Abstract
The experimental transfer of the vanA gene cluster from Enterococcus faecalis to Staphylococcus aureus has raised fears about the occurrence of such genetic transfer in clinical isolates of methicillin-resistant staphylococci. Recently, infections by a S. aureus strain carrying the enterococcal vancomycin resistance vanA gene cluster were reported. The possible emergence and dissemination of these strains is a serious health threat and makes optimization of prevention strategies and fast detection methods absolutely necessary. In the present study, we developed a PCR protocol for simultaneous detection of enterococcal vanA and vanB genes , the staphylococcal methicillin and mupirocin resistance markers mecA and ileS-2, and identification of S. aureus. As no vancomycin-resistant S. aureus isolates were available for our study, we used mixtures of enterococcal and staphylococcal colonies that harbored the different resistance markers to show that these genes could be detected simultaneously. This protocol could be used to facilitate the detection and identification of predictable S. aureus or methicillin-resistant strains carrying vanA or vanB.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Staphylococcus aureus is one of the most common causes of nosocomial and community-acquired infections worldwide [2, 6]. Since the emergence of methicillin-resistant strains of S. aureus (MRSA), the glycopeptide vancomycin has been the antibiotic of choice for serious MRSA infections [5]. In 1996, a S. aureus strain with intermediate resistance to vancomycin (VISA) (vancomycin MIC= 8μg /ml) was first isolated from a patient in Japan [10]. Shortly afterward, VISA strains were isolated in USA, Europe and other Asian countries [9, 24], arousing considerable concern regarding the emergence of S. aureus strains for which there will be no effective therapy. Characterization of these VISA strains indicates that the mechanisms of resistance are complex and involve changes in cell wall content and composition [3, 22]. In addition, the appearance of a powerful glycopeptide resistance mechanism among clinical isolates of enterococci, associated with the vanA and vanB gene clusters [4, 13, 15, 21], and the demonstration of experimental transfer of the vanA gene cluster from Enterococcus faecalis to S. aureus [16] have raised fears about the occurrence of such genetic transfer in clinical isolates of methicillin-resistant staphylococci. This type of transfer has already been reported in clinical isolates of Streptococcus bovis (vanB) [20] and Bacillus circulans (vanA) [12].
In June 2002, the world's first reported clinical infection due to S. aureus with high resistance to vancomycin (VRSA) (vancomycin MIC>128 μg/ml) was diagnosed in a patient in the USA [23]. A second isolate was reported in October of the same year [14]. These isolates contain the vanA genes from enterococci and the methicillin-resistance gene mecA. The possible emergence and dissemination of VRSA strains is a serious health threat and makes it absolutely necessary to optimize prevention strategies and fast detection methods.
In the present study, we developed a multiplex PCR protocol that allows the simultaneous detection of the two most widespread, transferable vancomycin resistance genes, vanA and vanB, and the methicillin resistance gene mecA, as well as the identification of S. aureus (femB). In addition, since mupirocin has been suggested as a reasonable adjuvant agent to prevent staphylococcal infections, and high resistance is already widespread [5, 18, 19], detection of the ileS-2 gene was also included.
Material and methods
Bacterial strains and DNA extraction
As there were no vancomycin-resitant S. aureus isolates available for our study, mixtures of enterococcal and staphylococcal colonies that harbored the different resistance markers were used to show that these genes could be detected simultaneously. Bacterial strains and susceptibility testing methods were previously described [17, 18]. The following strains were used: E. faecalis V583 (vancomycin-resistant, VanB), E. faecalis ATCC 29212 (vancomycin-susceptible), E. faecium BM4147 (vancomycin-resistant, VanA), S. aureus ATCC 29213 (methicillin- and mupirocin-susceptible), S. aureus SEIMC (methicillin-resistant and mupirocin-susceptible), S. aureus isolate 242 (methicillin- and mupirocin-resistant) and Staphylococcus epidermidis 8859-65 (methicillin- and mupirocin-resistant). For DNA extraction, bacteria were grown overnight on BHI agar plates. One colony from a Staphylococcus strain and one from a Enterococcus strain were mixed and resuspended in 25 μl of sterile distilled water, and heated at 100 °C for 15 min. A 5-μl aliquot of this suspension was directly used as template for PCR amplification.
