Abstract
The aim of this study was to develop a rapid detection assay to identify methicillin-resistant Staphylococcus aureus by simultaneous testing for the mecA, nuc, and femB genes using the loop-mediated isothermal amplification (LAMP) method. LAMP primers were designed using online bio-software (http://primerexplorer.jp/e/), and amplification reactions were performed in an isothermal temperature bath. The products were then examined using 2% agarose gel electrophoresis. MecA, nuc, and femB were confirmed by triplex TaqMan real-time PCR. For better naked-eye inspection of the reaction result, hydroxy naphthol blue (HNB) was added to the amplification system. Within 60 min, LAMP successfully amplified the genes of interest under isothermal conditions at 63 °C. The results of 2% gel electrophoresis indicated that when the Mg2+ concentration in the reaction system was 6 μmol, the amplification of the mecA gene was relatively good, while the amplification of the nuc and femB genes was better at an Mg2+ concentration of 8 μmol. Obvious color differences were observed by adding 1 μL (3.75 mM) of HNB into 25 μL reaction system. The LAMP assay was applied to 128 isolates cases of methicillin-resistant Staphylococcus aureus, which were separated from the daily specimens and identified by Vitek microbial identification instruments. The results were identical for both LAMP and PCR. LAMP offers an alternative detection assay for mecA, nuc, and femB and is faster than other methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an important human pathogen that produces a variety of toxins and causes a wide range of infections, including skin abscesses, necrotizing pneumonia, joint infections, and endocarditis [14, 22, 24, 28]. Recently, MRSA has emerged as an important pathogen in public health, causing significant morbidity [1] and often displaying multidrug resistance [3, 18]. Rapid detection of MRSA is imperative for both treatment and implementation of infection control policies to prevent disease spread and outbreaks [3, 6, 21].
Molecular methods such as polymerase chain reaction (PCR) and real-time PCR have been used for rapid MRSA identification [8, 12, 19]. Loop-mediated isothermal amplification (LAMP), developed by the Japanese researcher Notomi, is a novel gene amplification method that can complete DNA amplification under isothermal conditions with several outstanding advantages, including simplicity, rapid response, high sensitivity, and cost effectiveness [16]. LAMP eliminates the need for expensive PCR equipment or expertise. Using four specific primers that span six distinct sequences of a target gene, the entire procedure can be completed in less than 60 min by incubating all reagents in a single tube. To efficiently test the reaction result, a DNA intercalating dye, such as SYBR green, propidium iodide, or Picogreen, is added to the products after the reaction [13, 17]. Because the assay requires opening the tubes, it is associated with an increased risk of nucleic acid aerosol pollution as in the case of gel electrophoresis. Motoki et al. reported a simpler colorimetric assay using the Mg2+ ion concentration indicator hydroxy naphthol blue(HNB), where the color changes from violet to sky blue [7]. The LAMP detection sensitivity was equivalent to that of the assay using SYBR green, and the positive/negative result could be easily judged using the naked eye.
MecA gene is a specific drug-resistant MRSA gene that plays a decisive role in drug resistance [2, 4, 20], and detect the nuc gene helps in the rapid identification of S. aureus from clinical specimens [23]. FemB can be used as a marker to differentiate S. aureus from coagulase-negative Staphylococci [11].
Here, we report the combined use of three LAMP assays for identification of MRSA as follows: an assay targeting mecA to identify methicillin resistance, an assay targeting nuc to distinguish S. aureus from other Staphylococcus spp, and an assay targeting femB for coagulase-positive Staphylococci.
Materials and Methods
Bacterial Strains
This study was performed at the Navy General Hospital from January to December 2014. The standard control strain used in this study was the methicillin-resistant S. aureus American Type Culture Collection (ATCC) strain 33591. The MRSA strains tested in this study were collected from the clinical microbiological laboratory at the Navy General Hospital.
