Abstract
Acanthopagrus butcheri completes its entire life history within estuaries and coastal lakes of southern Australia, although adults occasionally move between estuaries via the sea. Consequently, it is expected that populations of A. butcheri in different estuaries will be genetically distinct, with the magnitude of genetic divergence increasing with geographic isolation. However, previous genetic studies of A. butcheri from southeast Australia yielded conflicting results; allozyme variation exhibited minimal spatial structuring (θ = 0.012), whereas mitochondrial DNA distinguished the majority of populations analyzed (θ = 0.263) and genetic divergence was positively correlated with geographic isolation. This discrepancy could reflect high male gene flow, which impacts nuclear but not mitochondrial markers. Here we estimated allele frequencies at five nuclear microsatellite loci across 11 southeast Australian populations (595 individuals). Overall structuring of microsatellite variation was weaker (θ = 0.088) than that observed for mitochondrial DNA, but was able to distinguish a greater number of populations and was positively correlated with geographic distance. Therefore, we reject high male gene flow and invoke a stepping-stone model of infrequent gene flow among estuaries for both sexes. Likewise, management of A. butcheri within the study range should be conducted at the scale of individual or geographically proximate estuaries for both sexes. The lack of allozyme structuring in southeast Australia reflects either the large variance in structuring expected among loci under neutral conditions and the low number of allozymes surveyed or a recent colonization of estuaries such that some but not all nuclear loci have approached migration-drift equilibrium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Patterns of molecular variation among groups of individuals separated spatially or temporally are extensively employed to test for contemporary gene flow, as this knowledge is beneficial for studies of ecology and evolution, as well as for the assessment of conservation priorities (e.g., Garber et al., 2004; Beacham et al., 2005). However, different classes of molecular markers may provide conflicting interpretations of gene flow, owing to differences in their underlying characteristics (mutations rates, mode of inheritance, function, effective population size). Consequently, it is desirable to apply several classes of molecular markers when studying gene flow in a given taxon (Arnaud-Haond et al., 2003; Hoarau et al., 2004), in addition to nonmolecular techniques (e.g., tagging, telemetry, otolith chemistry).
The black bream Acanthopagrus butcheri (Munro) is an estuarine sparid (Pisces: Sparidae) occurring throughout the southern half (27°40′–43°40′S) and almost entire longitudinal range of Australia (Allen et al., 2002). This is an important commercial and recreational fishery species (Kailola et al., 1993; Coutin, 2000), and its dependence on estuarine environments confers elevated conservation concern (Hodgkin, 1994). Fisheries in some estuaries have also experienced recent collapses (Blackwood River, Sarre and Potter, 2000; Gippsland Lakes, Coutin and Conron, 2006). Technologies for the captive breeding and rearing of A. butcheri have recently been developed with the aim of future production for human consumption (Doupé et al., 2005), and progeny have also been used to augment natural populations (Lenanton et al., 1999). Consequently, knowledge of stock structuring in this species is desirable for the future management and conservation of this resource (Carvalho and Hauser, 1995).
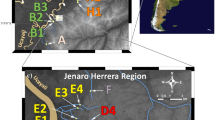
Acanthopagrus butcheri is a rare example of a fish that completes its entire lifecycle within estuaries (Potter and Hyndes, 1999). Movement between estuaries appears infrequent based on tagging studies, but has been documented for estuaries over 50 km apart (Butcher and Ling, 1962; Gorman, 1965; P.C. Coutin, unpublished). Consequently, populations in different estuaries are expected to be genetically divergent from one another (Bilton et al., 2002; Watts and Johnson, 2004), and represent distinct stocks for management purposes. However, in contrast to expectations based on ecology and tagging, studies of allozymes (nuclear genome) among southeast Australian populations of A. butcheri revealed low spatial structuring of variation; allele frequencies at three polymorphic loci were homogeneous throughout much of southeast Australia, with the exception of two peripheral populations (Figure 1; Farrington et al., 2000; Burridge et al., 2004). Conversely, variation in mitochondrial DNA (mtDNA) exhibited significant heterogeneity throughout the same range (Figure 1; Burridge et al., 2004), consistent with expectations.
Acanthopagrus butcheri sampling localities from estuaries in southeast Australia. Numbers in parentheses represent sample sizes of individuals scored for microsatellite variation during this study (and mtDNA variation of sites analyzed for the first time: Glenelg, Painkalac, and Thomson). Allozyme allele and mitochondrial haplotype frequencies are depicted below sites analyzed. (Allozyme data: Farrington et al., 2000; Burridge et al., 2004. Mitochondrial data: Burridge et al., 2004; this study). Populations that are homogeneous for allele of haplotype frequencies are either encircled or linked by dashed lines.
From a management and conservation perspective, the previous genetic results for A. butcheri must be interpreted cautiously. Although the mtDNA structuring is strong and consistent with expectations, the predominantly maternal inheritance of this marker precludes any inference of male population structuring (Birky et al., 1989). Given the lack of allozyme structuring, it is possible that A. butcheri exhibits high male gene flow among estuaries, and hence the overexploitation of one estuary may have implications beyond that locality. The possibility of male-biased dispersal in fishes has only recently received attention, but has been documented in several species. Some rock-dwelling cichlids exhibit male-biased dispersal, as adjacent females have higher average relatedness than adjacent males (Knight et al., 1999). Male-biased dispersal has also been documented for salmonids, based on either tagging studies (Hutchings and Gerber, 2002) or microsatellite analyses and assignment tests (Bekkevold et al., 2004; Fraser et al., 2004). Male-biased dispersal has also been postulated for discrepancies between nuclear autosomal and mtDNA population structuring of other taxa (e.g., Rassmann et al., 1997; Lyrholm et al., 1999). Given that A. butcheri exhibits protogynous hermaphrodism in some estuaries (Rowland and Snape, 1994), male-biased dispersal could also result from size-biased dispersal. Consequently, rigorous assessment of A. butcheri population structuring requires a more thorough survey of autosomal variation to assess male gene flow. Previous surveys were constrained by the low polymorphism of allozymes; of more than 50 enzyme systems surveyed in A. butcheri, only LDH*, MDH*-2, and GPI*-1 exhibited scorable polymorphism (Chaplin et al., 1998; Farrington et al., 2000).
