Abstract
Pathology as a common diagnostic test of cancer is an invasive, time-consuming, and partially subjective method. Therefore, optical techniques, especially Raman spectroscopy, have attracted the attention of cancer diagnosis researchers. However, as Raman spectra contain numerous peaks involved in molecular bounds of the sample, finding the best features related to cancerous changes can improve the accuracy of diagnosis in this method. The present research attempted to improve the power of Raman-based cancer diagnosis by finding the best Raman features using the ACO algorithm. In the present research, 49 spectra were measured from normal, benign, and cancerous breast tissue samples using a 785-nm micro-Raman system. After preprocessing for removal of noise and background fluorescence, the intensity of 12 important Raman bands of the biological samples was extracted as features of each spectrum. Then, the ACO algorithm was applied to find the optimum features for diagnosis. As the results demonstrated, by selecting five features, the classification accuracy of the normal, benign, and cancerous groups increased by 14% and reached 87.7%. ACO feature selection can improve the diagnostic accuracy of Raman-based diagnostic models. In the present study, features corresponding to ν(C–C) αhelix proline, valine (910–940), νs(C–C) skeletal lipids (1110–1130), and δ(CH2)/δ(CH3) proteins (1445–1460) were selected as the best features in cancer diagnosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nowadays, there exist different types of cancers with complex causes and cures, which affect human health. Among such cancers, breast cancer is one of the most common cancers in women annually, leading to thousands of deaths. Early detection is useful for the treatment of breast cancer [1].
Screening mammography followed by histopathological diagnosis is used to identify and characterize breast lesions. The treatment in many cases is breast-conserving surgery. The surgery aims to preserve as much healthy tissue as possible while removing the tumor thoroughly. Therefore, an intra-operative guidance tool is needed to assess large tissue areas and detect lesions in real-time [2].
Therefore, many different studies have been carried out on the earlier, faster, and more accurate detection of this type of cancer; nevertheless, histopathology remains the gold standard for diagnosis. Despite its strengths, this method has its weaknesses including being invasive, prolonged response time, and its dependency on the pathologist’s experience and skill. Therefore, recently, different techniques such as optical coherence tomography (OCT), white light reflectance (WLR), auto-fluorescence, and Raman spectroscopy have been proposed to solve these problems. OCT and WLR rely on the visualization of changes in tissue structure. These techniques provide little or no information about the molecular composition of tissue and, therefore, generally provide low specificity. Auto-fluorescence imaging has shown to improve diagnostic sensitivity. Nonetheless, the specificity of this technique is low too [2].
Raman spectroscopy, which analyzes molecular vibrations, can provide high molecular specificity. Any changes from healthy tissue to cancer are reflected in their Raman spectra. This technique can characterize biological tissues in vivo or in vitro noninvasively and without any need to prepare the tissue. These specifications facilitate the translation of the technique to the clinic. Moreover, many anatomical locations can be assessed in vivo by the use of optical fibers in combination with Raman spectroscopy. Researchers in the assessment of different cancers have utilized this method [2,3,4,5,6].
Raman spectroscopy is a method that relies on inelastic scattering of monochromatic light usually coming from a laser source. When monochromatic light penetrates a sample, some of it scatters, either in the same frequency of the incident light (Rayleigh scattering) or in different frequencies (Raman scattering). The frequency difference between the incident and scattered light depends on the vibration frequency of the sample’s molecular bonds. Therefore, Raman spectroscopy can provide a unique fingerprint for each material. This is a technique to identify different materials, including biological samples [7].
Cancer-related cellular and molecular changes cause differences in measured Raman spectra. In the present research, we aimed to find the best changes relevant to cancer in Raman spectra as discriminating features to improve the diagnosis of malignant (cancer) and benign neoplasm. Therefore, we developed a model to discriminate normal, benign, and cancerous samples of breast tissue and subsequently optimized the model by removing useless features using the ant colony optimization (ACO) technique.