The isolates were previously isolated from patients sputum (n = 72), drainage fluid (n = 1), pus (n = 11), urine (n = 4), venous blood (n = 14), secreta (n = 25), and catheter (n = 1). All MRSA isolates were streaked on blood and mannitol salt agars and incubated overnight at 35 °C. Isolates that showed β-hemolysis on blood agar and appeared as yellow colonies in mannitol salt agar were subjected to coagulase and cefoxitin disk diffusion tests. The isolates was confirmed as a MRSA strain if it produced a positive result in the coagulase test and had a zone diameter of less than 22 mm on a cefoxitin disk diffusion test prior to real-time PCR.
Preparation of DNA Template
Crude DNA was extracted from overnight growth cultures of all isolates on 5% sheep blood agar by directly boiling the cell lysate. Briefly, a loopful of bacterial culture was mixed with 100 μL of DNA extraction buffer (Da An Gene, China).The suspension was boiled at 100 °C for 10 min and centrifuged at 13,000 rpm for 5 min. The supernatant was used as a DNA template for LAMP and PCR analyses.
Triplex TaqMan Real-Time PCR Primers and Probes
A 105 bp region of mecA, a 154 bp region of nuc, and a 183 bp region of femB were tested using a triplex TaqMan real-time PCR assay. mecA/nuc/femB-specific PCR primers and TaqMan fluorescent probes were selected using the Beacon Designer 7 bio-software. FAM, HEX, and ROX markers were used to label the fluorescent probe at the 5′ end, and the 3′ end was labeled with BHQ1 (Table 1) (AuGCT DNA-SYN Biotechnology, China).
Triplex TaqMan Real-Time PCR Detection of mecA, nuc, and femB
Triplex TaqMan real-time PCR targeting the mecA, nuc, and femB genes was performed on 128 clinically isolated MRSA samples. Briefly, the PCR reactions were performed in a 20 μL volume containing 10 μL of 2 × SuperReal PreMix (Probe) mix (TIGEN, China), 1 μL of the primer pair (for the mecA, nuc, and femB genes), 0.5 μL of the fluorescence probe (for mecA, nuc, and femB), 1 μL of bacterial genomic DNA, and 7.5 μL of ddH2O. PCR was performed using a SLAN-96P Real-time PCR System (Shanghai Hongshi, China). The PCR conditions were maintained at 95 °C for 5 min for initial denaturation followed by 40 cycles of 94 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. Following PCR, 5 μL aliquots of each sample were subjected to electrophoresis on 2% agarose gel to validate their identities. The bands were visualized using a gel documentation system (Heima, China).
LAMP Primers
LAMP primers were designed for the highly conserved regions of the mecA, nuc, and femB genes from Staphylococcal strains using the Primer Explorer V4 software (http://primerexplorer.jp:81/lam/) LAMP primers included two outer primers, F3 and B3, and two inner primers, FIP and BIP (Table 2) (AuGCT DNA-SYN Biotechnology, China).
LAMP Assays
Optimization of the LAMP assay was performed on ATCC 33591. The LAMP reaction was performed in a 25 μL reaction mixture containing 2.5 μL of 10× Bst buffer, 0.8/1.2 μL of MgSO4 (25 mM), 1.6 μL of dNTPs (10 mM), 4μL of betaine (5 M), 1μL of HNB (3.75 mM),1.6 μM each of FIP and BIP, 0.2 μM each of F3 and B3,1μL of Bst DNA polymerase(8 IU), 1μL of template, and 3.8μL/3.2μL of ddH2O. The reaction mixtures were incubated at 63 °C for 60 min and heated to 80 °C for 15 min to terminate the reaction. The amplification products were detected using 2% agarose gel electrophoresis to observe the band under UV illumination.
Results
Triplex TaqMan Real-Time PCR Testing mecA, nuc, and femB
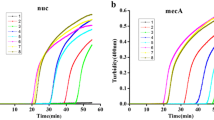
We directly detected the mecA, nuc, and femB genes of ATCC 33591 using the triplex assay in a single tube. Initially, each gene was detected with a single-target TaqMan PCR assay (Fig. 1a) to obtain the specific PCR amplification curves for these three genes. The PCR products were analyzed by 2% agarose gel electrophoresis to observe the specificity of the amplification bands (Fig. 1b). Then, each gene was detected with a triplex TaqMan real-time PCR assay. The PCR curves of mecA and nuc using a single-target assay (blue) were consistent with the triplex-target assay (red). The femB PCR curve was slightly lower, but an effective amplification curve was still obtained (Fig. 1c).