The aim of this study was to determine whether male gene flow in A. butcheri has been sufficient to homogenize nuclear genetic variation among southeast Australian estuaries. To address this question, we determined allele frequencies at five microsatellite loci among 11 populations (minimum sample of 40 individuals). Microsatellites are likely to represent a more accessible source of polymorphism than allozymes, and previous studies employing these markers have revealed population structuring not apparent from the analysis of allozymes (e.g., Bentzen et al., 1996; Shaw et al., 1999a,b; Wirth and Bernatchez, 2001), probably owing to the greater statistical power associated with higher allelic diversity (Goudet et al., 1996). Our null hypothesis is that male gene flow among estuaries is sufficient to homogenize microsatellite allele frequencies over large spatial scales in southeast Australia, equivalent to that observed for allozymes. We also surveyed estuaries intermediate to those previously distinguished by mtDNA, in an effort to address the finer spatial extent of population structuring (i.e., Glenelg, Painkalac, Thomson; Figure 1).
Materials and Methods
Study Design
At least 40 individuals from each of 11 estuaries were genotyped at five microsatellite loci. We utilized eight of the samples analyzed by Farrington et al. (2000) and Burridge et al. (2004), plus three samples collected at estuaries intermediate to those previously distinguished by mtDNA (Figure 1). The three new samples were analyzed for microsatellite and mtDNA variation, but not allozyme variation given the widespread homogeneity of allele frequencies already documented at these loci in southeast Australia (Farrington et al., 2000; Burridge et al., 2004) and tissue constraints (ethanol-preserved fin clips).
Microsatellite Analysis
Total DNA was isolated from tissue samples using a high-salt precipitation method (Crandall et al., 1999). Five microsatellite loci developed for species of sparid were analyzed (Table 1). Three loci were derived from A. butcheri (Yap et al., 2000). The remaining loci were derived from other sparids, Pagrus auratus (Adcock et al., 2000) and A. schlegeli (Jeong et al., unpublished), and were screened for polymorphism in A. butcheri for the first time herein. Loci were polymerase chain reaction (PCR) amplified with conditions comprising 1 × PCR buffer, 0.2 mM dNTPs, and 0.2 units of Taq DNA polymerase (Invitrogen), with MgCl2 concentrations and primer sequences provided in Table 1. For all loci except pAb2A5, dye label incorporation followed Schuelke (2000); the forward primer was 5′ appended with an 18 bp M13 sequence (TGTAAAACGACGGCCAGT), and employed at 0.03 μM, while the reverse primer and a FAM- or HEX-labeled M13 primer were employed at 0.5 μM. This method is more economic than individually labeling a primer for each locus (Schuelke, 2000). Thermal cycling conditions for each locus except pAb2A5 were 3 min at 94°C, followed by 8 cycles of 94°C for 30 s, annealing temperature (Table 1) for 30 s, and 72°C for 1 min, followed by 33 cycles as before but with annealing at 53°C, and then a final extension of 72°C for 5 min. Locus pAb2A5 was amplified using a FAM-labeled forward primer and a reverse primer, both at 0.5 μM, and with thermal cycling conditions as above except that annealing was at 64°C throughout. PCR products were separated on a 6% denaturing polyacrylamide gel using an ABI 373 (Applied Biosystems) following the manufacturer's instructions. PCR product lengths were determined relative to the GS400 size standard (ABI).
Mitochondrial DNA Analysis
MtDNA analysis was conducted on the three samples not previously analysed for this marker (Glenelg, Painkalac, Thomson). An approximately 1100-bp segment of mtDNA that included the noncoding control region, the tRNAPhe gene, and part of the 12S rRNA gene was chosen for PCR- restriction fragment length polymorphism (PCR-RFLP) analysis. The PCR conditions and primers PT and PU of Jean et al. (1995) were employed. Two restriction enzymes (four-base recognition) were employed that digest this fragment at sites known to be polymorphic, NsiI and DpnII (Burridge et al., 2004). Restriction enzyme digests of PCR products from each individual were carried out for 1 h at 37°C using 8 μl of amplified DNA, 1–3 units of enzyme, and the appropriate concentration of reaction buffer. The digested samples were electrophoresed through 2.0% agarose gels followed by ethidium bromide staining, and visualized under UV illumination.
Data Analysis
Concordances of microsatellite genotype frequencies with those expected under Hardy-Weinberg equilibrium, and the independence of genotypes among loci, were examined using exact tests implemented by Genepop 3.1c (Raymond and Rousset, 1995), followed by sequential Bonferroni correction of P values (Rice, 1989). Non-neutrality of mitochondrial DNA variation was assessed via the Ewens-Watterson-Slatkin test (Slatkin, 1996) implemented using Arlequin 2.001 (Schneider et al., 2000).
Microsatellite allele and mitochondrial haplotype frequency homogeneity among samples was assessed by exact tests using Genepop, with microsatellite results combined across loci using Fisher's method, following Ryman and Jorde (2001). Variance of allele and haplotype frequencies among samples (F ST, Wright, 1978) was estimated by calculating θ (Weir and Cockerham, 1984) with FSTAT 2.9.3 (Goudet, 2001). Significance of observed θ values was estimated from 1000 permutations of alleles or haplotypes among samples. Values of G ST (Nei, 1987) for microsatellites and allozymes were also calculated using FSTAT, and converted into Hedrick's (2005) standardized measure of genetic differentiation (G′ST) to facilitate comparison between these nuclear markers.
The presence of a significant relationship between geographic isolation and the genetic divergence of samples was assessed using a Mantel test (10,000 randomizations), implemented by IBD 1.2 (Bohonak, 2002) and based on geographic distance and θ. Geographic distance was represented by the contemporary shoreline distance between estuaries, excluding the circumference of Port Phillip Bay for comparisons of localities east and west of the Yarra, given the comparatively narrow entrance to this bay (Figure 1).
Results
Microsatellite Variation
Each locus was polymorphic in all samples except locus pAb1H1 in Onkaparinga, Surrey, and Thomson, and locus pAb2D11 in Surrey (Appendix). Allelic diversity was substantially higher than that observed for allozymes across samples in common, although heterozygosities were similar (Table 2). Genotype frequencies did not differ from Hardy-Weinberg expectations following Bonferroni correction with the exception of locus Asc21–218 at Gippsland (P = 0.0029), and locus pAb2D11 at Hopkins (P = 0.0016). There was no evidence for genotypic disequilibrium among loci following Bonferroni correction (P > 0.0235).