Simultaneous detection of vanA, vanB, femB, mecA and ileS-2 by PCR
The primers used in this study are listed in Table 1. Multiplex PCR assays were carried out with the DNA suspension obtained from the Staphylococcus/Enterococcus mixtures. We ensured that our PCR protocol was adequate for the individual amplification of each DNA fragment: vanB (1,145 bp), vanA (732 bp), femB (651 bp), ileS-2 (456 bp) and mecA (310 bp) (not shown). Multiplex PCR was then done using the following mixtures: E. faecalis V583 (vanB)/S. aureus 242 (femB, mecA, ileS-2), E. faecium BM4147 (vanA)/S. aureus 242 (femB, mecA, ileS-2), E. faecalis ATCC 29212/S. aureus ATCC 29213 (femB), E. faecalis V583 (vanB)/S. aureus ATCC 29213 (femB), E. faecium BM4147 (vanA)/S. aureus ATCC 29213 (femB), E. faecalis V583 (vanB)/S. aureus SEIMC (femB, mecA), E. faecium BM4147 (vanA)/S. aureus SEIMC (femB, mecA), E. faecalis V583 (vanB)/S. epidermidis (mecA, ileS-2), E. faecium BM4147 (vanA)/S. epidermidis (mecA, ileS-2). The PCR conditions used were modifications of protocols previously described by us [17, 18]. A 5-µl aliquot of the DNA suspension was added to 28.5 μl of PCR mixture consisting of 1× reaction buffer [16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8)], 0.2 mM concentration of each of the four dNTPs (Promega Corp., Madison, Wis.), 3 mM MgCl2, 1.5 μM of each femB primer, 0.5 μM of each vanA and vanB primer, 0.4 μM of each ileS-2 primer and 0.4 μM of each mecA primer, and 1.25 U of Taq DNA polymerase (Bioline, London, UK). The femB primers were used to identify S. aureus strains, and the mecA, ileS-2, vanA and vanB primers to detect the respective resistance markers. In order to reduce the formation of nonspecific extension products, the protocol included a "hot-start". DNA amplification was carried out in a GeneAmp PCR system 2700 (Applied Biosystems, Foster City, Calif., USA) with the following thermal cycling profile: an initial denaturation step at 94 °C for 5 min was followed by 10 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 64 °C for 30 s, and extension at 72 °C for 45 s), and another 25 cycles of amplification (denaturation at 94 °C for 30 s, annealing at 50 °C for 45 s, and extension at 72 °C for 2 min), ending with a final extension step at 72 °C for 10 min. After PCR amplification, 5 μl were removed and subjected to agarose gel electrophoresis (2% agarose, 1× TBE, 100 V) to estimate the sizes of the amplification products by comparison with a 100-bp DNA ladder (Roche Diagnostics, Mannheim, Germany) .The gel was stained with ethidium bromide and the amplicons were visualized under a UV light.
Results and discussion
The reaction conditions for the multiplex PCR assay were optimized to ensure that each target sequence was satisfactorily amplified. The primers used in this study differ in annealing temperatures, which increased the possibility of occurrence of unwanted bands that originated from nonspecific amplification. Therefore, two rounds of amplification with different annealing temperatures were carried out. Multiplex PCR with targets that differ widely in size often favors amplification of the shorter target over the longer ones [18]. Thus, to ensure amplification of each target, different primer concentrations, template DNA preparations, and MgCl2 concentrations were assayed, and those described above were chosen.
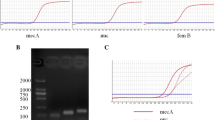
Figure 1 shows the results obtained with the optimized multiplex PCR assay as described in Material and methods; amplification of vanA, vanB, femB, ileS-2 and mecA targets produced distinct bands corresponding to their respective molecular size that were easily recognizable. vanA or vanB could be detected in mixtures containing femB (lanes 4 and 5), femB and mecA (lanes 6 and 7), and femB, mecA, and ileS-2 (lanes 1 and 2). vanA, vanB, ileS-2 and mecA were also simultaneously detected in the presence of the S. epidermidis (femB–) DNA. No amplification of the antibiotic resistance markers was observed in the control without DNA (lane 10) or in the mixture containing two susceptible strains (E. faecalis ATCC 29212 and S. aureus ATCC 29213) (lane 3). The entire protocol for the multiplex PCR assay, including the DNA extraction and electrophoresis, can be completed in less than 5 h.
Agarose gel electrophoresis of multiplex PCR amplification products from different enterococcal/staphylococcal mixes. Lanes: M 100-bp DNA ladder (Roche Diagnostics, Mannheim, Germany), 1 Enterococcus faecalis V583 (vanB1)/Staphylococcus aureus 242 (femB, mecA, ileS-2), 2 Enterococcus faecium BM4147 (vanA)/S. aureus 242 (femB, mecA, ileS-2), 3 E. faecalis ATCC 29212/S. aureus ATCC 29213 (femB), 4 E. faecalis V583 (vanB1)/S. aureus ATCC 29213 (femB), 5 E. faecium BM4147 (vanA)/S. aureus ATCC 29213 (femB), 6 E. faecalis V583 (vanB1)/S. aureus SEIMC (femB, mecA), 7 E. faecium BM4147 (vanA)/S. aureus SEIMC (femB, mecA), 8 E. faecalis V583 (vanB1)/S. epidermidis (mecA, ileS-2), 9 E. faecium BM4147 (vanA)/S. epidermidis (mecA, ileS-2), 10 control without a DNA template
Since vancomycin is frequently the drug of choice for the treatment of infections caused by now common MRSA, the recent appearance of a fully vancomycin-resistant strain of S. aureus has confronted us with the possible existence of non-tractable infections. The protocol described here could be used to facilitate the detection and identification of S. aureus or MRSA strains carrying vanA or vanB.