Primers and probes for the triplex TaqMan PCR assay. a Using the mecA, nuc, and femB primers as well as a fluorescent probe, individual reactions were performed in a single tube. b The PCR product bands were consistent with the expected molecular weight, confirming PCR product specificity. c Three sets of primers and fluorescent probes reacted in one PCR tube
Establishment of the LAMP Method
The LAMP assays for mecA, nuc, and femB were optimized using ATCC 33591. The amplified DNA fragments were detected by 2% agarose gel electrophoresis, and the bands were observed under UV illumination (Fig. 2a, where the MRSA genomic DNA ladder is in the lane with bands and the with water negative control template had no band). In addition, we added 1μL of HNB (150 μM) into the LAMP reaction system, wherein the dye color changed from violet to sky blue (Fig. 2b). These LAMP assays were applied to detect clinical MRSA strains and to produce better amplification. Selected mecA testing results from the clinical MRSA isolates are shown in Fig. 2c.
Establishing the LAMP assay and improving its response to naked-eye inspection. a Optimized LAMP reaction conditions. b Upon adding HNB into the LAMP reaction system, the color changed when a positive amplification product was detected. c Application of LAMP technology for testing genes from clinical isolates
Comparing Test Results of the Triplex TaqMan Real-Time PCR and LAMP Assays
Data for the triplex TaqMan real-time PCR assay were collected utilizing FAM (mecA), HEX (nuc), and ROX (femB) as the reporter dyes to minimize the potential for emission spectrum overlap. Clinical MRSA isolates (n = 128) were tested by triplex TaqMan real-time PCR, and a total of 91 strains that the traditional method identified as MRSA were mecA positive. We then applied the LAMP method for the same clinical MRSA isolates (n = 128) and also acquired 91 mecA-positive isolates, matching the real-time PCR results. Nuc and femB were determined to be positive via triplex TaqMan real-time PCR and LAMP testing, as shown in Table 3.
Discussion
Staphylococcus aureus is one of the important pathogenic bacteria that are commensal to the human nares and skin [22, 23]. Infections caused by methicillin-resistant S. aureus (MRSA) strains have exhibited an increasing trend in numerous countries and regions [15]. To reduce the harm caused by MRSA, it is necessary to establish a fast and accurate gene detection method. Thus far, there are several methods for identifying MRSA, including routine standard procedures (colony morphology, Gram staining, and testing of catalase, hyaluronidase, and coagulase), the Vitek 2 automated system, the API-Staph kit, immunological assays, mass spectrometry, and PCR (regular PCR as well as quantitative PCR) [5, 22, 25]. However, loop-mediated isothermal amplification (LAMP) technology is a nucleic acid detection method that requires less stringent experimental conditions. Since the invention of LAMP technology, it has been applied to the detection of Yersinia pseudotuberculosis, Salmonella, Pseudomonas aeruginosa, and Listeria monocytogenes [5, 9, 26, 27, 29]. However, due to products that would differ in size, LAMP is not suitable for the detection of multiple genes in the same system. Therefore, we developed three LAMP assays targeting mecA, nuc, and femB to replace a single or triplex TaqMan real-time PCR assay.