Exact tests rejected microsatellite allele frequency homogeneity for the majority of sample comparisons (Table 3), with exceptions between several adjacent localities (Port Adelaide-Onkaparinga, Glenelg-Surrey, Hopkins-Painkalac, Painkalac-Thomson, Gippsland-Sydenham). Exclusion of locus Asc21–218 during comparisons against the Gippsland sample (deviating from Hardy-Weinberg expectations) did not alter the significance of allele frequency heterogeneity. The exclusion of pAb2D11 for comparisons against the Hopkins sample (deviating from Hardy-Weinberg expectations) did alter the significance of heterogeneity; Surrey, Yarra, and Gippsland were no longer significantly different from Hopkins. However, Hopkins was distinguished from all samples except Painkalac by analysis of microsatellite genotype—as opposed to allele—frequency homogeneity (P < 0.05 following Bonferroni correction), and this approach does not require Hardy-Weinberg equilibrium. mtDNA analysis also distinguished Hopkins from the majority of samples (Table 3). Structuring of microsatellite variation across all 11 populations (θ) was 0.088, and significantly greater than zero (P < 0.001).
Mitochondrial DNA Variation
Eight haplotypes were observed, all populations were polymorphic, and haplotype frequencies in each sample (Appendix) were consistent with selective neutrality according to the Ewens-Watterson-Slatkin test (P > 0.059). Genetic homogeneity was rejected for the majority of pairwise sample comparisons (Table 3). Nonrejection of homogeneity mostly involved comparisons of adjacent or second-most-adjacent samples, although the Yarra sample could not be distinguished from those collected across a wider range (Thomson to Glenelg, Table 3). Structuring of mtDNA variation across all 11 samples (θ) was 0.185, and was significantly greater than zero (P < 0.001) (previously 0.263 across eight populations, P < 0.001, Burridge et al., 2004).
Isolation by Distance
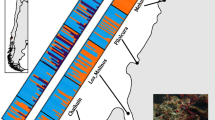
A significant positive relationship with geographic distance was observed for θ microsatellites and θ mtDNA (Figure 2; r microsatellites = 0.6920, P < 0.003; r mtDNA = 0.5766, P < 0.005). Inspection of the plot points revealed that the significant relationships were not due to exceptional divergence of peripheral populations, which could be reflective of vicariance, but rather a steady increase in genetic divergence with geographic distance.
Plots of θ against geographic distance (shoreline distance between sample sites) for molecular markers employed to assess population structuring in Acanthopagrus butcheri from southeast Australia. Top: θ derived from each class of molecular marker (allozyme, mitochondrial DNA, microsatellite) across all sites for which data were collected. (Allozyme data: Farrington et al., 2000; Burridge et al., 2004. Mitochondrial data: Burridge et al., 2004; this study). Middle and lower: θ for individual microsatellite loci.
Discussion
High Male Gene Flow?
The null hypothesis of this study was that gene flow among estuaries has been sufficient to homogenize microsatellite allele frequencies in A. butcheri over large spatial scales in southeast Australia. Given that strong spatial structuring of mtDNA variation has been observed for A. butcheri in this region (Burridge et al., 2004; this study), a lack of structuring in nuclear markers cannot be explained by female gene flow, but rather may be reflective of male movements. However, based on a survey of five microsatellite loci we reject our null hypothesis; microsatellites distinguished the majority of A. butcheri samples from one another. Only some of the populations that were adjacent to each other were genetically indistinguishable (Port Adelaide-Onkaparinga, Glenelg-Surrey, Hopkins-Painkalac, Painkalac-Thomson, Gippsland-Sydenham). Whenever mtDNA distinguished populations, microsatellites were in agreement, such that interpretation of population structuring can be extended to both sexes. In addition, microsatellites also distinguished several populations that could not be separated based on mtDNA haplotype frequencies. Most notedly, this represented the distinction of the Yarra population from a broad range of samples (between Glenelg and Gippsland), but also other sets of populations (i.e., Hindmarsh from sites further west; Hopkins vs. Painkalac; Thomson vs. Hopkins and Gippsland) (Table 3).
Isolation by Distance
The presence of significant positive relationships between genetic divergence and geographic isolation for both microsatellite and mtDNA variation in A. butcheri indicates low levels of contemporary gene flow within southeast Australia that is predominantly restricted to adjacent estuaries, consistent with the one-dimensional stepping stone model of Kimura and Weiss (1964). Isolation by distance relationships has been observed for estuarine fishes in other parts of Australia (Chenoweth and Hughes, 1997; Chenoweth et al., 1998; Jerry and Baverstock, 1998). There is also such a relationship in A. butcheri from Western Australia, based on reanalysis of the data presented by Chaplin et al. (1998) (not shown). It appears that isolation by distance relationships, and stepping-stone gene flow among demes may apply to the majority of Australian fishes that predominantly occupy estuaries.
Interestuary Movement
It has been previously hypothesized that periods of high river discharge facilitate gene flow between estuaries for A. butcheri (Burridge et al., 2004). There have been observations of individuals occurring out at sea, but still proximate to a river mouth, during periods of high discharge (Lenanton, 1977; Holt, 1978; Sherwood and Backhouse, 1982; Lenanton et al., 1999), and individuals may employ nearshore coalescing floodwater plumes to move between estuaries (e.g., Grimes and Kingsford, 1996), or undertake such movement in fully marine conditions which they can readily tolerate (Norriss et al., 2002). There is no evidence for interestuary gene flow mediated by egg or larval stages in A. butcheri, as none have been recorded out at sea. Spawning in estuaries open to the ocean also occurs during periods of low river discharge and is preceded by an upstream migration, followed by larval and juvenile development in the upper and mid reaches of estuaries (Sherwood and Backhouse, 1982; Newton, 1996; Haddy and Pankhurst, 1998).
Lack of Allozyme Structuring
The comparative homogeneity of allele frequencies at protein loci across southeast Australian populations could reflect balancing selection, given that these molecules are functionally significant and estuaries exhibit temporal and spatial heterogeneity in temperatures, salinities, and dissolved oxygen concentrations (Hodgkin, 1994). While several statistical tests exist for the presence of selection at allozymes, their application to A. butcheri is constrained by the low number of polymorphic loci and alleles per locus, and estimated F ST < 0 (see Lewontin and Krakauer, 1973; Baer, 1999; Arnaud-Haond et al., 2003; Guinand et al., 2004). Other studies have inferred selection by contrasting allozyme population structuring against molecular markers thought immune to direct selection, such as microsatellites (e.g., Karl and Avise, 1992; Pogson et al., 1995; Estoup et al., 1998; Lemaire et al., 2000; Buonaccorsi et al., 2001; De Innocentiis et al., 2001; Dufresne et al., 2002; Arnaud-Haond et al., 2003). However, an appreciation is required for the broad variance in neutral structuring expected among loci (Lewontin and Krakauer, 1973; McDonald et al., 1996; Bierne et al., 2003), and the fact that the low homozygosity and higher mutation rates typical of microsatellites can constrain estimates of F ST to small values, particularly when migration rates are low (Goudet et al., 1996; Hedrick, 1999; Balloux et al., 2000; Buonaccorsi et al., 2001).