Materials and methods
In this study, 49 Roman spectra were measured from 11 normal, cancerous, and benign samples. Then, interfering factors including noise and background fluorescence were removed using range independent algorithm (RIA) [8]. Next, the intensity of 12 important Raman bands of the biological samples was extracted as discriminating features of each Raman spectrum (Table 1). Finally, the ACO was applied to find the best of the resultant 12 features for diagnosis.
Samples and spectra
A set of breast tissue samples consisting of three cancerous (invasive ductal carcinoma), three normal (obtained from the margin of tumors), and five benign (fibrocystic change) samples was borrowed from the pathology lab of Kashan’s Shahid Beheshti Hospital in the state of fixed in formalin solution (10% neutral buffered formaldehyde in water). Taken out from formalin for a few minutes, the samples were measured by Raman spectroscopy. Then, considering the size of the samples, between three to six spectra were measured in terms of different features.
A Senterra-Bruker micro-Raman spectroscope with a ×50 lens was used in this research. This spectroscope with 785-nm wavelength and 10-mW power diode laser measures the spectra in 500–3200-cm−1 interval with a resolution less than 3 cm−1.
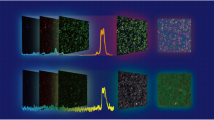
After spectroscopy, tissue samples were taken back to the formalin solution and sent to the pathology lab, and the remainder of the histopathology procedure comprising tissue processing (including dehydration, clearing, and impregnation), embedding in paraffin, sectioning by microtome, staining by hematoxylin and eosin, and finally, slide examination under a light microscope and diagnosis of the disease were conducted by an expert pathologist. The pathologist’s diagnosis was attached to the spectra obtained from each sample as the label of class in the dataset. This procedure of sample preparation for spectroscopy and histopathology is shown in Fig. 1 with some sample photographs of underlying steps.
Finally, 49 spectra including 17 from cancerous, 14 from normal, and 18 from benign samples were obtained. The raw spectra were processed using MATLAB 7 software.
Preprocessing
The purpose of preprocessing is to remove interfering factors such as noise and background fluorescence from the Raman spectra. At first, the resolution increased to 1 cm−1 by spline interpolation for correct detection of peak locations. Then, background fluorescence was removed using RIA introduced by Krishna in 2012 [8].
In RIA, the spectrum is cut into the required wavenumber range and then extrapolated in both ends using least square linear fitting. Then, two Gaussian peaks with suitable heights and widths are added to both sides of the extrapolation. Finally, the resulting spectrum is smoothed iteratively. In each iteration, the minimum of the smoothed and original spectrum is retained. The algorithm is continued until the accurate retrieval of the two added Gaussian peaks [8].
In the present study, the RIA algorithm was used in the 500–3200 wavenumber range. The height of the added Gaussian peaks was twice the maximum height of the spectrum and their FWHM was equal to 40 cm−1. Moreover, a zero-order Savitzky-Golay (SG) smoothing filter with a span of 20 spectral points was selected. Finally, normalization was done with respect to the intensity of 1655 cm−1 (Amide I) band that was clearly observed in all the spectra; the ratio of the other Raman bands to this band is widely used as a discriminating feature for cancer diagnosis [3].
Feature extraction
After preprocessing, a dataset containing 2700 features for every 49 spectra in the range of 500–3200 was prepared. The 13 most important bands of the biological samples were determined (Table 1), and the height of the spectral peaks in these bands was extracted as a feature. As mentioned before, the 12th band was considered as the normalizing band.
Feature selection using the ant colony optimization algorithm
The ant colony optimization (ACO) is a metaheuristic method that is used to find the best path in a weighted graph using artificial ants. The ants move on the graph stochastically, but with bias produced by a pheromone model. The pheromone guides ants to the shortest path incurring the lowest cost (the best) solution.