References
Anthony RM, Connor AM, Power EGM, French GL (1999) Use of the polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur J Clin Microbiol Infect Dis 18:30–34
Archer GL (1998) Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis 26:1179–1181
Avison MB, Bennett PM, Howe RA, Walsh TR (2002) Preliminary analysis of the genetic basis for vancomycin resistance in Staphylococcus aureus strain Mu50. J Antimicrob Chemother 49:255–260
Cetinkaya Y, Falk P, Mayhall CG (2000) Vancomycin-resistant enterococci. Clin Microbiol Rev 13:686–707
Chambers HF (1997) Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev 10:781–791
Chambers HF (2001) The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis 7:178–182
Dutka-Malen S, Evers S, Courvalin P (1995) Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol 33:24–27
Geha DJ, Uhl JR, Gustaferro CA, Persing DH (1994) Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol 32:1768–1772
Hamilton-Miller JM (2002) Vancomycin-resistant Staphylococcus aureus: a real and present danger? Infection 30:118–124
Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC (1997) Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother 40:135–136
Kobayashi N, Wu H, Kojima K, Taniguchi K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I (1994) Detection of mecA, femA and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol Infect 113:259–266
Ligozzi M, Lo Cascio G, Fontana R (1998) VanA gene cluster in a vancomycin-resistant clinical isolate of Bacillus circulans. Antimicrob Agents Chemother 42:2055–2059
Méndez-Álvarez S, Pérez-Hernández X, Claverie-Martín F (2000) Glycopeptide resistance in enterococci. Int Microbiol 3:71–80
Miller D, Urdaneta V, Weltman A (2002) Public health dispatch: Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb Mortal Wkly Rep 2002 51:902
Murray BE (2000) Vancomycin-resistant enterococcal infections. N Engl J Med 342:710–721
Noble WC, Virani Z, Cree RGA (1992) Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett 93:195–198
Pérez-Hernández X, Méndez-Álvarez S, Claverie-Martín F (2002) A PCR assay for rapid detection of vancomycin resistant enterococci. Diag Microbiol Infect Dis 42:273–277
Pérez-Roth E, Claverie-Martín F, Villar J, Méndez-Álvarez S (2001) Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol 39:4037–4041
Pérez-Roth E, Claverie-Martín F, Batista N, Moreno A, Méndez-Álvarez S (2002) Mupirocin resistance in methicillin-resistant Staphylococcus aureus clinical isolates in a Spanish hospital. Co-application of multiplex PCR assay and conventional microbiology methods. Diag Microbiol Infect Dis 43:123–128
Poyart C, Pierre C, Quesne G, Pron B, Berche P, Trieu-Cuot P (1997) Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob Agents Chemother 41:24–29
Rice LB (2001) Emergence of vancomycin-resistant enterococci. Emerg Infect Dis 7:183–187
Sieradzki K, Pinho MG, Tomasz A (1999) Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J Biol Chem 274:18942–18946
Sievert DM, Boulton ML, Stoltman G, Johnson D, Stobierski MG, Downes FP, Somsel PA, Rudrik JT, Brown W, Hafeez W, Lundstrom T, Flanagan E, Johnson R, Mitchell J, Chang S (2002) Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb Mortal Wkly Rep 51:565–567
Srinivasan A, Dick JD, Perl TM (2002) Vancomycin resistance in staphylococci. Clin Microbiol Rev 15:430–438
Acknowledgements
We thank Dr. Ninivé Batista, Microbiology Service, Nuestra Señora de Candelaria University Hospital, Santa Cruz de Tenerife, and Dr. Nadia Liassine, Central Laboratory of Bacteriology, University Hospital, Geneva, for providing bacterial strains. This work was supported by Spanish Fondo de Investigación Sanitaria grants PI 01/0636 and PI 01/3150 from the Instituto de Salud Carlos III (Spain), and by Consejería de Educación, Cultura y Deportes grant 2001/020 from the Canarian Autonomous Government. ERT and SMA were supported by FUNCIS PI 40/00 and FIS contract 99/3060, respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ramos-Trujillo, E., Pérez-Roth, E., Méndez-Álvarez, S. et al. Multiplex PCR for simultaneous detection of enterococcal genes vanA and vanB and staphylococcal genes mecA, ileS-2 and femB . Int Microbiol 6, 113–115 (2003). https://doi.org/10.1007/s10123-003-0118-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-003-0118-z