Here we standardized measures of genetic differentiation at microsatellites and allozymes relative to theoretical maximums derived from expected heterozygosities (G′ST; Hedrick, 2005). However, the structuring observed across allozymes was not significantly lower than that for microsatellites for samples in common to both marker types (Table 2; U 0.05(1),3,5 = 12, P > 0.10). Therefore, we cannot reject selective neutrality of the allozyme variation; the difference in structuring between allozymes and microsatellites may simply reflect the broad variance in structuring expected among loci under neutral conditions and an inadequate sampling of protein loci. Alternatively, a recent colonization history could explain the lack of allozyme structuring.
Estuaries are generally ephemeral over geological and evolutionary timescales (Schubel and Hirschberg, 1978; Hodgkin, 1994; Kench, 1999; Bilton et al., 2002), and the coastline in the vicinity of Bass Strait (separating Tasmania from southeast Australia) has been particularly disrupted during glacial/interglacial cycles, most recently during the Holocene marine transgression (Kench, 1999). Consequently, if contemporary estuaries were colonized only recently, they may not have yet attained migration-drift equilibrium (Slatkin, 1987; Hutchinson and Templeton, 1999 Castric and Bernatchez, 2003). While the presence of a isolation by distance relationship for microsatellites suggests the attainment of migration-drift equilibrium for nuclear loci (Slatkin, 1987; Hutchinson and Templeton, 1999), an inspection of plots for individual loci reveal that Asc21-218 and pAb2D11 are mostly responsible for this pattern, whereas pAb1H1, pAb2A5, and Pma1n exhibit comparatively uniform and low θ regardless of spatial scale (Figure 2). Therefore, colonization of southeast Australian estuaries may have been sufficiently recent that some, but not all nuclear loci have attained migration-drift equilibrium and reveal population structuring, regardless of whether they are microsatellites or allozymes. The fact that each of the same three allozymes revealed population structuring of variation in A. butcheri among Western Australian estuaries (LDH* θ= 0.096, MDH-2* θ = 0.268, GPI-1* θ = 0.054, all P < 0.001 based on a reanalysis of data from Chaplin et al. 1998) could reflect differences in colonization times among southeast and Western Australian estuaries.
Conservation Implications
The microsatellite data support mtDNA and tagging study inferences of low contemporary gene flow, which is predominantly restricted to adjacent estuaries. Consequently, separate management of distinct populations (i.e., stocks) is desirable for the avoidance of regional overexploitation given that replenishment of depleted stocks by immigrants is likely to be slow (Carvalho and Hauser, 1995). In addition, given the varied environmental conditions likely to be encountered among estuaries, the possibility of local adaptation by A. butcheri populations cannot be discounted (Bilton et al., 2002). While differences in growth rates among some populations (Sarre and Potter, 2000) have been ascribed to environmental rather than genetic variation (Partridge et al., 2004), this does not in itself preclude local adaptation for other traits, nor indicate that translocation of individuals among distinct populations is without nongenetic risks (e.g., disease transmission).
While genetic data have not distinguished several of the estuaries we sampled, it should be noted that the absence of genetic divergence among samples does not necessarily indicate that such populations are panmictic or receive migrants at such a rate to rapidly recover from overexploitation (Carvalho and Hauser, 1995; Waples, 1998). It is worth noting that our interpretation of nuclear autosomal population structuring would be very different (lower) had we not sampled loci pAb2D11 or Asc21–218, and additional loci could reveal even finer spatial structuring of variation. This study illustrates the importance of surveying many loci for studies of population structuring, be they allozymes or microsatellites (Arnaud-Haond et al., 2003; Hoarau et al., 2004).
References
GJ Adcock JH Bernal Ramírez L Hauser P Smith GR Carvalho (2000) ArticleTitleScreening of DNA polymorphisms in samples of archived scales from New Zealand snapper J Fish Biol 56 1283–1287 Occurrence Handle10.1006/jfbi.2000.1242
GR Allen SH Midgley M Allen (2002) Field Guide to the Freshwater Fishes of Australia Western Australian Museum Perth)
S Arnaud-Haond F Bonhomme F Blanc (2003) ArticleTitleLarge discrepancies in differentiation of allozymes, nuclear and mitochondrial DNA loci in recently founded Pacific populations of the pearl oyster Pinctada margaritifera J Evol Biol 16 388–398 Occurrence Handle10.1046/j.1420-9101.2003.00549.x
CF Baer (1999) ArticleTitleAmong-locus variation in F ST: fish, allozymes and the Lewontin-Krakauer test revisited Genetics 152 653–659
F Balloux H Brünner N Lugon-Moulin J Hausser J Goudet (2000) ArticleTitleMicrosatellites can be misleading: an empirical and simulation study Evolution 54 1414–1422 Occurrence Handle10.1554/0014-3820(2000)054[1414:MCBMAE]2.0.CO;2
TD Beacham DE Hay KD Le (2005) ArticleTitlePopulation structure and stock identification of Eulachon (Thaleichthys pacificus), an anadromous smelt, in the Pacific northwest Mar Biotechnol 7 363–372 Occurrence Handle10.1007/s10126-004-4075-0
