Abstract
Mechanisms related to ecological or sexual selection have favoured sympatric speciation events in African and Central American lake cichlids. Allopatric divergence is the predominant speciation process observed in Amazonia, although, to the best of our knowledge, no study to date has attempted to determine whether speciation processes could exist under sympatric conditions in Amazonian cichlids. The Apistogramma agassizii species complex is an excellent model for investigating the existence of sympatric divergence events in the Amazon, as it shares many common life history characteristics with African Haplochromine cichlids in which sympatric speciation mechanisms are well documented. The genetic structure of A. agassizii was analysed by genotyping 889 individuals with ten microsatellite loci, collected from 26 sites distributed among small streams in 11 micro-basins in a very small portion of the Peruvian Amazon. It revealed 22 genetic populations identified according to panmictic criteria (FIS estimator) and strongly differentiated: FST estimator (0.034 to 0.356). Such a strong genetic structuring on such small geographical areas has never been demonstrated before in an Amazonian fish. Several of these populations may have diverged sympatrically and repeatedly in small stream networks. The results are discussed with respect to divergence processes, including sympatric speciation, which may be associated with the observed genetic structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the mechanisms involved in the initiation and persistence of genetic divergence in speciation processes is a central concern in evolutionary and ecological studies (Funk, et al., 2006; Maan & Seehausen, 2010; Seehausen & Magalhaes, 2010; Marques et al., 2017). The identification of these mechanisms and their role in speciation processes is key to understanding the driving force of biodiversity. Haplochromine cichlids from the African great lakes and particularly from Lake Victoria have been a model of choice for studying these mechanisms in vertebrates (Kornfield & Smith, 2000; Schluter, 2000; Kocher, 2004; Salzburger & Meyer, 2004; Meier et al., 2017). Theoretical and empirical evidence have accumulated on the prominent role of interactions between ecological and sexual selection in the rapid evolution of cichlids in the African great lakes (Gavrilets, 2004; Salzburger, 2009; Wagner, et al., 2012; Poelstra et al., 2018), but also for those living in Central American lakes (Wilson et al., 2000; Barluenga et al., 2006; Machado-Schiaffino et al., 2017). These interactions, when they promote linkage disequilibrium between genes involved in adaptation (e.g. trophic and visual adaptation mechanisms) and those involved in mating, facilitate the evolution of reproductive isolation (Seehausen et al., 2008; Wagner et al., 2012; Richards et al., 2019). The genomic potential (gene pool), through the rearrangement and recombination of ancestral polymorphism, would also play an important role in speciation processes and would have conditioned and accelerated the most spectacular radiation observed to date, in Haplochromines cichlids of Lake Victoria (McGee et al., 2020).
The role of geographical conditions (allopatric, parapatric, sympatric) in divergence and speciation processes are often difficult to define in nature because they are actually distributed along a continuum (Rice & Hostert, 1993; Fitzpatrick et al., 2008). The conditions relating to sympatry are the most difficult in practice. Their definition was originally based on an exclusively geographical criterion, and populations or species found in the same geographical area were then considered sympatric (Poulton, 1904). However, on a small geographical scale, spatial structuring in the distribution of populations can limit gene flow and condition their divergence. A later approach considered that the speciation process operates from a panmictic population, or deme, splitting into new demes that can diverge to form new species (Fitzpatrick et al., 2008), but it only relied on population genetic mechanisms, without taking into account the effect of geography. In addition to the notion of “deme”, the distribution of populations through their intrinsic dispersal capacities, or “normal cruising range of individuals”, was subsequently proposed (Mallet et al., 2009). Lately, in an effort to standardise these notions, Richards et al. (2019) have proposed that a genetic divergence event could have taken place under sympatric conditions if four criteria are met: (1) the demes (panmictic populations) form a monophyletic clade (originating from the same ancestral and exclusive deme), (2) their distribution areas (or “cruising range”) must overlap widely (geographical criterion), (3) the populations must not be the result of secondary contacts, (4) they must be reproductively isolated. Our study is based on this conceptual framework.
The environmental conditions in which Amazonian cichlids live are markedly different from those of the African or Central American lake cichlids. The Amazon is the world largest hydrological basin comprising a fractal network of thousands of interconnected watercourses ranging from kilometres-wide rivers to tiny forest brooks, larges floodplains and myriads of temporary or permanently connected lakes (Sioli, 1984; Goulding et al., 2003). A further complexity comes from the chemically contrasted natures of the waters flowing through this network: from Andean-born, nutrient-rich and basic white waters to the nutrient-poor, acidic black- or neutral clear- waters (Sioli, 1984). These environmental conditions are accompanied by highly variable levels of connectivity between ecosystems and a marked seasonality rhythmed by the flood pulse (Junk et al., 1989; Junk & Wantzen, 2004). The Amazon basin thus offers innumerable ecological niches that could be favourable to the appearance of adaptive radiations, sympatric and ecological speciation mechanisms, as observed in African rifts and Central American lake environments.
More than a third of the 258 cichlid species described for the Amazon basin (Dagosta & Pinna, 2019) belong to the genus Apistogramma (Cichliformes: Cichlidae). Endemic to South America, it currently totals 94 valid species, usually small (often < 8 cm), mainly distributed in the Amazon basin (Römer, 2000, 2006). The dwarf cichlids of the genus Apistogramma offer the opportunity to test the existence of speciation or sympatric divergence events in the Amazon. Indeed, they share many similarities with the Haplochromine cichlids of the African great lakes: high morphological variability, marked sexual dimorphism, colour polymorphism associated with differential mate choice, parental care, high endemicity, and trophic adaptations (Römer, 2000, 2006; Römer & Beseinherz, 2005; Ready et al., 2006; Engelking et al., 2010). Whereas most species within this genus have extremely small distribution areas, the morphological species Apistogramma agassizii has the widest geographical distribution in the Amazon basin. It has colonised many aquatic environments in fluvial or lacustrine systems, can live in all types of water (white, clear, black), although it is found preferentially in areas where plant debris accumulate. This species shows a marked sexual dimorphism, a significant colour polymorphism on a local and regional scale and provides intense parental care for its offspring (Römer, 2000, 2006). For the Peruvian Amazon, in a small portion of the Loreto region alone, Estivals et al. (2020) have shown that A. agassizii is actually composed of at least 3 biological species reproductively isolated by prezygotic barriers through preferential mate choice and the barrier effect of rivers. These three species occur in an extremely reduced geographical area compared to the range originally described for A. agassizii. The authors suggest that the important genetic variations within each of these three species may in fact be composed of diverging populations, or even constitute a species complex. Here, the geographical distribution of the genetic polymorphism of A. agassizii Sp1, the most widely distributed species within this complex, was analysed with the objectives of (1) assessing the link between the genetic structuring of the species identified as A. agassizii Sp1 and the organisation of the hydrographic network into micro-basins and streams, and (2) identifying the geographical conditions that may have favoured the observed genetic structure.
