Abstract
Antimicrobial resistance (AMR) in Clostridioides difficile remains a significant threat to global healthcare systems, not just for the treatment of C. difficile infection (CDI), but as a reservoir of AMR genes that could be potentially transferred to other pathogens. The mechanisms of resistance for several antimicrobials such as metronidazole and MLSB-class agents are only beginning to be elucidated, and increasingly, there is evidence that previously unconsidered mechanisms such as plasmid-mediated resistance may play an important role in AMR in this bacterium. In this review, the genetics of AMR in C. difficile will be described, along with a discussion of the factors contributing to the difficulty in clearly determining the true burden of AMR in C. difficile and how it affects the treatment of CDI.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antimicrobial resistance (AMR) is a major threat to global public health that requires urgent action across all government sectors and society [1]. Clostridioides (Clostridium) difficile is a significant AMR pathogen, ranked by the US Centers for Disease Control and Prevention in 2013 as an urgent AMR threat costing the US healthcare system ~ USD1 billion annually [2]. Despite some improvements over the last decade, C. difficile remains an urgent threat, with 223,900 cases of C. difficile infection (CDI), and 12,800 deaths in the USA in 2017 [3].
One of the features of C. difficile that enables it to flourish when other bacteria have perished is its extensive repertoire of AMR, which includes resistance to lincomycin and clindamycin, aminoglycosides, tetracyclines, macrolides, cephalosporins, penicillins and fluoroquinolones [4]. The highest rates of resistance were to ciprofloxacin, levofloxacin, erythromycin and fusidic acid [5]. As with anaerobes in general, C. difficile typically has higher rates of resistance to older-generation antimicrobials than newer, e.g. ciprofloxacin (2nd generation, 99% resistant) vs moxifloxacin (4th generation, 36% resistance) [6]. Paradoxically, CDI is both induced by antimicrobials and then treated with antimicrobials: Overall exposure to antimicrobials was associated with a 60% increased risk of CDI [7]. Some antimicrobials are more commonly associated with inducing CDI than others, including cephalosporins, clindamycin, carbapenems, trimethoprim/sulphonamides, erythromycin, broad-spectrum β-lactams, ampicillin/amoxicillin and later generations of fluoroquinolones [4, 6,7,8,9,10].
In particular, antimicrobials that reduce the anaerobic microflora of the gut (e.g. Bacteroides spp. and Bifidobacterium spp.) while sparing certain facultative anaerobes (e.g. Enterococcus spp.) appear to be strong inducers of CDI [11]; 3rd-generation cephalosporins have the highest attributable risk, whereas clindamycin has the highest relative risk of inducing CDI [7]. Other antimicrobials such as tetracyclines and aminoglycosides are not strongly associated with the induction of CDI despite frequent resistance [7, 11]. The association of macrolides with CDI induction is unclear, with research both for [10] and against [7, 9] and their use in combination therapy potentially masking associations [9, 11]. Resistance mechanisms in C. difficile include chromosomal resistance genes, mobile genetic elements (MGEs), alterations to metabolic pathways and antimicrobial targets, and biofilms [6], with some strains showing multidrug resistance (MDR), defined as resistance to three or more antimicrobial agents (see Tables 1 and 2). In this review, the genetics of AMR in C. difficile will be described, along with a discussion of the factors contributing to the difficulty in clearly determining the true burden of AMR in C. difficile and what it means for CDI.
Resistance to antimicrobials used to treat CDI
Of particular concern is the development of resistance to recommended treatment drugs. Since the removal of metronidazole as a recommended 1st line treatment in North America and Europe, vancomycin and fidaxomicin are now recommended for both mild and severe cases of CDI [12, 13] and, while rare, resistance or treatment failure has been reported for both [5]. In recent years, increasing rates of treatment failure, reported inferior performance compared to vancomycin and reports of neurotoxicity have relegated metronidazole to use only when vancomycin and fidaxomicin are not available, and then only for single courses [13]. However, metronidazole remains recommended in Australasia [14] and by the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) for mild/moderate cases [15]. Rifaximin (a rifamycin) has proven efficacy and reduces recurrence [8] but is vulnerable to the rapid development of resistance, while a novel class antimicrobial, ridinilazole, is showing promising performance in ongoing phase III trials [16].