D Bekkevold MM Hansen KD Mensberg (2004) ArticleTitleGenetic detection of sex-biased dispersal in historical and contemporary populations of anadromous brown trout Salmo trutta Mol Ecol 13 1707–1712 Occurrence Handle10.1111/j.1365-294X.2004.02156.x
P Bentzen CT Taggart DE Ruzzante D Cook (1996) ArticleTitleMicrosatellite polymorphism and the population structure of Atlantic cod (Gadus morhua) in the northwest Atlantic Can J Fish Aquat Sci 53 2706–2721 Occurrence Handle10.1139/cjfas-53-12-2706
N Bierne C Daguin F Bonhomme P David P Borsa (2003) ArticleTitleDirect selection on allozymes is not required to explain heterogeneity among marker loci across a Mytilus hybrid zone Mol Ecol 12 2505–2510 Occurrence Handle10.1046/j.1365-294X.2003.01936.x
DT Bilton J Paula JDD Bishop (2002) ArticleTitleDispersal, genetic differentiation and speciation in estuarine organisms Est Coast Shelf Sci 55 937–952 Occurrence Handle10.1006/ecss.2002.1037
CW Birky P Fuerst T Maruyama (1989) ArticleTitleOrganelle gene diversity under migration, mutation, and drift: equilibrium expectations, approach to equilibrium, effects of heteroplastic cells, and comparison to nuclear genes Genetics 121 613–627
AJ Bohonak (2002) ArticleTitleIBD (isolation by distance): a program for analyses of isolation by distance J Hered 93 153–154 Occurrence Handle10.1093/jhered/93.2.153
MJ Brownstein JD Carpten JR Smith (1996) ArticleTitleModulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping BioTechniques 20 1004–1010
VP Buonaccorsi JR McDowell JE Graves (2001) ArticleTitleReconciling patterns of inter-ocean molecular variance from four classes of molecular markers in blue marlin (Makaira nigricans) Mol Ecol 10 1179–1196 Occurrence Handle10.1046/j.1365-294X.2001.01270.x
CP Burridge AC Hurt LW Farrington PC Coutin CM Austin (2004) ArticleTitleStepping stone gene flow in an estuarine-dwelling sparid from south-east Australia J Fish Biol 64 805–819 Occurrence Handle10.1111/j.1095-8649.2004.0347.x
AD Butcher JK Ling (1962) ArticleTitleBream tagging experiments in East Gippsland during April and May 1944 Vic Nat 78 256–264
GR Carvalho L Hauser (1995) Molecular genetics and the stock concept in fisheries GR Carvalho TJ Pitcher (Eds) Molecular Genetics in Fisheries Chapman and Hall London 55–80
V Castric L Bernatchez (2003) ArticleTitleThe rise and fall of isolation by distance in the anadromous brook charr (Salvelinus fontinalis Mitchill) Genetics 163 983–996
JA Chaplin GA Baudains HS Gill R McCulloch IC Potter (1998) ArticleTitleAre assemblages of black bream (Acanthopagrus butcheri) in different estuaries genetically distinct? Int J Salt Lake Res 6 303–321
SF Chenoweth JM Hughes (1997) ArticleTitleGenetic population structure of the catadromous Perciform: Macquaria novemaculeata (Percichthyidae) J Fish Biol 50 721–733
SF Chenoweth JM Hughes CP Keenan S Lavery (1998) ArticleTitleConcordance between dispersal and mitochondrial gene flow: isolation by distance in a tropical teleost, Lates calcarifer (Australian barramundi) Heredity 80 187–197 Occurrence Handle10.1046/j.1365-2540.1998.00292.x
P Coutin (2000) ArticleTitleBlack bream-1997 Fish Vic Assess Rep Ser 18 1–72
Coutin PC, Conron S (2006) “Black bream.” In: Fisheries Co-Management Council Annual Report 2004–05. (Parkville, Victoria: Fisheries Co-Management Council), pp 31–33
KA Crandall JW Fetzner SH Lawler M Kinnersley CM Austin (1999) ArticleTitlePhylogenetic relationships among the Australian and New Zealand genera of freshwater crayfishes Aust J Zool 47 199–214 Occurrence Handle10.1071/ZO99011
S Innocentiis ParticleDe L Sola S Cataudella P Bentzen (2001) ArticleTitleAllozyme and microsatellite loci provide discordant estimates of population differentiation in the endangered dusky grouper (Epinephelus marginatus) within the Mediterranean Sea Mol Ecol 10 2163–2175 Occurrence Handle10.1046/j.1365-294X.2001.01371.x
RG Doupé GA Sarre GJ Partridge AJ Lymbery GI Jenkins (2005) ArticleTitleWhat are the prospects for black bream Acanthopagrus butcheri (Munro) aquaculture in salt-affected inland Australia? Aquac Res 36 1345–1355 Occurrence Handle10.1111/j.1365-2109.2005.01350.x
F Dufresne E Bourget L Bernatchez (2002) ArticleTitleDifferential pattern of spatial divergence in microsatellite and allozyme alleles: further evidence for locus-specific selection in the acorn barnacle, Semibalanus balanoides? Mol Ecol 11 113–123 Occurrence Handle10.1046/j.0962-1083.2001.01423.x
A Estoup F Rousset Y Michalakis J-M Cornuet M Adriamanga R Guyomard (1998) ArticleTitleComparative analysis of microsatellite and allozyme markers: a case study investigating microgeographic differentiation in brown trout (Salmo trutta) Mol Ecol 7 339–353 Occurrence Handle10.1046/j.1365-294X.1998.00362.x
LW Farrington CM Austin P Coutin (2000) ArticleTitleAllozyme variation and stock structure in the black bream, Acanthopagrus butcheri (Munro) (Sparidae) in southern Australia: implications for fisheries management, aquaculture and taxonomic relationship with Acanthopagrus australis (Günther) Fish Manag Ecol 7 265–279 Occurrence Handle10.1046/j.1365-2400.2000.00178.x
DJ Fraser C Lippé L Bernatchez (2004) ArticleTitleConsequences of unequal population size, asymmetric gene flow and sex-biased dispersal on population structure in brook charr (Salvelinus fontinalis) Mol Ecol 13 67–80 Occurrence Handle10.1046/j.1365-294X.2003.02038.x