Methods
Study Sites and Field Methods
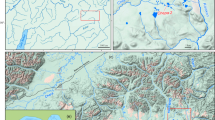
The biological species Apistogramma agassizii (Sp1) was sampled between 2015 and 2018 at 26 geographical sites spread over 11 different micro-basins (stream networks) (Fig. 1). Eight of the micro-basins are tributaries of the Ucayali River (A-H), two of the Marañon River (J,K) and one of the Amazon River (I). The sampling sites are all located outside the floodplain of the Ucayali, Marañon and Amazon rivers. The width of the sampled streams ranged approximately from 20 to 0.5 m with a depth of less than 2 m and covered a wide range of white, clear, black water types, according to Sioli (1984) (Figure SI 1). Fish were caught by professional aquarium fishers using beach seines or dip nets mainly in lentic areas where plant debris accumulated. Fish caught in the same stream and along a transect of less than 100 m were considered to belong to the same sampling site. A total of 889 individuals were collected (mean per site = 34, σ = 12). At the camp, all individuals were anaesthetised with eugenol following the protocol of Chanseau et al., (2002) until respiratory arrest, then preserved in a tube with its own identification code and filled with 96% ethanol. The ethanol in each of the tubes was renewed after 24 h, before conservation in the collection of the “Laboratorio de Biología y Genética Molecular del Instituto de Investigaciones de la Amazonía Peruana” (LBGM-IIAP, Iquitos, Peru).
Elevation map of: a in red, the area where Apistogramma agassizii is expected in Amazonia, b the complete study area, Loreto (Peru) with the 26 sample sites, c the sampling area in Jenaro Herrera region. Each stream network (micro-basin) connected to Ucayali, Marañon or Amazon rivers are identified by one letter from A to K and a colour code. Each sample site is identified by a number and preceded by the letter and the colour of the corresponding micro-basin
Genotyping
DNA from the samples was extracted from 10 mg of tail fin using a modified protocol from Doyle & Doyle (1987). The 10 microsatellite loci (SSRs) were organised and amplified in 4 multiplexes using the Qiagen Multiplex polymerase chain reaction kit and the primers defined by Quérouil et al. (2015) for Apistogramma. All selected SSRs correspond to highly polymorphic dinucleotide motifs. For more details of the DNA extraction methods, amplification conditions and genotyping consult Estivals et al. (2020), as they are identical to those presented there. The ten SSR markers were genotyped on 889 individuals and each run was composed of samples from at least two different geographical locations and positive controls consisting of reference genotypes to correct for possible shift problems in allelic size assignment. The negative controls used in the amplifications were analysed for possible contamination.
Allele sizes were obtained using the Geneious 9.1.8 program (Kearse et al., 2012). For each locus, when the chromatic signal of an allele was difficult to interpret, a new amplification and reading was performed. Individuals with missing values for more than three loci were removed from the analyses.
Data Analysis
Population Structure
The program STRUCTURE version 2.3.4 (Pritchard et al., 2000) was used as it allows to minimise deviation from panmixia to define clusters of individuals independently from their geographic location and then to evaluate the admixture between geographic populations, most of which are close to each other.
The chosen model accepted admixture (i.e. gene flow), correlated allele frequencies and did not take into account the geographical origin of individuals. In order to search for panmictic units integrating the largest possible number of individuals, the analysis was performed in several steps. The first step was performed on all individuals. The ‘optimal’ K value was determined from the ΔK measure (Evanno et al., 2005). The deviations from panmixia within these clusters were estimated by calculating the theta value of the Weir & Cockerham (1984) FIS and considered significant when P < 0.025. At the end of this first step, the clusters which did not show significant deviations from panmixia were considered as representative of genetic populations (demes). In a second step, a new STRUCTURE analysis was carried out independently for each cluster with remaining significant deviation to panmixia. This process was repeated until obtaining panmictic clusters or until STRUCTURE was no longer able to partition the analysed cluster.
For each STRUCTURE analysis 15 independent iterations were performed for each partitioning K (K from 1 to 27), with a burn-in period of 100,000, followed by a number of Markov chain Monte Carlo (MCMC) repetitions of 500,000. At each step analysis, each individual was placed in the cluster for which it had the highest percentage assignment value (inferred ancestry).
Allelic and Genetic Diversity
Allelic and genetic diversities were evaluated for each cluster highlighted by structure, considering: total number of alleles (k), allelic richness (Ar), total number of private alleles (priv) and their proportion (priv/k), expected heterozygosity (He), unbiased expected heterozygosity (Hn.b.) and observed heterozygosity (Hobs.), using the GENETIX software (Belkhir et al., 2004) and the ‘PopGenKit’ R package (Paquette, 2012).
Population Descriptors: Measuring Differentiation
The degrees of genetic differentiation between clusters were calculated for each pair of clusters from the FST theta estimator (Weir & Cockerham, 1984) using the GENETIX program (Belkhir et al., 2004). Then, genetic distances (Nei, 1972) were calculated for each pair of clusters from which an unrooted dendrogram was constructed using the Neighbour Joining method. The robustness of the nodes of the dendrogram was tested by a bootstrap test following 1000 draws. Genetic distance calculations were performed using the R package ‘adegenet’ (Jombart, 2008).
Isolation by Distance
The hypothesis of isolation by distance between clusters predominant at one site, was tested using their genetic distances (Rousset, 1997) calculated with the program Genetix (Belkhir et al., 2004) and their geographical distances (km), measured by following the streamlines calculated from ArcMap version 10.4.1 (ArcMap™, 1995–2015) and ImageJ version 1.49 (Rasband, 1997–2012). A Mantel test 1967(1967) was first performed on the entire dataset from 104 replicates using the mantel.randtest function of the ‘ade4’ package (Dray & Dufour, 2007; Bougeard & Dray, 2018; Chessel et al., 2004; Dray et al., 2007). A Mantel correlogram test (package ‘vegan’, Oksanen et al., 2015), was then performed to search for the presence of isolation by distance at different geographical scales, defining kilometre classes at the intra-micro-basin (0–5 km) and inter-micro-basin (5–82 km, 82–200 km, 200–400 km) levels, considering their positioning on the hydrographic network (Ucayali, Marañon, Amazon).
Genetic Relatedness/Kinship
Genetic relatedness (kinship) was searched for in the clusters for which a deviation from panmixia could not be rejected at the end of the STRUCTURE analyses. The null hypothesis (absence of relatedness) was tested by permutations (1000), by comparing the average value of genetic identity (Mathieu et al., 1990) obtained on all pairs of comparisons between individuals belonging to the tested cluster, with the distribution of values of genetic identity expected in an unstructured population, using the identix version 1.1 program (Belkhir et al., 2002).
Results
Genetic Structure
Four steps and 16 STRUCTURE analyses were required to identify the set of genetic clusters closest to panmixia and that best approximate the demes (or genetic populations) (Fig. 2, Table SI 1). At the end of the first step analysis, STRUCTURE identified 6 clusters showing a deviation to panmixia (Fig. 2, Table SI 1). After the second step analysis, carried out independently on these six clusters, STRUCTURE identified a total of 16 clusters with six of them showing a deviation to panmixia. In the third step analysis, carried out on each of these six new clusters showing a deviation to panmixia, STRUCTURE identified a sub structuration in four clusters for which a total of 10 new clusters were identified. Among these 10 new clusters, 3 clusters still showed a deviation to panmixia at the end of the step analysis. At the end of the fourth step analysis carried out on the 3 last clusters showing a deviation to panmixia, STRUCTURE was no longer able to identified a sub structuration. A total of 22 clusters were then identified, with 17 for which panmixia could not be rejected (FIS estimator min = − 0.093, p < 0.072; FIS estimator max = 0.059 p < 0.045; at significance level 0.025 (Table SI 1). The remaining 5 clusters (3, 11, 13, 15 and 18) all have significant heterozygote deficits (Table SI 1).