Methicillin resistance in S. aureus is primarily mediated by mecA, which encodes the low-affinity penicillin-binding protein 2a or 2′ (PBP2a or PBP2′). However, mecA is also widely distributed among coagulase-negative Staphylococci (CNS) and is associated with CNS methicillin resistance [28]. Thus, detecting mecA alone cannot discriminate between MRSA and methicillin-resistant CNS. However, the femB gene locus is distant from the mecA gene on the chromosome, and femB is involved in cell wall pentaglycine side chain and interpeptide bridge formation [10]. FemB is highly conserved in S. aureus but not found in CNS [25]. In this experiment, we generated primers and the fluorescence probes for mecA, nuc, and femB, and analyzed all isolates. By adjusting the molar concentration of the primers and probes, the amplification using three pairs of primers and three probes simultaneously in a reaction system was similar to the amplification of each individual gene. All isolates were detected by triplex TaqMan real-time PCR. The results indicated that mecA was only detected in 72.7% of the isolated strains that were identified as MRSA by traditional methods. The positive rates of nuc and femB detection were 100 and 98.4%, respectively. Utilizing the results of triplex TaqMan real-time PCR as the reference, we employed LAMP to investigate all isolates. The detection results of the LAMP method were 72.7% mecA, 100% nuc, and 98.4% femB, which were 100% consistent with the triplex TaqMan real-time PCR. Compared to the triplex TaqMan real-time PCR, the entire process of the LAMP method has less stringent experimental condition requirements and its specificity and accuracy are equal to those of the triplex TaqMan real-time PCR method.
In conclusion, the combined use of the mecA, nuc, and femB-LAMP assays will be beneficial for coagulase-positive MRSA differentiation. Because of its simplicity and ease of performance without the need for sophisticated instrumentation, this process can be easily adapted for any microbiology laboratory as a rapid molecular bench to assay.
References
Addicks JP, Gotting M, Jensen AM et al (2010) MRSA—current aspects of resistance, pathology, epidemiology and therapy. Versicherungsmedizin 62:183–188
Chambers HF, Deleo FR (2009) Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi:10.1038/nrmicro2200
Davis MF, Peterson AE, Julian KG et al (2013) Household risk factors for colonization with multidrug-resistant Staphylococcus aureus isolates. PLoS ONE 8:e54733. doi:10.1371/journal.pone.0054733
Fishovitz J, Hermoso JA, Chang M et al (2014) Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life 66:572–577. doi:10.1002/iub.1289
French GL (2009) Methods for screening for methicillin-resistant Staphylococcus aureus carriage. Clin Microbiol Infect 15(Suppl 7):10–16. doi:10.1111/j.1469-0691.2009.03092.x
Geiger K, Brown J (2013) Rapid testing for methicillin-resistant Staphylococcus aureus: implications for antimicrobial stewardship. Am J Health Syst Pharm 70:335–342. doi:10.2146/ajhp110724
Goto M, Honda E, Ogura A et al (2009) Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 46:167–172. doi:10.2144/000113072
Hirvonen JJ (2014) The use of molecular methods for the detection and identification of methicillin-resistant Staphylococcus aureus. Biomark Med 8:1115–1125. doi:10.2217/bmm.14.60
Kim HJ, Kim HS, Lee JM et al (2016) Rapid detection of Pseudomonas aeruginosa and Acinetobacter baumannii Harboring bla(VIM-2), bla(IMP-1) and bla(OXA-23) genes by using loop-mediated isothermal amplification methods. Ann Lab Med 36:15–22. doi:10.3343/alm.2016.36.1.15
Kobayashi N, Wu H, Kojima K et al (1994) Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol Infect 113:259–266
Liu ZZ, Xiong YL, Fan XJ et al (2011) The correlation between expression level of femB and resistance phenotype of methicillin-resistant Staphylococcus aureus (MRSA). Sichuan Da Xue Xue Bao Yi Xue Ban 42:661–665
Montazeri EA, Khosravi AD, Jolodar A et al (2015) Identification of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from burn patients by multiplex PCR. Burns 41:590–594. doi:10.1016/j.burns.2014.08.018
Mori Y, Kanda H, Notomi T (2013) Loop-mediated isothermal amplification (LAMP): recent progress in research and development. J Infect Chemother 19:404–411. doi:10.1007/s10156-013-0590-0