AF Garber MD Tringali KC Stuck (2004) ArticleTitlePopulation structure and variation in red snapper (Lutjanus campechanus) from the Gulf of Mexico and Atlantic coast of Florida as determined from mitochondrial DNA control region sequence Mar Biotechnol 6 175–185 Occurrence Handle10.1007/s10126-003-0023-7
Gorman TB (1965) Seaward movement of black bream. Aust Fish News 24, 9
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet, J. (1995). FSTAT (v.1.2): a computer program to calculate f-statistics. J Hered 86, 485–486
J Goudet M Raymond T Meeüs Particlede R Rousset (1996) ArticleTitleTesting differentiation in diploid populations Genetics 144 1933–1940
CB Grimes MJ Kingsford (1996) ArticleTitleHow do riverine plumes influence fish larvae-do they enhance recruitment? Mar Fresh Res 47 191–208 Occurrence Handle10.1071/MF9960191
B Guinand C Lemaire F Bonhomme (2004) ArticleTitleHow to detect polymorphisms undergoing selection in marine fishes? A review of methods and case studies, including flatfishes J Sea Res 51 167–182 Occurrence Handle10.1016/j.seares.2003.10.002
JA Haddy NW Pankhurst (1998) ArticleTitleAnnual change in reproductive condition and plasma concentrations of sex steroids in black bream, Acanthopagrus butcheri (Munroe) (Sparidae) Mar Fresh Res 49 389–397 Occurrence Handle10.1071/MF97239
PW Hedrick (1999) ArticleTitlePerspective: highly variable loci and their interpretation in evolution and conservation Evolution 53 313–318 Occurrence Handle10.2307/2640768
PW Hedrick (2005) ArticleTitleA standardized genetic differentiation measure Evolution 59 1633–1638 Occurrence Handle10.1554/05-076.1
G Hoarau AM-T Piquet HW Veer Particlevan der AD Rijnsdorp WT Stam JL Olsen (2004) ArticleTitlePopulation structure of plaice (Pleuronectes platessa L.) in northern Europe: a comparison of resolving power between microsatellites and mitochondrial DNA data J Sea Res 51 183–190 Occurrence Handle10.1016/j.seares.2003.12.002
EP Hodgkin (1994) Estuaries and coastal lagoons LS Hammond RN Synnot (Eds) Marine Biology Longman South Melbourne 315–332
Holt CP (1978) The biology of three teleost species in the Swan River estuary. BSc Honours Thesis. Perth: Murdoch University
JA Hutchings L Gerber (2002) ArticleTitleSex-biased dispersal in a salmonid fish Proc Roy Soc Lond B 269 2487–2493 Occurrence Handle10.1098/rspb.2002.2176
DW Hutchinson AR Templeton (1999) ArticleTitleCorrelation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability Evolution 53 1898–1914 Occurrence Handle10.2307/2640449
C-T Jean S-C Lee C-F Hui C-T Chen (1995) ArticleTitleVariation in mitochondrial DNA and phylogenetic relationships of fishes of the subfamily Sparinae (Perciformes: Sparidae) in the coastal waters of Taiwan Zool Stud 34 270–280
DR Jerry PR Baverstock (1998) ArticleTitleConsequences of a catadromous life-strategy for levels of mitochondrial DNA differentiation among populations of the Australian bass, Macquaria novemaculeata Mol Ecol 7 1003–1013 Occurrence Handle10.1046/j.1365-294x.1998.00418.x
PJ Kailola MJ Williams PC Stewart RE Reichelt A McNee C Geieve (1993) Australian fisheries resources Bureau of Resource Science, Department of Primary Industries and Energy, and the Fisheries Research and Development Corporation Canberra
SA Karl JC Avise (1992) ArticleTitleBalancing selection at allozyme loci in oysters: implications from nuclear RFLPs Science 256 100–102 Occurrence Handle10.1126/science.1348870
PS Kench (1999) ArticleTitleGeomorphology of Australian estuaries: review and prospect Aust J Ecol 24 367–380 Occurrence Handle10.1046/j.1442-9993.1999.00985.x
M Kimura GH Weiss (1964) ArticleTitleThe stepping stone model of population structure and the decrease of genetic correlation with distance Genetics 49 561–576
ME Knight MJH Oppen Particlevan HL Smith C Rico GM Hewitt GF Turner (1999) ArticleTitleEvidence for male-biased dispersal in Lake Malawi cichlids from microsatellites Mol Ecol 8 1521–1527 Occurrence Handle10.1046/j.1365-294x.1999.00740.x
C Lemaire G Allegrucci M Naciri L Bahri-Sfar H Kara F Bonhomme (2000) ArticleTitleDo discrepancies between microsatellite and allozyme variation reveal differential selection between sea and lagoon in the sea bass (Dicentrarchus labrax)? Mol Ecol 9 457–467 Occurrence Handle10.1046/j.1365-294x.2000.00884.x
RCJ Lenanton (1977) ArticleTitleAspects of the ecology of fish and commercial crustaceans of the Blackwood River estuary Western Australia Fish Res Bull (Western Australian Marine Research Laboratories, Department of Fisheries and Fauna) 19 1–72
RCJ Lenanton SG Ayvazian CJ Dibden G Jenkins GA Sarre (1999) The use of stock enhancement to improve the catch rates of black bream Acanthopagrus butcheri (Munro) for Western Australian Recreational Fishers BR Howell E Moksness T Svåsand (Eds) Stock Enhancement and Sea Ranching Fishing News Books Oxford 219–230
RC Lewontin J Krakauer (1973) ArticleTitleDistribution of gene frequency as a test of the theory of selective neutrality of polymorphisms Genetics 74 175–195
T Lyrholm O Leimar B Johanneson U Gyllensten (1999) ArticleTitleSex-biased dispersal in sperm whales: contrasting mitochondrial and nuclear genetic structure of global populations Proc Roy Soc Lond B 266 347–354 Occurrence Handle10.1098/rspb.1999.0644
JH McDonald BC Verrelli LB Geyer (1996) ArticleTitleLack of geographic variation in anonymous nuclear polymorphisms in the American oyster, Crassostrea virginica Mol Biol Evol 13 1114–1118
M Nei (1987) Molecular Evolutionary Genetics New York Columbia University Press
GM Newton (1996) ArticleTitleEstuarine ichthyoplankton ecology in relation to hydrology and zooplankton dynamics in a salt wedge estuary Mar Fresh Res 47 99–111 Occurrence Handle10.1071/MF9960099