Illustration of the 4 steps-analysis with STRUCTURE and the histograms of the corresponding clusters according to de higher value of ΔK. Q = mean ancestry to cluster. The red stars below clusters indicate deviation to panmixia. ∑ = sum of clusters found for each step analysis. Colour code is independent between each step analysis
Geographical Distribution of Clusters
Clusters have a geographic distribution that is highly dependent on micro-basins and their streams (Fig. 3). Indeed, at least 76% and up to 100% of the individuals within a cluster come from a single micro-basin: 12 clusters out of 22 are endemic to a single micro-basin and the remaining 10 are overwhelmingly present in one micro-basin (median = 100; mean 95.6, σ = 7.2). When multiple clusters are endemic or predominant in a single micro-basin, each cluster predominates in a single stream that harbours at least 64% and up to 100% of the total cluster size (median = 94; mean 90.1, σ = 9.7) (Table 1). Of the 26 streams sampled, 18 have at least 2 clusters.
Allelic and Genetic Diversity
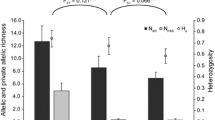
The numbers of individuals in the 22 clusters are heterogeneous, ranging from 16 to 106 with a mean value of 40 and a standard deviation of 19 (Table 2). Allelic richness is also variable, ranging from 3.3 to 10.0 (mean = 5.9, σ = 1.9) approximately. Private alleles are present in low frequencies and generally correspond to rare alleles. Cluster 14, in addition to having the highest allelic richness (≈ 10.0), has the highest number of private alleles. Clusters 1 and 2 show the lowest genetic diversity compared to the other clusters.
Genetic Differentiation
The FST values between cluster pairs range from 0.034 to 0.356, all of which are significant (p-value < 0.001). The median value of the theta estimator of FST is high: 0.151 (Table 3). Clusters 1 and 2 are the two most differentiated and correspond to individuals from micro-basins J (cluster 2) and K (clusters 1), both tributaries of the Marañon River.
Genetic Relationship Between Clusters (Genetic Distance)
The dendrogram of genetic distances reveals an organisation of clusters by both micro-basins and geographic distances (Fig. 4, note that colour codes are different from those of Fig. 3). The nodes are all supported by bootstrap values between 6 and 100. The grouping of clusters 1 and 2 is supported by a bootstrap value of 100 and gathers all individuals sampled in micro-basins K and J (tributaries of the Marañon) plus 2 individuals from micro-basins B and G.
Dendrogram of genetic distances between clusters using Nei (1972) genetic distance and a Neighbour joining construction. Numbers in dark blue, within the dendrogram, correspond to clusters. Only node values greater than 60 are shown. The pie charts illustrate in which sampling sites the clusters were identified and in which proportion. Each sampling site is identified by its own colour code but different from Fig. 3. Each large coloured triangle corresponds to a micro-basin (A to K)
Clustering of clusters 14, 12, 9, 8, and 13 corresponds primarily to individuals sampled in micro-basins A (31/35), B (98/123), F (38/39), H (41/47), and I (60/60), respectively. These micro-basins are tributaries of the Ucayali (A, B, F and H) and the Amazon (I). Clusters 13 and 14 are among the least genetically differentiated (FST = 0.092), yet they mainly gather individuals from the most geographically distant micro-basins.
Clusters 19, 20, 21, and 22 form a set supported by a bootstrap value of 64, which primarily collects individuals sampled in micro-basin C (C1 = 42/77; C2 = 25/30; C3 = 26/30; C1 = 27/77, respectively). Stream C1 gathers 42 of the individuals from cluster 19 (N = 43) and 27 of the individuals from cluster 22 (N = 28): hence these two clusters predominate and coexist sympatrically in the same small stream (Fig. 3, Figure SI 1).
The grouping of clusters 3, 4, 5, 6 and 7 is supported by a bootstrap value of 65 and gathers mainly individuals sampled in micro-basin D (D3 = 50/52; D2 = 12/12; D4 = 34/40; D5 = 35/39; D1 = 27/30, respectively; Fig. 4).
Clusters 16, 17, and 18 form a set supported by a bootstrap value of 64 and groups mainly individuals sampled in micro-basin E (E4 = 33/38, E2 = 26/26; E3 = 26/32, respectively).
Clusters that are endemic or predominant in a micro-basin form a single genetic set. This means that clusters such as in C, D or E are genetically closer to each other than they are to clusters in other micro-basins.
Nevertheless, cluster 15, which gathers all the individuals sampled in stream E1, does not group together with the other endemic or predominant clusters of micro-basin E. Similarly, clusters 10 and 11, which correspond mainly to individuals sampled in stream G2 (42 individuals of cluster 11; N = 43) and G1 (23 individuals of cluster 10; N = 28), do not group together.
Isolation by Distance
A significant (p-value = 0.003) isolation by distance was observed only for the smallest geographic distance class (0–5 km; Table 4), which corresponds to intra-micro-basin level. For all other comparisons, isolation by distance was rejected (p-value = 0.385).
Genetic Relatedness/Kinship
Among the 5 clusters with significant heterozygote deficits at the end of the analysis (3, 11, 13, 15 and 18), only clusters 11 and 15 could correspond to genetic relatedness (11: p-value = 0.006; 15: p-value = 0.005).
Discussion
Cluster Reality/Signification?
The identified clusters respond to both a geographical and biological logic. Indeed, the information carried by the clusters is consistent with the geographical distribution of individuals by stream or by micro-basins. Most of the clusters (17 out of 22) did not show significant deviations from panmixia. They would thus correspond to geographical units in which individuals reproduce randomly, forming reproductive units representing genetic populations in A. agassizii and that we will consider hereafter as being the representation of demes (Fitzpatrick et al., 2008). For five (3, 11, 13, 15 and 18) out of the 22 clusters, the deviation from panmixia could not be rejected. Two of them (cf. clusters 11 and 15) would correspond to family structures (related individuals). However, the significant heterozygote deficits observed in these “clusters” could be the consequence of a mixing of different demes (Wahlund effect, 1928) not detectable by STRUCTURE owing to the small size of the data matrix to analyse. This hypothesis could be tested by increasing our sampling sites (including new streams) within the micro-basins.
The demes are all endemic or predominant to a micro-basin or to a stream within a micro-basin. If we consider only individuals from demes endemic to a stream and those that represent the predominant fraction at a given site, it totals 797 individuals out of 889 analysed. The average assignment value obtained for these individuals for their respective clusters is 0.923 (σ = 0.11) while that obtained for the remaining, minority fraction at one site (92 individuals) is 0.74 (σ = 0.175). This lower value of the assignment rate could be explained by current or past gene flow (introgressions) between some demes.