In the present problem, the ACO algorithm produces a large population of artificial ants that look for the best subset of features to distinguish classes in a high dimensional feature space (12 features). In this research study, each artificial ant was attributed to a unique subset of features. The artificial ants interact via virtual chemical pheromone distributed on the features. The pheromones changed dynamically in each iteration and reinforced themselves using positive feedback. For removing redundant features, an evaporation constant was applied in such a way that the effect of pheromone decreased evenly over time.
The ACO algorithm iteratively executed a loop including three central elements [10].
-
(1)
Creating ants for each subset of features proportional to the trace of pheromone on that subset
Spectral features were assigned to artificial ants by the following transition probability function:
where τi(t) is the amount of pheromone for the ith spectral feature in time (t), \( {\eta}_i^{\beta } \) identifies background information (ACO allows adding information to background search to improve the result), and α and β are pheromone and background information weights. Therefore, the ants are more likely to choose such spectral features provided the background information or the amount of pheromone is high.
In the first step, the value of all pheromone was equal to 1; therefore, each ant was able to choose spectral variables with proper probability according to background information.
-
(2)
Evaluating the performance of each ant (i.e., evaluating the classification accuracy of each subset of features)
In the present study, the QDA classifier was used and its performance was measured by each ant using the leave one out method. Accordingly, in each implementation, one spectrum was put aside and QDA was trained with the rest of the spectra. Then, the performance of the retained spectrum was evaluated. This process continued until all the Raman spectra were classified.
-
(3)
Updating pheromone trace by evaporation constant and classifier performance
The amount of the pheromone τi for each spectral feature was updated according to the following equation:
in which ρ is a constant between 0 and 1 and simulates the pheromone evaporation rate and Δτi is related to the accuracy of the ants’ classification. It should be mentioned that there was a slight difference between various versions of ACO, mostly related to the pheromone update process. In this study, the following formula was used to calculate Δτi; here, Ei is the classifier error (1—classification accuracy).
Classification accuracy is the ratio of the number of truly classified instances (spectra) to the number of total instances.
Over the ACO steps, the best ant was selected as the elite ant. Thus, Raman features with the best classification accuracy were allowed to increase pheromone, while the pheromone in the rest of the ants gradually evaporated.
These three phases were repeated step by step until obtaining the best classification accuracy.
The ACO parameters used in the present study are shown in Table 2.
In order to find the optimum subset of features, the ACO algorithm was applied 40 times with NF (number of features) elements, where NF changed from 1 (the smallest subset) to 12 (the whole set).
Results
Figure 2 shows the mean of spectra after preprocessing in the three classes: cancerous, normal, and benign. The dotted line is for the cancerous, continuous line for the normal, and dashed line for the benign class. The horizontal axis refers to the wave number and the vertical axis indicates the intensity.
Table 3 shows the NF selected features using ACO from NF = 1 to NF = 12. The features were numbered from 1 to 12 as F1 to F12. The features selected in all the 40 repetitions were shown by ✓, while the features never selected were shown by ×; for the other features, the number of their selections was reported. Evidently, the best results were related to the 5- and 7-feature subsets having a minimum classification error equal to 0.1224.
The confusion matrix of classification visualizes the performance of classification. Each row of the confusion matrix represents the predicted class of instances while the columns represent their actual class. The (i,j) element of this matrix is the number of instances belonging to class j and is classified as class i. Subsequently, the elements on the main diagonal of the matrix (i = j) represent true classified instances.
Table 4 shows the results of the classification in three different states including without ACO and with the best 5- and 7-element subsets. Dark columns indicate the number of correctly classified spectra in each class. In the first state (without ACO), the accuracy of the diagnosis equaled 0.73. In the 5- and 7-feature states, the diagnosis accuracy increased to 0.87. Therefore, it is seen that diagnosis accuracy increased by 14% while the number of features reduced.
In addition, the confusion matrices of 12-, 5-, and 7-feature states are shown in Tables 5, 6, and 7, respectively.
The diagram of processing including the results is shown in Fig. 3.