JV Norriss JE Tregonning RJC Lenanton G Sarre (2002) ArticleTitleBiological synopsis of the black bream, Acanthopagrus butcheri (Munroe) (Teleostei: Sparidae) in Western Australia with reference to information from other southern states Fish Res Report (Western Australian Marine Research Laboratories) 93 1–48
GJ Partridge GA Sarre NG Hall GI Jenkins J Chaplin IC Potter (2004) ArticleTitleComparisons between the growth of Acanthopagrus butcheri cultured from broodstock from two estuarine populations that are reproductively isolated and differ markedly in growth rate Aquaculture 231 51–58 Occurrence Handle10.1016/j.aquaculture.2003.08.005
GH Pogson KA Mesa RG Boutilier (1995) ArticleTitleGenetic population structure and gene flow in the Atlantic cod Gadus morhua: a comparison of allozyme and nuclear RFLP loci Genetics 139 375–385
IC Potter GA Hyndes (1999) ArticleTitleCharacteristics of the ichthyofaunas of southwestern Australian estuaries, including comparisons with holarctic estuaries and estuaries elsewhere in temperate Australia: a review Aust J Ecol 24 395–421 Occurrence Handle10.1046/j.1442-9993.1999.00980.x
K Rassmann D Tautz F Trillmich C Gliddon (1997) ArticleTitleThe microevolution of the Galapagos marine iguana Amblyrhynchus cristatus assessed by nuclear and mitochondrial genetic analyses Mol Ecol 6 437–452 Occurrence Handle10.1046/j.1365-294X.1997.00209.x
M Raymond F Rousset (1995) ArticleTitleGENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism J Hered 86 248–249
WR Rice (1989) ArticleTitleAnalysing tables of statistical tests Evolution 43 223–225 Occurrence Handle10.2307/2409177
SJ Rowland R Snape (1994) ArticleTitleLabile protogynous hermaphroditism in the black bream, Acanthopagrus butcheri (Munro) (Sparidae) Proc Linn Soc NSW 114 225–232
N Ryman PE Jorde (2001) ArticleTitleStatistical power when testing for genetic differentiation Mol Ecol 10 2361–2373 Occurrence Handle10.1046/j.0962-1083.2001.01345.x
GA Sarre IC Potter (2000) ArticleTitleVariation in age compositions and growth rates of Acanthopagrus butcheri (Sparidae) among estuaries: some possible contributing factors Fish Bull 98 785–799
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver. 2.001: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland http://lgb.unige.ch/arlequin/)
JR Schubel DJ Hirschberg (1978) Estuarine graveyards, climatic change, and the importance of the estuarine environment ML Wiley (Eds) Estuarine Interactions Academic Press New York 285–303
M Schuelke (2000) ArticleTitleAn economic method for the fluorescent labelling of PCR fragments Nat Biotech 18 233–234 Occurrence Handle10.1038/72708
PW Shaw GJ Pierce PR Boyle (1999) ArticleTitleSubtle population structuring within a highly vagile marine invertebrate, the veined squid Loligo forbesi, demonstrated with microsatellite DNA markers Mol Ecol 8 407–417 Occurrence Handle10.1046/j.1365-294X.1999.00588.x
PW Shaw C Turan JM Wright M O'Connell GR Carvalho (1999) ArticleTitleMicrosatellite DNA analysis of population structure in Atlantic herring (Clupea harengus), with direct comparison to allozyme and mtDNA RFLP analyses Heredity 83 490–499 Occurrence Handle10.1038/sj.hdy.6885860
JE Sherwood GN Backhouse (1982) ArticleTitleHydrodynamics of salt wedge estuaries-implications for successful spawning in black bream (Acanthopagrus butcheri) Warrnambool Inst Adv Educ, Faculty Applied Sci Tech, Res Report 82/3 1–5
M Slatkin (1987) ArticleTitleGene flow and the geographic structure of natural populations Science 236 787–792 Occurrence Handle10.1126/science.3576198
M Slatkin (1996) ArticleTitleA correction to the exact test based on the Ewens sampling distribution Genet Res 68 259–260 Occurrence Handle10.1017/S0016672300034236
RS Waples (1998) ArticleTitleSeparating the wheat from the chaff-patterns of genetic differentiation in high gene flow species J Hered 89 438–445 Occurrence Handle10.1093/jhered/89.5.438
RJ Watts MS Johnson (2004) ArticleTitleEstuaries, lagoons and enclosed embayments: habitats that enhance population subdivision of inshore fishes Mar Fresh Res 55 641–651 Occurrence Handle10.1071/MF04051
BS Weir CC Cockerham (1984) ArticleTitleEstimating F-statistics for the analysis of population structure Evolution 38 1358–1370d Occurrence Handle10.2307/2408641
T Wirth L Bernatchez (2001) ArticleTitleGenetic evidence against panmixia in the European eel Nature 409 1037–1040 Occurrence Handle10.1038/35059079
Wright S (1978) “Variability within and among populations.” In: Evolution and the Genetics of Populations, Vol. 4 (Chicago: The University of Chicago Press)
ES Yap PBS Spencer JA Chaplin IC Potter (2000) ArticleTitleThe estuarine teleost, Acanthopagrus butcheri (Sparidae), shows low levels of polymorphism at five microsatellite loci Mol Ecol 9 2155–2234 Occurrence Handle10.1046/j.1365-294X.2000.105337.x
Acknowledgments
Funding for the project was provided via a School of Life and Environmental Sciences grant to CPB. Prof. Chris Austin (Charles Darwin University) initiated genetic research of this species at Deakin University. We thank Patrick Coutin (Primary Industries Research Victoria) and Adrian Arkinstall, VICTAG Coordinator, Australian National Sportfishing Association, for assistance with the collection of samples. Devon Keeney (Otago University) and past and present members of the Molecular Ecology and Biodiversity Laboratory, Deakin University, made comments that improved the manuscript. Peter Unmack (University of Oklahoma) generously provided the template for Figure 1. The experiments described herein comply with the current Australian laws, and were approved by the Animal Ethics Committee of Deakin University.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Frequencies of microsatellite alleles and mitochondrial haplotypes at 11 populations of Acanthopagrus butcheri from southeast Australia