Deme Structure in the Study Area
Differentiation values (FST) between the 22 demes vary between 0.034 and 0.356 (median = 0.151; mean = 0.157, σ = 0.064). These very high values are comparable to those found by Amado et al., (2011) but at an interspecific level in cichlids of the genus Symphysodon (range 0.02–0.38; mean = 0.242, σ = 0.110) or comparable to those found by Willis et al. (2015) between geographically distant populations in the cichlid species Cichla temensis (Cichliformes: Cichlidae) (0.157 vs. 0.186). Furthermore, it should be noted that this genetic structuring into 22 demes is observed in a much more restricted geographical area (49,000 km2) than those of the previous two studies, carried out at the scale of the Amazon and Orinoco basins (Fig. 5).
Role of Geographical Factors on Genetic Differentiation
It is interesting to note that the genetic distance between the 22 demes distributed in the different streams is not correlated with their geographical distance. This lack of correlation could be explained by the presence of barriers to dispersal in the study area. Indeed, the demes of micro-basins J (cluster 2) and K (cluster 1) on the left bank of the Marañon correspond to the most differentiated from all the others demes, even those geographically close to them but located in micro-basins distributed on the right bank of the Ucayali and the Amazon. The courses of the large rivers could therefore play the role of barrier to dispersal and to gene flow in A. agassizii between the left bank of the Marañon and the right bank of the Ucayali and Amazon rivers.
Moreover, even when the micro-basins are located on the same bank and geographically close, all the demes are endemic or predominant in a single micro-basin. Comparable to what was proposed by Wallace (1876) for terrestrial animal species, the course of large rivers could therefore also play the role of a structural or ecological barrier to the dispersal of fishes, here A. agassizii, even between micro-basins located on the same bank.
Additionally, we have observed that some streams partially dried out during the dry season, which could entail severe population decrease or trigger local extinctions of genetic clusters. These environmental fluctuations could therefore lead to multiple bottlenecks or founding effects, driving genetic differentiation under genetic drift effect. The effect of genetic drift is even more pronounced when the distances between demes are large (geographical isolation between demes within a micro-basin) or when geographical barriers limit gene flow (role of barrier rivers between micro-basins).
Evidence of a Sympatric Divergence Process in Cichlids in the Amazon?
In each of the micro-basins C, D and E in the Jenaro Herrera region, the demes are more closely related genetically to each other than to other micro-basins. This means that for these micro-basins, the endemic or predominant demes in the same micro-basin would have diverged from the same ancestral deme. It seems very unlikely that on three occasions, the predominant demes within these micro-basins would have diverged into different micro-basins (that our sampling failed to collect) and then that they would have met following secondary contacts in the same micro-basin. The interpretation of our observations by multiple divergence events (from a same ancestral deme, but different in each micro-basin) having occurred independently in the three micro-basins seems to be the most likely interpretation. Furthermore, for the demes corresponding to clusters 19 and 22, quasi-endemic to stream C1, as well as for the demes that have an overlapping “normal cruising range of individuals”, the observed differentiation seems to have operated in perfect sympatry.
The presence of strongly differentiated demes between micro-basins, between streams within the same micro-basin and within the same stream, suggests the existence of pre-zygotic or post-zygotic barriers favouring divergence between these demes. Otherwise, gene flow between demes would oppose the forces of divergence and tend towards genetic homogenisation. However, the evolution of intrinsic postzygotic isolation among fish species usually takes millions of years (review in Seehausen & Wagner, 2014). It is also a rare phenomenon in cichlid species, where divergence is usually maintained through prezygotic barriers such as behavioural mate choice (Seehausen et al., 1997; Wilson et al., 2000; Römer & Beisenherz, 2005). Behavioural mate choice has been widely demonstrated in African (Seehausen et al., 1997; Danley & Kocher, 2001; Wagner et al., 2012) and Central American cichlid species (Barlow & Siri, 1997; Wilson et al., 2000; Barluenga & Meyer, 2004), but also in many species of Apistogramma (Römer & Beisenherz, 2005; Ready et al., 2006; Engelking et al., 2010), including A. agassizii (Estivals et al., 2020). Moreover, during mating experiments, it was demonstrated that females of Apistogamma species choose males of their own morph far more precisely when males of closely or sympatric species, populations, or even colour morphs are presented (Römer & Beisenherz, 2005; Römer, 2006; Engelking et al., 2010; Estivals et al., 2020). In spite of results showing that this mechanism may not be completely fixed (Römer et al., 2014), it therefore seems likely that pre-zygotic barriers through mate choice play an important role in the maintenance and divergence of A. agassizii demes in sympatry.
Interestingly, for each stream network with white water types (B, I, J, and K), only one predominant cluster was identified, whereas in most stream networks with clear or black water, several predominant clusters were identified. Seehausen et al. (1997) showed that when the turbidity of the environment increases, sexual selection between closely related species no longer occurs. Similarly here, reproductive isolation between demes could therefore be due to mate choice and its strength could be directly conditioned by environmental conditions (i.e. water transparency) of the habitat. Differential selection between habitats could have initiated the differentiation process between demes in contact (Gavrilets, 2004). However, we cannot reject the hypothesis that disruptive ecological selection, without mate choice, could be involved in reproductive isolation (Schluter, 2000; Van Doorn et al., 2009; Elmer et al., 2010). An interaction between sexual and ecological selection ‘facilitating’ the evolution towards reproductive isolation is also possible (Seehausen et al., 2008; Wagner et al., 2012; Richard et al. 2019; McGee et al. 2020).
In addition to the criterion of sharing the same geographical area, the demes present in micro-basins C, D and E meet the three conditions required for sympatric speciation (Mallet et al., 2009): (1) they would have diverged from an ancestral deme, (2) they would not result from secondary contacts, but would be distributed where they formed, and (3) they would be reproductively isolated.
Deme Structure in the Micro-basins
The high values of genetic differentiation (FST) observed in the Jenaro Herrera region (up to 0.223) have never been observed before in such a small region of the Amazon for a fish species. In micro-basins C, D and E, the geographical distances between the sampled streams do not exceed 5 km following the course of the river networks (mean (C,D,E) = 2.77 km, σ = 0.98). The genetic differentiation observed between the predominant demes within streams is correlated with geographical distance, which may reflect poor dispersal abilities of the demes.
Between streams within the same micro-basin, the extreme FST values observed are of the same order of magnitude as those observed between demes of Apistogramma gephyra (Cichliformes: Cichlidae) (FST = 0.128) or Apistogramma pertensis (Cichliformes: Cichlidae) (FST = 0.228) that are at least 55 km apart and are thought to have diverged since the Middle or Late Pleistocene, as a result of the separation of their original watersheds (Leitão et al., 2017). The differentiation values observed in A. agassizii are therefore extremely strong both at the scale of the micro-basin and between micro-basins and despite the proximity of their confluences (a few hundred meters for C, D and E), their demes did not mix.
It is worth emphasizing that the demes corresponding to clusters 19 and 22 are both abundant in stream C1, with 42 individuals for cluster 19 (N = 43) and 27 individuals for cluster 22 (N = 28). They are genetically differentiated (FST = 0.051; p < 0.001), but are more closely genetically related to each other than to other demes, suggesting they could be sister demes originating from a common ancestor deme. The individuals in stream C1 were sampled over a fraction of the stream less than 100 m long and ~ 1 m width (Figure SI 1), so if there was even partial gene flow between these two demes, one would expect them to homogenise rapidly. There is therefore a barrier to reproduction between these two perfectly sympatric demes. According to Mayr´s (1942) species definition, “Species are groups of actually or potentially interbreeding natural populations, which are isolated from other such groups”, these demes would constitute biological species.
