Qian P-Y (2005) Growth and survival of juvenile barnacle Balanus amphitrite: interactive effects of cyprid energy reserve and habitat. Mar Ecol Prog Ser 299, 229–237
Todd CD, Keough MJ (1994) Larval settlement in hard substratum epifaunal assemblages: a manipulative field study of the effects of substratum filming and the presence of incumbents. J Exp Mar Biol Ecol 181, 159–187
Trapido-Rosenthal HG, Morse DE (1986) Availability of chemosensory receptors is down-regulated by habituation of larvae to a morphogenetic signal. PNAS 83, 7658–7662
Turner EJ, Zimmer-Faust RK, Palmer MA, Luckenbach M (1994) Settlement of oyster (Crassostrea virginica) larvae: effects of water flow and a water-soluble chemical cue. Limnol Oceanogr 39, 1579–1593
Wahl M (1997) Living attached: aufwuchs, fouling, epibiosis. In: Fouling Organisms of the Indian Ocean: Biology and Control Technology. Nagabushanam R, Thompson M, eds. (New Delhi: Oxford & IBH) pp 31–84
Weiner R, Sledjeski D, Quintero E, Coon S, Walch M (1993) Periphytic bacteria cue oyster larvae to set on fertile benthic biofil larvae by microbial biofilm cues. Biofouling 12, 81–93
Wheeler GL, Tait K, Taylor A, Brownlee C, Joint I (2006) Acyl-homoserine lactones modulate the settlement of zoospores of the marine alga Ulva intestinalis via a novel chemokinetic mechanism. Plant Cell Environ 29, 608–616
Whelan A, Regan F (2006) Antifouling strategies for marine and riverine sensors. J Environ Monit 8, 880–886
Wimpenny J (1996) Ecological determinants of biofilm formation. Biofouling 10, 43–63
Yan LM, Boyd KG, Adams DR, Burgess JG (2003) Biofilm-specific cross-species induction of antimicrobial compounds in bacilli. Appl Environ Microbiol 69, 3719–3727
Yebra DM, Kiil S, Dam-Johansen K (2004) Antifouling technology-past, present and future steps towards efficient and environmentally friendly antifouling coatings. Prog Org Coatings 50, 75–104
Zhao B, Zhang S, Qian, P -Y (2003) Larval settlement of the silver- or goldlip pearl oyster Pinctada maxima (Jameson) in response to natural biofilms and chemical cues. Aquaculture 220, 883–901
Zimmer-Faust RK, Tamburri MN (1994) Chemical identity and ecological implications of a waterborne, larval settlement cue. Limnol Oceanogr 39, 1075–1087
Zobell CE, Allen EC (1935) The significance of marine bacteria in the fouling of submerged surfaces. J Bacteriol 29(3), 239–251
Acknowledgments
We thank Dr. J. Pechenik for his comments on the manuscript. This work was supported by research grants (CAS-CF03/04, HKUST 6402/05M, COMRRDA04/05.SC01, CA04/05.SC01) to P.Y. Qian.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qian, PY., Lau, S.C.K., Dahms, HU. et al. Marine Biofilms as Mediators of Colonization by Marine Macroorganisms: Implications for Antifouling and Aquaculture. Mar Biotechnol 9, 399–410 (2007). https://doi.org/10.1007/s10126-007-9001-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-007-9001-9