Locus/allele | Port Adelaide | Onkaparinga | Hindmarsh | Glenelg | Surrey | Hopkins | Painkalac | Thomson | Yarra | Gippsland | Sydenham |
|---|---|---|---|---|---|---|---|---|---|---|---|
Asc21–218 | (124) | (106) | (120) | (142) | (74) | (108) | (80) | (100) | (100) | (108) | (82) |
222 | – | – | – | – | – | – | – | – | 0.010 | 0.019 | – |
234 | – | – | – | – | – | 0.009 | – | – | – | – | – |
240 | 0.024 | 0.019 | – | 0.197 | 0.216 | 0.102 | 0.188 | 0.140 | 0.090 | 0.093 | 0.183 |
244 | 0.016 | 0.009 | – | – | – | 0.009 | 0.013 | 0.050 | – | – | – |
246 | 0.016 | – | 0.017 | 0.014 | 0.095 | 0.065 | 0.050 | 0.070 | 0.100 | 0.176 | 0.195 |
248 | 0.032 | 0.019 | – | 0.092 | 0.108 | 0.019 | – | 0.010 | 0.010 | 0.019 | 0.085 |
250 | 0.903 | 0.943 | 0.933 | 0.627 | 0.446 | 0.731 | 0.650 | 0.560 | 0.730 | 0.537 | 0.463 |
252 | – | – | 0.017 | 0.007 | – | 0.009 | – | 0.020 | 0.010 | 0.065 | 0.049 |
254 | 0.008 | 0.009 | 0.033 | 0.063 | 0.135 | 0.046 | 0.087 | 0.110 | 0.050 | 0.083 | 0.012 |
256 | – | – | – | – | – | 0.009 | 0.013 | 0.040 | – | 0.009 | 0.012 |
Pab1H1 | (110) | (106) | (116) | (140) | (56) | (104) | (80) | (96) | (100) | (108) | (84) |
151 | – | – | – | – | – | – | – | – | – | – | 0.012 |
153 | – | – | – | – | – | – | 0.013 | – | – | – | 0.024 |
155 | – | – | – | 0.007 | – | – | – | – | – | 0.009 | 0.012 |
157 | – | – | – | – | – | – | – | – | – | – | 0.012 |
159 | 0.009 | – | – | 0.007 | – | – | – | – | – | – | – |
161 | 0.964 | 1.000 | 0.974 | 0.964 | 1.000 | 0.981 | 0.988 | 1.000 | 0.970 | 0.870 | 0.869 |
163 | – | – | – | 0.007 | – | 0.010 | – | – | – | 0.019 | 0.024 |
165 | 0.027 | – | 0.009 | 0.014 | – | – | – | – | – | 0.046 | 0.012 |
167 | – | – | – | – | – | 0.010 | – | – | 0.020 | 0.037 | 0.024 |
169 | – | – | 0.017 | – | – | – | – | – | 0.010 | 0.019 | 0.012 |
Pab2D11 | (126) | (132) | (120) | (144) | (80) | (108) | (80) | (100) | (102) | (108) | (84) |
122 | – | – | – | 0.021 | – | – | – | – | – | – | – |
126 | 0.968 | 0.955 | 0.900 | 0.938 | 1.000 | 0.843 | 0.837 | 0.870 | 0.990 | 0.306 | 0.369 |
128 | – | – | – | 0.028 | – | 0.046 | 0.063 | 0.100 | 0.010 | 0.130 | 0.119 |
130 | 0.032 | 0.045 | 0.100 | 0.014 | – | 0.111 | 0.100 | 0.030 | – | 0.537 | 0.500 |
132 | – | – | – | – | – | – | – | – | – | 0.019 | 0.012 |
134 | – | – | – | – | – | – | – | – | – | 0.009 | – |
Pab2A5 | (126) | (132) | (120) | (144) | (78) | (108) | (78) | (100) | (102) | (108) | (84) |
108 | – | – | – | – | – | – | – | – | 0.010 | – | – |
110 | – | – | – | – | – | 0.046 | – | – | – | 0.056 | 0.071 |
114 | – | – | – | – | – | 0.009 | – | – | – | 0.019 | – |
116 | 0.460 | 0.598 | 0.758 | 0.701 | 0.731 | 0.741 | 0.603 | 0.620 | 0.667 | 0.713 | 0.690 |
118 | – | – | 0.025 | – | – | – | – | – | – | – | – |
120 | 0.540 | 0.402 | 0.217 | 0.299 | 0.269 | 0.204 | 0.397 | 0.380 | 0.314 | 0.213 | 0.238 |
122 | – | – | – | – | – | – | – | – | 0.010 | – | – |
Pma1n | (124) | (128) | (114) | (142) | (70) | (108) | (80) | (92) | (98) | (108) | (84) |
151 | – | – | – | – | 0.029 | – | – | – | – | – | – |
153 | 0.008 | – | – | – | – | – | – | – | – | – | 0.012 |
157 | 0.742 | 0.742 | 0.518 | 0.796 | 0.714 | 0.685 | 0.637 | 0.587 | 0.612 | 0.639 | 0.595 |
159 | – | – | – | 0.028 | 0.029 | 0.028 | 0.013 | 0.033 | 0.031 | 0.056 | 0.024 |
161 | 0.105 | 0.070 | 0.193 | 0.035 | 0.114 | 0.074 | 0.087 | 0.087 | 0.061 | 0.046 | 0.083 |
163 | – | – | 0.035 | 0.007 | 0.014 | 0.037 | 0.038 | 0.033 | 0.020 | 0.120 | 0.119 |
165 | – | 0.016 | – | – | – | – | – | – | – | 0.009 | 0.024 |
167 | – | – | – | 0.007 | – | 0.046 | 0.013 | 0.011 | 0.041 | 0.037 | 0.012 |
169 | – | – | – | – | – | 0.019 | – | – | – | 0.009 | 0.036 |
171 | – | – | 0.009 | – | – | 0.009 | – | – | 0.010 | 0.028 | 0.036 |
173 | 0.145 | 0.164 | 0.237 | 0.127 | 0.071 | 0.093 | 0.213 | 0.228 | 0.224 | 0.056 | 0.060 |
175 | – | 0.008 | 0.009 | – | – | 0.000 | – | – | – | – | – |
177 | – | – | – | – | – | 0.009 | – | 0.011 | – | – | – |
179 | – | – | – | – | 0.029 | – | – | 0.011 | – | – | – |
MtDNA | (52) | (54) | (52) | (72) | (40) | (52) | (40) | (48) | (51) | (53) | (41) |
AA | 0.904 | 0.870 | 0.712 | 0.542 | 0.350 | 0.308 | 0.525 | 0.250 | 0.412 | 0.226 | 0.268 |
AB | 0.077 | 0.037 | 0.173 | 0.042 | – | 0.096 | – | 0.042 | 0.078 | 0.076 | 0.073 |
AC | – | – | – | – | – | 0.038 | – | – | – | – | – |
BA | – | – | – | – | 0.050 | 0.077 | – | – | 0.059 | – | – |
BB | 0.019 | 0.093 | 0.077 | 0.347 | 0.525 | 0.192 | 0.200 | 0.250 | 0.294 | 0.075 | – |
BD | – | – | 0.038 | 0.028 | 0.025 | – | – | – | – | – | – |
CA | – | – | – | – | – | 0.039 | – | – | – | 0.019 | – |
CB | – | – | – | 0.042 | 0.050 | 0.250 | 0.275 | 0.458 | 0.157 | 0.604 | 0.659 |
Rights and permissions
About this article
Cite this article
Burridge, C.P., Versace, V.L. Population Genetic Structuring in Acanthopagrus butcheri (Pisces: Sparidae): Does Low Gene Flow Among Estuaries Apply to Both Sexes?. Mar Biotechnol 9, 33–44 (2007). https://doi.org/10.1007/s10126-006-6023-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-006-6023-7