What Would Fall Under Allopatric, Parapatric and Sympatric Condition in the Study Area?
We do not know the extent of the distribution areas of the observed demes, nor those of their overlaps. However, we do know the geographical location of the individuals sampled from the 22 demes, and we can therefore understand their “normal cruising range of individuals” (Mallet, 2008) in relation to the geographical conditions in which they evolve. According to Mallet et al. (2009)’s definition of allopatry: “Where groups of populations are separated by uninhabited space across which dispersal and gene flow occurs at very low frequency”, micro-basins could correspond to distinct geographical units between which gene flow is very reduced, similar to allopatric or ‘micro-allopatric’ conditions. The genetic divergence between demes of different micro-basins could depend on ecological and physical constraints (barrier role of a river to dispersion inter-banks or along the same bank) which would favour genetic differentiation phenomena by vicariance.
In river systems, the dispersal of individuals tends to be asymmetrical, following the direction of the current (Hanfling & Weetman, 2006; Crispo et al., 2006). In a few cases dry season anadromous as well as catadromous migratory movements of Apistogramma species have been reported by different observers as well (summarized in Römer, 2000, 2006). Some fish species, particularly Apistogramma, may disperse by taking refuge in rafts of floating vegetation, dispersing passively over long distances (Schiesari et al., 2003). However, between micro-basins we do not observe any gradient of genetic polymorphism in the differentiation of the demes or in their number: on the contrary, each identified deme is endemic or predominant in a single micro-basin and no movement of dispersion can clearly be demonstrated between micro-basins. Under the assumption that the demes were mostly formed where they are now, there may be an ecological barrier to their dispersal. The morphological species A. agassizii prefers shallow areas with little current (lentic) where plant debris and especially dead leaves accumulate (Römer, 2000). This type of habitat is mostly found in small streams, such as those in micro-basins and in lakes. On the contrary, large rivers correspond to lotic and deep environments that are not very favourable to the presence of A. agassizii. It has been shown in cichlids from the African great lakes that a habitat discontinuity of only a few tens of metres can sometimes be sufficient to isolate two demes (Rico & Turner, 2002). The reduction in gene flow between micro-basins, whose mouths in the main river are separated by only a few hundred metres following a shoreline dispersal, could also be reinforced in lotic environments by inappropriate habitats and higher predation in large rivers (Dodrill et al., 2016; Deacon et al., 2018).
Sympatric Conditions
Within a micro-basin we would be under the conditions of sympatry as described by Mallet et al. (2009): “where individuals are physically capable of encountering one another with moderately high frequency. Populations may be sympatric if they are ecologically segregated, as long as a fairly high proportion of each population encounters the other along ecotones; and they may be sympatric, yet breed at different seasons”. Indeed, habitats suitable for the presence of A. agassizii were observed along the networks of streams that make up these micro-basins. Each sampled stream is represented by a single collection site that groups individuals sampled over a section of stream of only about 100 m. Our representation of the distribution of demes in the streams and a fortiori in the micro-basins is therefore very fragmented. However, although the endemic or predominant demes identified in the micro-basins are usually predominant in a single stream, they are also often present in other streams of the same micro-basin, which extends their actual distribution (“normal cruising range of individuals” according to Mallet et al., 2009) outside the stream where they are predominant. The cruising ranges of the demes may therefore overlap between streams of the same micro-basin, which supports the hypothesis of sympatry.
Apistogramma, the Model for Amazonian Cichlids?
In this Amazonian cichlid species, we show here a complex genetic structure, compatible with multiple divergence events that could have taken place in different geographical situations, from allopatry to sympatry and under the constraint of different ecological characteristics ranging from those of large rivers to those of small streams. Although speciation processes in Amazonian ichthyofauna are thought to be mainly based on allopatric conditions (Hubert et al., 2007; Albert & Reis, 2011; Dias et al., 2013), part of this biodiversity could also be based on sympatric speciation. In addition to sharing many characteristics with the Haplochromines of the African great lakes, A. agassizii presents an important genetic structuring that tends to be organised according to different geographical scales, thus allowing us to study the influence of geographical, but also ecological and historical conditions on the divergence process. The important levels of differentiation observed between demes and their maintenance in sympatry raise questions about the status of the species Apistogramma agassizii. Our observations reinforce the hypothesis that A. agassizii represents a complex of species and demes in the process of divergence at diverse geographical scales (Estivals et al., 2020). It seems very likely that comparable studies on other species of the same genus, such as Apistogramma bitaeniata (Cichliformes: Cichlidae) and Apistogramma cacatuoides (Cichliformes: Cichlidae), which have a wide distribution in the Amazon basin, could bring such a genetic structuring to light and thus would reinforce the hypothesis of sympatric speciation. The potential number of biological species grouped in the morphological species A. agassizii, and more broadly in the genus Apistogramma, could then be by several hundreds of species (Ready et al., 2006; Estivals et al., 2020). This might also be true for other highly diversified cichlid taxa such as Crenicichla, for which sympatric divergence has been suggested in the Neotropics (Burress et al., 2018; Piálek et al., 2012; Říčan et al., 2016). In order to unravel the mechanisms involved in their evolution, a particular effort should be made to study mechanisms related to ecological and sexual selection that have played an important role in the rapid evolution of cichlids in the African great lakes and Central American lakes (Van Doorn & Weissing, 2001; Gavrilets, 2004; Salzburger, 2009; Wagner et al., 2012; Machado-Schiaffino et al., 2017). Additionally, several recent studies have shown that secondary gene flow, hybridization and introgressions events are common in cichlids (Malinsky et al., 2015; Meier et al., 2017; Brock & Wagner, 2018; Poelstra et al., 2018; Richards et al., 2018), and are thought to have conditioned and accelerated species divergence (McGee et al., 2020). Given the paradigm constituted by the cichlids of African and Central American lakes, the genus Apistogramma thus appears to be an excellent model for the study of specific and adaptive radiation events in cichlids at the scale of Amazonian dendritic networks.
Data Availability
Data are available from the authors upon reasonable request.
References
Albert, J., & Reis, R. (2011). Historical Biogeography of Neotropical Freshwater Fishes. New York: University of California Press. https://doi.org/10.1525/california/9780520268685.001.0001
Amado, M., Farias, I., & Hrbek, T. (2011). A Molecular perspective on systematics, taxonomy and classification amazonian discus fishes of the genus Symphysodon. International Journal of Evolutionary Biology, 2011, 1–16. https://doi.org/10.4061/2011/360654
ArcGIS® software by Esri. ArcGIS® and ArcMap™ are the intellectual property of Esri (1995–2015)
Amado, M., Farias, I., & Hrbek, T. (2011). A molecular perspective on systematics, taxonomy and classification Amazonian discus fishes of the genus Symphysodon. International Journal of Evolutionary Biology, 2011, 360654. https://doi.org/10.4061/2011/360654
Barlow, G. W., & Siri, P. (1997). Does sexual selection account for the conspicuous head dimorphism in the Midas cichlid? Animal Behaviour, 53(3), 573–584. https://doi.org/10.1006/anbe.1996.0332.