Vancomycin
Vancomycin is a glycopeptide antimicrobial that inhibits cell wall synthesis in Gram-positive bacteria by inhibiting peptidoglycan biosynthesis. Minimum inhibitory concentration (MIC) breakpoints vary from 2 mg/L (EUCAST) to 4 mg/L (CLSI) [17, 18]. A common vancomycin resistance mechanism in many bacteria is van gene clusters which alter the D-Ala-D-Ala peptidoglycan terminus and reduce its affinity for vancomycin [19]. Despite the presence of certain van genes and orthologs, mechanisms of vancomycin resistance in C. difficile remain poorly understood. The vanG gene cluster found in ~ 85% of C. difficile is related to the van operon that results in vancomycin resistance when induced in Enterococcus faecalis [20], but induction with vancomycin does not appear to confer resistance in C. difficile [11, 19]. However, a recent study demonstrated mutations in the VanS/R sensor kinase/response regulator triggered constitutive expression of the previously silent VanG, conferring vancomycin resistance in C. difficile ribotype (RT) 027 strains [21]. Several transposon (Tn) elements, e.g. Tn1549, have been identified in C. difficile, but unlike the original E. faecalis elements, these transposons lack a functional vanB [6]. An overview of AMR transposons in C. difficile is provided in Table 2.
A susceptible isolate of C. difficile carrying a phenotypically silent vanB homologue has been identified, suggesting the potential for transfer of other vancomycin resistance genes [21]. The vanB operon was located on a ~ 42 kb Tn916-like conjugative Tn with significant similarity to Tn1549 from vancomycin-resistant enterococci (VRE) [22]. The vanRB gene, known to be involved in regulating the expression of vanB, was fragmented, and no in vitro effect on vancomycin resistance was observed [22]. The vanZ ortholog CD1240 conferred low-level teicoplanin resistance, but not vancomycin resistance, and was not induced by glycopeptide antimicrobials, rather by host peptide LL-37 [19]. Teicoplanin is not routinely recommended, nor is it approved, for use in the USA; however, it is used in Europe and appears to result in lower recurrence rates than vancomycin [19]. Resistant isolates with mutations in other gene families have been found, including murG, a peptidoglycan glycosyltransferase involved in cell wall maturation [6, 21, 23]. Biofilm may also play a role, as vancomycin induces biofilm formation and biofilms are resistant to high concentrations of vancomycin [6], up to a 12-fold increase in MIC over planktonic cells [23].
Plasmids may have a role in vancomycin resistance; the acquisition of plasmid pX18-498 reduced susceptibility to vancomycin eightfold in C. difficile [17]. In vivo testing in mice showed decreased susceptibility to vancomycin and increased disease severity [17]. The plasmid may have a role in cell wall integrity, as cells containing the plasmid did not rupture in the presence of vancomycin while plasmid-lacking cells did. Genetic analysis identified a differentially expressed amidase gene of the family responsible for susceptibility to cell wall targeting antimicrobials in Mycobacterium tuberculosis [17].
Vancomycin achieves high concentrations in faeces; however, this does not appear to correlate to concentrations in mucosa nor clinical outcomes [5, 17], despite the notion that the high faecal concentration may overcome increased MICs [6]. These high concentrations may exacerbate negative outcomes as examples of the Eagle effect, where higher drug concentrations kill fewer bacteria, have been found for vancomycin against C. difficile. Vancomycin concentrations of up to 2048 mg/L were survivable in vitro by four strains of C. difficile in one study by Jarrad et al., despite a bactericidal concentration of 4 mg/L [24]. Thus, high doses may disproportionately affect the normal gut microbiota without additional effect on C. difficile, promoting recurrence [24].
Reports have also found no link between clinical outcomes and vancomycin resistance [5], further clouding the causes of treatment failure. Reduced susceptibility to vancomycin has been found in several strains of C. difficile, RTs 027, 001, 017, 078 and 356/607 [6, 25]; however, the clinical implications of this are unclear. In one study, two isolates had reduced susceptibility to vancomycin, yet the patients responded well to vancomycin treatment [26]. In this case, reduced susceptibility was determined using the EUCAST breakpoint (> 2 mg/L), with the isolates having MICs of 3 mg/L, which would still be considered susceptible with CLSI breakpoints [26]. This highlights the need for the standardisation of methodology.