Napolitano LM (2008) Early appropriate parenteral antimicrobial treatment of complicated skin and soft tissue infections caused by methicillin-resistant Staphylococcus aureus. Surg Infect 9(Suppl 1):s17–s27. doi:10.1089/sur.2008.063.supp (Larchmt)
Navarro MB, Huttner B, Harbarth S (2008) Methicillin-resistant Staphylococcus aureus control in the 21st century: beyond the acute care hospital. Curr Opin Infect Dis 21:372–379. doi:10.1097/QCO.0b013e3283013add
Notomi T, Okayama H, Masubuchi H et al (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63
Notomi T, Mori Y, Tomita N et al (2015) Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 53:1–5. doi:10.1007/s12275-015-4656-9
Oteo J, Belen Aracil M (2015) Molecular characterization of resistance mechanisms: methicillin resistance Staphylococcus aureus, extended spectrum beta-lactamases and carbapenemases. Enferm Infecc Microbiol Clin 33(Suppl 2):27–33. doi:10.1016/S0213-005X(15)30012-4
Palavecino EL (2014) Rapid methods for detection of MRSA in clinical specimens. Methods Mol Biol 1085:71–83. doi:10.1007/978-1-62703-664-1_3
Pozzi C, Waters EM, Rudkin JK et al (2012) Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog 8:e1002626. doi:10.1371/journal.ppat.1002626
Sturenburg E (2009) Rapid detection of methicillin-resistant Staphylococcus aureus directly from clinical samples: methods, effectiveness and cost considerations. Ger Med Sci 7:Doc06. doi:10.3205/000065
Su J, Liu X, Cui H et al (2014) Rapid and simple detection of methicillin-resistance Staphylococcus aureus by orfX loop-mediated isothermal amplification assay. BMC Biotechnol 14:8. doi:10.1186/1472-6750-14-8
Sudhaharan S, Vanjari L, Mamidi N et al (2015) Evaluation of LAMP assay using phenotypic tests and conventional PCR for detection of nuc and mecA genes among clinical isolates of Staphylococcus spp. J Clin Diagn Res 9:DC06–09. doi:10.7860/JCDR/2015/13962.6315
Tacconelli E, De Angelis G, de Waure C et al (2009) Rapid screening tests for meticillin-resistant Staphylococcus aureus at hospital admission: systematic review and meta-analysis. Lancet Infect Dis 9:546–554. doi:10.1016/S1473-3099(09)70150-1
Towner KJ, Talbot DC, Curran R et al (1998) Development and evaluation of a PCR-based immunoassay for the rapid detection of methicillin-resistant Staphylococcus aureus. J Med Microbiol 47:607–613
Wang Y, Wang Y, Luo L et al (2015) Rapid and sensitive detection of Shigella spp. and Salmonella spp. by multiple endonuclease restriction real-time loop-mediated isothermal amplification technique. Front Microbiol 6:1400. doi:10.3389/fmicb.2015.01400
Wu R, Liu X, Guo B et al (2014) Development of double loop-mediated isothermal amplification to detect Listeria monocytogenes in food. Curr Microbiol 69:839–845. doi:10.1007/s00284-014-0661-1
Zervos M (2008) Treatment options for uncomplicated community-acquired skin and soft tissue infections caused by methicillin-resistant Staphylococcus aureus: oral antimicrobial agents. Surg Infect 9(Suppl 1):s29–s34. doi:10.1089/sur.2008.065.supp (Larchmt)
Zhang H, Feng J, Xue R et al (2014) Loop-mediated isothermal amplification assays for detecting Yersinia pseudotuberculosis in milk powders. J Food Sci 79:M967–M971. doi:10.1111/1750-3841.12436
Funding
This work was supported by the National Natural Science Foundation (No. 81401311) and the Capital Characteristic Clinical Application Research (WU JIEPING Foundation No. Z141107006614009). This work was also supported by the Navy General Hospital Innovation Cultivation Foundation (No. CXPY201412).
Author’s Contributions
CQY and ZQY conceived the study, collected and analyzed the data, and drafted the manuscript. GJW and LYJ conceived the project and provided technical support for data collection and analysis. CCG conceived the study and revised the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no competing interests.
Ethical Approval
The subjects of this work are bacteria that were separated and purified from clinical samples. This article does not contain any studies with human or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Chen, C., Zhao, Q., Guo, J. et al. Identification of Methicillin-Resistant Staphylococcus aureus (MRSA) Using Simultaneous Detection of mecA, nuc, and femB by Loop-Mediated Isothermal Amplification (LAMP). Curr Microbiol 74, 965–971 (2017). https://doi.org/10.1007/s00284-017-1274-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-017-1274-2