Barluenga, M., & Meyer, A. (2004). The Midas cichlid species complex: Incipient sympatric speciation in nicaraguan cichlid fishes? Molecular Ecology, 13(7), 2061–2076. https://doi.org/10.1111/j.1365-294X.2004.02211.x
Barluenga, M., Stolting, K. N., Salzburger, W., Muschick, M., & Meyer, A. (2006). Sympatric speciation in nicaraguan crater lake cichlid fish. Nature, 439, 719–723. https://doi.org/10.1038/nature04325.
Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N., & Bonhomme, F. (2004). Genetix, logiciel sous windows TM pour la génétique des populations. Laboratoire Génome, populations, interactions CNRS UMR 5000. Université de Montpellier II.
Belkhir, K., Castric, V., & Bonhomme, F. (2002). identix, a software to test for relatedness in a population using permutation methods. Molecular Ecology Notes, 2(4), 611–614. https://doi.org/10.1046/j.1471-8286.2002.00273.x.
Bougeard, S., & Dray, S. (2018). Supervised multiblock analysis in R with the ade4 package. Journal of Statistical Software, 86(1), 1–17. https://doi.org/10.18637/jss.v086.i01
Brock, C. D., & Wagner, C. E. (2018). The smelly path to sympatric speciation? Molecular Ecology, 27(21), 4153–4156. https://doi.org/10.1111/mec.14845.
Burress, E. D., Piálek, L., Casciotta, J. R., Almirón, A., Tan, M., Armbruster, J. W., & et, & Říčan, O. (2018). Island- and lake-like parallel adaptive radiations replicated in rivers. Proceedings of the Royal Society B: Biological Sciences, 285(1870), 20171762. https://doi.org/10.1098/rspb.2017.1762
Chanseau, M., Bosc, S., Galiay, E., & Oules, G. (2002). L’utilisation de l’huile de clou de girofle comme anesthésique pour les smolts de saumon atlantique (Salmo salar L) et comparaison de ses effets avec ceux du 2-phenoxyethanol. Bulletin Français de la Pêche et de la Pisciculture. https://doi.org/10.1051/kmae:2002054
Chessel, D., Dufour, A. B., & Thioulouse, J. (2004). The ade4 package-I- one-table methods. R News, 4, 5–10.
Crispo, E., Bentzen, P., Reznick, D. N., Kinnison, M. T., & Hendry, A. P. (2006). The relative influence of natural selection and geography on gene flow in guppies. Molecular Ecology, 15(1), 49–62. https://doi.org/10.1111/j.1365-294X.2005.02764.x.
Dagosta, F., & de Pinna, M. (2019). The fishes of the Amazon: Distribution and biogeographical patterns, with a comprehensive list of species. Bulletin of the American Museum of Natural History, 431(1), 1–163. https://doi.org/10.1206/0003-0090.431.1.1
Danley, P. D., & Kocher, T. D. (2001). Speciation in rapidly diverging systems: Lessons from Lake Malawi. Molecular Ecology, 10(5), 1075–1086. https://doi.org/10.1046/j.1365-294x.2001.01283.x
Deacon, A. E., Jones, F. A. M., & Magurran, A. E. (2018). Gradients in predation risk in a tropical river system. Current Zoology, 64(2), 213–221. https://doi.org/10.1093/cz/zoy004
Dias, M. S., Cornu, J. F., Oberdorff, T., Lasso, C. A., & Tedesco, P. A. (2013). Natural fragmentation in river networks as a driver of speciation for freshwater fishes. Ecography, 36(6), 683–689. https://doi.org/10.1111/j.1600-0587.2012.07724.x.
Dodrill, M. J., Yard, M., & Pine Iii, W. E. (2016). Assessing predation risks for small fish in a large river ecosystem between contrasting habitats and turbidity conditions. American Midland Naturalist, 175(2), 206–221. https://doi.org/10.1674/0003-0031-175.2.206.
Doyle, J. J., & Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bulletin, 19, 11–15.
Dray, S., & Dufour, A. B. (2007). The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software, 22(4), 1–20. https://doi.org/10.18637/jss.v022.i04.
Dray, S., Dufour, A. B., & Chessel, D. (2007). The ade4 package - II: two-table and K-table methods. R News, 7(2), 47–52.
Elmer, K. R., Lehtonen, T. K., Kautt, A. F., Harrod, C., & Meyer, A. (2010). Rapid sympatric ecological differentiation of crater lake cichlid fishes within historic times. BMC Biology, 8, 60. https://doi.org/10.1186/1741-7007-8-60.
Engelking, B., Römer, U., & Beisenherz, W. (2010). Intraspecific colour preference in mate choice by female Apistogramma cacatuoides HOEDEMAN, 1951 (Teleostei: Perciformes: Cichlidae). Vertebrate Zoology, 60(3), 199–208.
Estivals, G., Duponchelle, F., Römer, U., García-Dávila, C., Airola, E., Deléglise, M., & Renno (2020). The amazonian dwarf cichlid Apistogramma agassizii (Steindachner, 1875) is a geographic mosaic of potentially tens of species: conservation implications. Conservation: Marine and Freshwater Ecosystems, 30(8), 1521–1539. https://doi.org/10.1002/aqc.3373.
Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14(8), 2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Fitzpatrick, B. M., Fordyce, J. A., & Gavrilets, S. (2008). What, if anything, is sympatric speciation? Journal of Evolutionary Biology, 21(6), 1452–1459. https://doi.org/10.1111/j.1420-9101.2008.01611.x.
Funk, D. J., Nosil, P., & Etges, W. J. (2006). Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proceedings of the National Academy of Sciences of the United States of America, 103(9), 3209–3213. https://doi.org/10.1073/pnas.0508653103.
Gavrilets, S. (2004). Fitness landscapes and the origin of species. Princeton University Press.
Goulding, M., Barthem, R., & Ferreira, E. J. G. (2003). The Smithsonian Atlas of the Amazon. Smithsonian Institution.
Hanfling, B., & Weetman, D. (2006). Concordant genetic estimators of migration reveal anthropogenetically enhanced source-sink population structure in the river sculpin, Cottus gobio. Genetics, 173(3), 1487–1501. https://doi.org/10.1534/genetics.105.054296.
Hubert, N., Duponchelle, F., Nuñez, J., García-Dávila, C., Paugy, D., & Renno, J. F. (2007). Phylogeography of the piranha genera Serrasalmus and Pygocentrus: Implications for the diversification of the neotropical ichthyofauna. Molecular Ecology, 16(10), 2115–2136. https://doi.org/10.1111/j.1365-294X.2007.03267.x
Jombart, T. (2008). Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics, 24(11), 1403–1405. https://doi.org/10.1093/bioinformatics/btn129.