The highest rates of vancomycin resistance have been found in North and South America [8], followed by Asia, a finding mirrored by the high levels of glycopeptide antimicrobial use in the USA and China [5]. Vancomycin was ineffective against 1.4% of isolates in a European study of 2698 isolates [27]. In a comparison of historical (1992–2014) and modern (2015–2019) isolates, resistance rates did not show a statistically significant increase [8]. However, a meta-review by Saha et al. did report a statistically significant increase in vancomycin resistance comparing pre- to post-2012, which correlated to an increase in global vancomycin usage [5]. This increase was dependant on the method of testing used, as including only studies performed with E-tests nullified the increase [5]. Vancomycin resistance was significantly higher in hypervirulent strains such as C. difficile RT027 than in other RTs [5], while RT018 and RT356 had increased mean MICs of vancomycin [28].
Metronidazole
Metronidazole is a nitroimidazole that inhibits DNA synthesis after its partial reduction by some anaerobic bacteria results in cytotoxicity. While metronidazole has been used as a first-line treatment for CDI for many years, it is no longer recommended by the Infectious Disease Society of America (IDSA) due to inferior cure rates for both initial and recurrent CDI compared to vancomycin [13]. Repeated or prolonged metronidazole use has also been linked to neurotoxicity [13, 29]. In addition, breakpoints for resistance vary significantly (EUCAST, 2 mg/L; CLSI, 32 mg/L) [8].
Metronidazole resistance mechanisms in C. difficile are just beginning to be understood but are likely multifactorial processes involving alterations to metabolism such as with nitroreductases, iron uptake, DNA repair or biofilm formation [6]. In other species, metronidazole resistance mechanisms include reduced growth and uptake rates, efflux [29], nitroimidazole reductases (encoded by nim genes), altered pyruvate–ferredoxin oxidoreductases (pfo or PFOR) and stress pathways [30], or mutations in hemN and ferric uptake regulator (fur) genes [31].
Iron plays a role in many reduction pathways associated with metronidazole, and changes in iron metabolism/homeostasis appear to be involved in resistance. PFOR (with iron–sulphur clusters) and ferredoxin are involved in the activation of metronidazole through reduction, and in C. perfringens, PFOR mutants were up to 100-fold more resistant [29]. Moura et al. analysed the genome of a C. difficile strain with stable metronidazole resistance (MIC 32 mg/L) and found that a metabolic pathway associated with pfo was likely altered, as the resistant isolate had a lower, more stable PFOR concentration than susceptible controls [31]. Ferritin was absent from the metronidazole-resistant strain during antimicrobial exposure, suggesting a role for iron storage [31]. A separate proteomic analysis found a reduction in iron uptake/transport proteins after metronidazole treatment and increased expression of Fur protein (regardless of metronidazole presence) in a resistant strain [32]. The resistant strain in question demonstrated fitness defects and required the inclusion of iron in media for growth [32].
Deshpande et al. constructed a highly mutable strain via deletion of DNA mismatch repair genes to study the evolution of resistance in vitro, demonstrating that resistance to metronidazole developed progressively, with MICs increasing from 2 to 16 mg/L [33]. Mutations first appeared in feoB1 (the main iron transporter) and then pfo, followed by xdh (a putative xanthine dehydrogenase), and finally, some variants with greater resistance (MIC = 64 mg/L) also had mutations in iscR, an iron–sulphur cluster regulator. While changes in nifj, xdh and iscR genes amplified the MIC further, they did not confer resistance without feoB1 loss, while the loss of feob1 alone resulted in low-level resistance [33].
Differential expression and/or increased concentrations of DNA repair protein RecA were detected in resistant strains on exposure to metronidazole [31, 32]. In other species, mutants with DNA repair defects (such as in RecA) were more susceptible to metronidazole [29]. Differential expression of oxidative stress and general stress response proteins was also found in resistant strains [32]. Mutations in thiamine biosynthesis (thiH) and glycerol-3-oxidoreductase (glyC) genes have also been associated with resistance in C. difficile [29], as has a C-terminal deletion at position 372 in hsmA in RT010 isolates [34]. Unfortunately, the role these changes may play in resistance remains elusive.