Discussion
In the present research, we were able to improve the Raman-based diagnosis accuracy of normal breast tissue and its neoplasia-related abnormalities (benign and cancerous tumor) using optimum feature selection by ACO.
Table 4 shows a diagnosis accuracy improvement from 73.4 to 87.7% before and after ACO feature selection, respectively. In addition to increasing the total diagnosis accuracy to more than 14%, according to Tables 5, 6, and 7, as shown in the distinctive increase in the diagnostic ratio of the ill-behaved normal class, the sensitivity and specificity of diagnosis increased in all the classes, Furthermore, this improvement in diagnosis power occurred simultaneously with reduction in the number of features that decreased the complexity of the diagnostic model. By reducing the number of features from 12 to 7 or 5, model complexity and consequently, its construction time decreased greatly, leading to the easier interpretation of the model.
As shown in Table 3, the features 2, 7, and 11 were selected for both the 5- and 7-feature states. These features refer to bands of proteins, and therefore, such bands are apparently important in diagnosing cancer. However, according to our test, only applying these three common features to the classifier decreases the efficiency of diagnosis.
Ant colony optimization among many other evolutionary-based optimization methods has shorter processing time and has been shown capable of exploiting mutual interactions among spectral variables according to their importance [10, 11]. Therefore, ACO has been chosen for spectral feature selection for dimension reduction, which is useful for real-time in vivo diagnosis. The present study proved its ability in reducing model complexity and simultaneously improving its discriminating power.
Conclusion
The present study showed that ACO feature selection can improve the diagnostic power of Raman-based cancer diagnosis. We reached the accuracy of 87.7% with only five features in the three discriminating classes of normal, benign, and cancerous samples of breast tissue.
References
Richards-Kortum R, Mahadevan-Jansen A, Ramanujam N (1996) Optical spectroscopy vs. the surgical suite [cancer detection]. IEEE Circuits and Devices Magazine 12:34–40
Santos IP et al (2017) Raman spectroscopy for cancer detection and cancer surgery guidance: translation to the clinics. Analyst 142(17):3025–3047. https://doi.org/10.1039/C7AN00957G
Mahadevan-Jansen A, Richards-Kortum RR (1996) Raman spectroscopy for the detection of cancers and precancers. J Biomed Opt 1:31–70
Raniero L et al (2011) In and ex vivo breast disease study by Raman spectroscopy. Theor Chem Accounts 130:1239–1247
Austin LA, Osseiran S, Evans CL (2016) Raman technologies in cancer diagnostics. Analyst 141:476–503
Wang W, Zhao J, Short M, Zeng H (2015) Real-time in vivo cancer diagnosis using raman spectroscopy. J Biophotonics 8(7):527–545
Lewis IR, Edwards H (2001) Handbook of Raman spectroscopy: from the research laboratory to the process line. CRC Press
Krishna H, Majumder SK, Gupta PK (2012) Range-independent background subtraction algorithm for recovery of Raman spectra of biological tissue. J Raman Spectrosc 43:1884–1894
Dehghani-Bidgoli Z, Baygi MHM, Kabir E, Malekfar R (2014) A comparative study between carcinoma and sarcoma using Raman spectroscopy. J Appl Spectrosc 80:893–898
Bergholt MS et al (2011) In vivo diagnosis of gastric cancer using Raman endoscopy and ant colony optimization techniques. Int J Cancer 128:2673–2680
Elbeltagi E, Hegazy T, Grierson D (2005) Comparison among five evolutionary-based optimization algorithms. Adv Eng Inform 19(1):43–53
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All the procedures performed in the study involving human participants were in accordance with the ethical standards of the Islamic Azad University Research Committee as well as with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all the participants included in the study.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Fallahzadeh, O., Dehghani-Bidgoli, Z. & Assarian, M. Raman spectral feature selection using ant colony optimization for breast cancer diagnosis. Lasers Med Sci 33, 1799–1806 (2018). https://doi.org/10.1007/s10103-018-2544-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10103-018-2544-3