Junk, W. J., Bayley, P. B., & Sparks, J. S. (1989). In D. P. Dodge (Ed.), The flood pulse concept in river floodplain systems. ( vol. 106). Canadian Special Publication of Fisheries and Aquatic Sciences. pp. 110–127
Junk, W. J., & Wantzen, K. M. (2004). In R. L. Welcomme, & T. Petr (Eds.), The flood pulse concept: new aspects, approaches and applications—An update: vol. Volume I (pp. 117–149). Food and Agriculture Organization and Mekong River Commission, FAO Regional Office for Asia and the Pacific.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P., & Drummond, A. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28(12), 1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kocher, T. D. (2004). Adaptive evolution and explosive speciation: the cichlid fish model. Nature Reviews Genetics, 5(4), 288–298. https://doi.org/10.1038/nrg1316.
Kornfield, I., & Smith, P. F. (2000). African cichlid fishes: model systems for evolutionary biology. Annual Review of Ecology Evolution and Systematics, 31, 163–196. https://doi.org/10.1146/annurev.ecolsys.31.1.163.
Leitão, C. S. S., Santos, C. H. A., Souza, É. M. S., Vilarinho, G. C. C., Paula-Silva, M. N., Val, P., Val, A. L., & de Almeida-Val, V. M. F. (2017). Development and characterization of microsatellite loci in amazonian dwarf cichlids Apistogramma spp. (Perciformes: Cichlidae): Uncovering geological influence on amazonian fish population. Journal of Applied Ichthyology, 33(6), 1196–1199. https://doi.org/10.1111/jai.13490
Maan, M. E., & Seehausen, O. (2010). Mechanisms of species divergence through visual adaptation and sexual selection: perspectives from a cichlid model system. Current Zoology, 56(3), 285–299. https://doi.org/10.1093/czoolo/56.3.285.
Machado-Schiaffino, G., Kautt, A. F., Torres-Dowdall, J., Baumgarten, L., Henning, F., & Meyer, A. (2017). Incipient speciation driven by hypertrophied lips in Midas cichlid fishes? Molecular Ecology, 26(8), 2348–2362. https://doi.org/10.1111/mec.14029.
Malinsky, M., Challis, R. J., Tyers, A. M., Schiffels, S., Terai, Y., Ngatunga, B. P., Miska, E. A., Durbin, R., Genner, M. J., & Turner, G. F. (2015). Genomic islands of speciation separate cichlid ecomorphs in an east african crater lake. Science, 350(6267), 1493–1498. https://doi.org/10.1126/science.aac9927.
Mallet, J. (2008). Hybridization, ecological races, and the nature of species: empirical evidence for the ease of speciation. Philosophical Transactions of the Royal Society of London Series B, 363(1506), 2971–2986. https://doi.org/10.1098/rstb.2008.0081.
Mallet, J., Meyer, A., Nosil, P., & Feder, J. L. (2009). Space, sympatry and speciation. Journal of Evolutionary Biology, 22(11), 2332–2341. https://doi.org/10.1111/j.1420-9101.2009.01816.x.
Mantel, N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Research, 27(2), 209–220.
Marques, D. A., Lucek, K., Haesler, M. P., Feller, A. F., Meier, J. I., Wagner, C. E., Excoffier, L., & Seehausen, O. (2017). Genomic landscape of early ecological speciation initiated by selection on nuptial colour. Molecular Ecology, 26(1), 7–24. https://doi.org/10.1111/mec.13774.
Mathieu, E., Autem, M., Roux, M., & Bonhomme, F. (1990). Épreuves de validation dans l’analyse de structures génétiques multivariées: comment tester l’équilibre panmictique? Revue de Statistique Appliquée, 38, 47–66.
Mayr, E. (1942). Systematics and the origin of species. Harvard University Press.
McGee, M., Borstein, S., Meier, J., Marques, D., Mwaiko, S., Taabu, A., Kishe, M. A., O’Meara, B., Bruggmann, R., Excoffier, L., & Seehausen, O. (2020). The ecological and genomic basis of explosive adaptive radiation. Nature, 586, 75–79. https://doi.org/10.1038/s41586-020-2652-7.
Meier, J. I., Marques, D. A., Mwaiko, S., Wagner, C. E., Excoffier, L., & Seehausen, O. (2017). Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nature Communications, 8, 1–11. https://doi.org/10.1038/ncomms14363.
Nei, M. (1972). Genetic distance between populations. The American Naturalist, 106(949), 283–292. https://doi.org/10.1086/282771.
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P., Minchin, P., O’Hara, B., Simpson, G. L., Solymos, P., Stevens, M. H. H., Szoecs, E., & Wagner, H. (2015). Vegan: Community Ecology Package. R Package Version, 2(2-1), 1–2.
Paquette, S. R. (2012). PopGenKit: useful functions for (batch) file conversion and data resampling in microsatellite datasets. R Package Version 1.0. Molecular Ecology Notes, 2, 611–614. https://doi.org/10.1046/j.1471-8278.2002.00273.x.
Piálek, L., Říčan, O., Casciotta, J., Almirón, A., & Zrzavý, J. (2012). Multilocus phylogeny of Crenicichla (Teleostei: Cichlidae), with biogeography of the C. lacustris group: species flocks as a model for sympatric speciation in rivers. Molecular Phylogenetics and Evolution, 62(1), 46–61. https://doi.org/10.1016/j.ympev.2011.09.006.
Poelstra, J. W., Richards, E. J., & Martin, C. H. (2018). Speciation in sympatry with ongoing secondary gene flow and a potential olfactory trigger in a radiation of Cameroon cichlids. Molecular Ecology, 27(21), 4270–4288. https://doi.org/10.1111/mec.14784.
Poulton, E. B. (1904). What is a species? Proceedings of the Entomological Society of London, 1903, lxxvii–cxvi.
Pritchard, J. K., Stephens, M., & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. https://doi.org/10.1093/genetics/155.2.945.
Quérouil, S., Diaz, V., García-Dávila, A., Römer, C., U., & Renno, J. F. (2015). Development and characterization of polymorphic microsatellite markers in neotropical fish of the genus Apistogramma (Perciformes: Labroidei: Cichlidae). Journal of Applied Ichthyology, 31(S4), 52–56. https://doi.org/10.1111/jai.12975.
Rasband, W. S., 1997–2012: ImageJ.US National Institutes of Health, Bethesda.
Ready, J. S., Sampaio, I., Schneider, H., Vinson, C., Santos, D., T., & Turner, G. F. (2006). Colour forms of amazonian cichlid fish represent reproductively isolated species. Journal of Evolutionary Biology, 19(4), 1139–1148. https://doi.org/10.1111/j.1420-9101.2006.01088.x.
Říčan, O., Piálek, L., Dragová, K., & Novák, J. (2016). Diversity and evolution of the middle american cichlid fishes (Teleostei: Cichlidae) with revised classification. Vertebrate Zoology, 66, 1–102.
Rice, W. R., & Hostert, E. E. (1993). Laboratory experiments on speciation: what have we learned in forty years? Evolution, 47(6), 1637–1653. https://doi.org/10.1111/j.1558-5646.1993.tb01257.x.
Richards, E. J., Poelstra, J. W., & Martin, C. H. (2018). Don’t throw out the sympatric speciation with the crater lake water: fine-scale investigation of introgression provides equivocal support for causal role of secondary gene flow in one of the clearest examples of sympatric speciation. Evolution letters, 2(5), 524–540. https://doi.org/10.1002/evl3.78.
Richards, E. J., Servedio, M. R., & Martin, C. H. (2019). Searching for sympatric speciation in the genomic era. Bioessays, 41(7), 1–10. https://doi.org/10.1002/bies.201900047
Rico, C., & Turner, G. F. (2002). Extreme microallopatric divergence in a cichlid species from Lake Malawi. Molecular Ecology, 11(8), 1585–1590. https://doi.org/10.1046/j.1365-294X.2002.01537.x.
Römer, U. (2000). Cichlid atlas. In natural history of south American dwarf cichlids (Vol. 1). Mergus Verlag.
Römer, U. (2006). Cichlid atlas. Natural history of south american dwarf cichlids. (Vol. 2). Mergus Verlag.
Römer, U., & Beisenherz, W. (2005). Intra- and interspecific mate choice of female Apistogramma cacatuoides (Teleostei: Cichlidae). Ichthyological Exploration of Freshwaters, 16(4), 339–345.
Römer, U., Engelking, B., & Beisenherz, W. (2014). Genetically determined mate choice can be influenced by learning in Apistogramma cacatuoides Hoedemann, 1951 (Teleostei, Cichlidae). Vertebrate Zoology, 64(2), 199–206.
Rousset, F. (1997). Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics, 145, 1219–1228. https://doi.org/10.1093/genetics/145.4.1219.
Salzburger, W. (2009). The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Molecular Ecology, 18(2), 169–185. https://doi.org/10.1111/j.1365-294X.2008.03981.x.
Salzburger, W., & Meyer, A. (2004). The species flocks of east african cichlid fishes: recent advances in molecular phylogenetics and population genetics. Naturwissenschaften, 91, 277–290. https://doi.org/10.1007/s00114-004-0528-6.
Schiesari, L., Zuanon, J., Azevedo-Ramos, C., Garcia, M., Gordo, M., Messias, M., & Vieira, E. (2003). Macrophyte rafts as dispersal vectors for fishes and amphibians in the Lower Solimes River, Central Amazon. Journal of Tropical Ecology, 19(3), 333–336. https://doi.org/10.1017/S0266467403003365.
Schluter, D. (2000). The ecology of adaptive radiation. Oxford University Press.
Seehausen, O., & Magalhaes, I. S. (2010). Geographical Mode and Evolutionary mechanism of ecological speciation in Cichlid Fish. In P. Grant & R. Grant (Eds.), In search of the causes of evolution (pp. 282–308). Princeton University Press.
Seehausen, O., Terai, Y., Magalhaes, I. S., Carleton, K. L., Mrosso, H. D., Miyagi, R., Van der Sluijs, I., Schneider, M. V., Maan, M. E., Tachida, H., Imai, H., -, & Okada, N. (2008). Speciation through sensory drive in cichlid fish. Nature, 455(7213), 620–626. https://doi.org/10.1038/nature07285.
Seehausen, O., Van Alphen, J. J. M., & Witte, F. (1997). Cichlid fish diversity threatened by eutrophication that curbs sexual selection. Science, 277(5333), 1808–1811. https://doi.org/10.1126/science.277.5333.1808.
Seehausen, O., & Wagner, C. E. (2014). Speciation in Freshwater Fishes. Annual review of ecology evolution and systematics, 45(1), 621–651. https://doi.org/10.1146/annurev-ecolsys-120213-091818
Sioli, H. (1984). The Amazon and its main affluents: Hydrography, morphology of the river courses, and river types. In H. Sioli (Ed.), The Amazon: Limnology and landscape ecology of a mighty tropical river and its basin (pp. 127–165). Dr W. Junk Publishers.
Steindachner, F. (1875). Beiträge der Kenntniss der Chromiden des Amazonenstromes. Sitzungsberichte der kaiserlichen Akademie der Wissenschaften. Mathematischnaturwissenschafliche Classe, 71, 61–137.
Van Doorn, G. S., Edelaar, P., & Weissing, F. J. (2009). On the origin of species by natural and sexual selection. Science, 326(5960), 1704–1707. https://doi.org/10.1126/science.1181661.
Van Doorn, G. S., & Weissing, F. J. (2001). Ecological versus sexual selection models of sympatric speciation: A synthesis. Selection, 2(1–2), 17–40. https://doi.org/10.1556/Select.2.2001.1-2
Wagner, C. E., Harmon, L. J., & Seehausen, O. (2012). Ecological opportunity and sexual selection together predict adaptive radiation. Nature, 487(7407), 366–369. https://doi.org/10.1038/nature11144.
Wagner, C. E., McCune, A. R., & Lovette, I. J. (2012). Recent speciation between sympatric Tanganyikan cichlid colour morphs. Molecular Ecology, 21(13), 3283–3292. https://doi.org/10.1111/j.1365-294X.2012.05607.x.
Wahlund, S. (1928). Zusammensetzung von populationen und korrelationserscheinungen vom standpunkt der vererbungslehre aus betrachtet. Hereditas, 11(1), 65–106. https://doi.org/10.1111/j.1601-5223.1928.tb02483.x.
Wallace, A. R. (1876). The geographical distribution of animals. Harper.
Weir, B. S., & Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38(6), 1358–1370. https://doi.org/10.2307/2408641.
Willis, S. C., Winemiller, K. O., Montaña, C. G., Macrander, J., Reiss, P., Farias, I. P., & Ortí, G. (2015). Population genetics of the speckled peacock bass (Cichla temensis), South America’s most important inland sport fishery. Conservation Genetics, 16, 1345–1357. https://doi.org/10.1007/s10592-015-0744-y.
Wilson, A. B., Noack-Kunnmann, K., & Meyer, A. (2000). Incipient speciation in sympatric Nicaraguan crater lake cichlid fishes: Sexual selection versus ecological diversification. Proceedings of the Royal Society B: Biological Sciences, 267(1458), 2133–2141. https://doi.org/10.1098/rspb.2000.1260
Acknowledgements
Guillain Estivals was supported by a scholarship of the Franco-Peruvian School of Life Sciences. Research was funded by and carried out within the framework of the International Mixt Laboratory - Evolution and Domestication of Amazonian Ichthyofauna (LMI-EDIA), Peruvian Amazon Research Institute - French National Research Institute for Sustainable Development - Gabriel René Moreno Autonomous University (IIAP-IRD-UAGRM). We would like to thank Philippe Cubry from UMR DIADE (Montpellier, France) for his advice on data analysis.
Author information
Authors and Affiliations
Contributions
Conceptualization: Guillain Estivals, Fabrice Duponchelle, Jean-François Renno. Developing methods: GE, FD, J-FR. Conducting the research: GE, FD, J-FR. Data analysis: GE, CM. Data interpretation: GE, FD, CM, J-FR. Preparation figures & tables: GE. Data interpretation, writing: GE, FD, CG-D, UR, CM, J-FR.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Estivals, G., Duponchelle, F., García-Dávila, C. et al. Exceptional Genetic Differentiation at a Micro-geographic Scale in Apistogramma agassizii (Steindachner, 1875) from the Peruvian Amazon: Sympatric Speciation?. Evol Biol 50, 1–17 (2023). https://doi.org/10.1007/s11692-022-09587-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-022-09587-9