Research into plasmid-mediated metronidazole resistance has been led by the work of Boekhoud et al. with their discovery of a high-copy number 7 kb plasmid (pCD-METRO) that conferred metronidazole resistance in C. difficile [30]. Figure 1 shows a representation of the pCD-METRO plasmid structure. This plasmid was found only in metronidazole-resistant strains (EUCAST breakpoint > 2 mg/L) and was internationally disseminated and present in both toxigenic and non-toxigenic strains. Surveillance in the Netherlands showed a prevalence of pCD-METRO of < 0.14%; however, the exact mechanism of resistance was unclear. The plasmid contained a small pseudogene with protein homology to nimB from Bacteroides fragilis, but this lacked the catalytic domain and induction via an inducible promoter did not confer resistance in the laboratory. A metallohydrase/oxidoreductase gene was also present but has not been further characterised. No reduction in growth rate was detected in the absence of metronidazole, and repeated passage on non-selective media failed to eliminate the plasmid from metronidazole-resistant strains. A guanine-cytosine content (GC%) of 41.6% did not match the C. difficile standard of ~ 28–30% and indicated acquisition via horizontal gene transfer from another unidentified organism [30].
Representation of the metronidazole-resistance plasmid pCD-METRO reported by Boekhoud et al. [30]. pCD-METRO plasmid with putative open reading frames (ORFs) shown in green on the outer circle and gene products listed beside. The inner-circle represents GC content, with above average in yellow, below average in purple
In isolates with the pCD-METRO plasmid, MIC values differed between RTs [30]. The plasmid was found in the initial stool sample along with a susceptible isolate, which differed by one core genome single-nucleotide polymorphism (cgSNP) to the resistant isolate found later, indicating the strains were clonal. This scenario was also identified later in a second case. Introduction of the pCD-METRO plasmid into a metronidazole-susceptible strain increased the MIC > 24-fold [30]. However, metronidazole resistance was still found in isolates lacking pCD-METRO [34], indicating alternate methods of resistance exist.
Metronidazole only reaches low concentrations in the colon and faeces [30], which may promote the development of resistance and play a role in treatment failures [6, 11], as Moura et al. demonstrated subinhibitory concentrations select and maintain isolates with increased MICs [4]. Heteroresistance may also play a role in treatment failure [35], as could other metronidazole-resistant microbiota in the gut by the inactivation or sequestration of metronidazole [30]. In many cases, metronidazole resistance did not correlate to treatment failure [5] which also occurred with metronidazole-susceptible isolates [30].
Susceptibility testing for metronidazole resistance is particularly inconsistent. Boekhoud et al. took 88 strains that two other clinical trials had characterised as having altered susceptibility and retested them using agar dilution and CLSI breakpoints/guidelines; they found that most strains were metronidazole-susceptible [30]. Inducible resistance subsequently lost during freeze–thaw cycles in handling was suggested as a possible explanation [30]. This is consistent with other reports that metronidazole resistance is lost after storage at low temperatures [32, 36, 37]. As samples are usually frozen for storage or transport before testing, metronidazole resistance may be higher than surveillance studies indicate [37].
Differences in media may also contribute to inconsistent findings, as two studies have demonstrated that heme appears to be essential for media-dependent metronidazole resistance. In one, strains classified as susceptible to metronidazole on brain–heart infusion (BHI) agar were resistant with higher MICs on Brucella blood agar (BBA) (measured by E-test) [30]. In a later study, MICs were increased up to 24-fold for strains grown on BBA/BHI blood agar and BHI agar supplemented with hemin [34]. Lynch et al. were able to generate a temperature-stable metronidazole resistant but reduced-fitness strain via a repeated passage on media with subinhibitory metronidazole concentrations [36]. This strain had mutations in the hemN (corporphyrinogm III oxidase) and thiH genes (involved in haem and thiamine biosynthesis, respectively), as well as in the fur gene [36]. Finally, testing methods also contribute to differences in characterisation, as the E-test may not distinguish isolates with reduced metronidazole susceptibility, with as many as 8% of metronidazole-resistant isolates misclassified [5, 38].
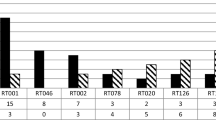
The highest rates of metronidazole resistance in C. difficile have been found in Asia, then North America [5] and Europe [8, 27]. Resistance remains rare but varied, with rates from 0 to 40% [28, 30], and seems more common in non-toxigenic strains, with 7–ninefold increased MIC values in non-toxigenic RT010 compared to toxigenic RT027, RT078 and RT001 [30]. One non-toxigenic clinical strain from China was reported with an MIC as high as 256 mg/L [4]. Saha et al. found a statistically significant decrease in resistance between pre-2012 to post-2012 studies, but these results were strongly impacted by testing methodology [5]. C. difficile RTs 027, 106, 001/072, 206, 010 and 356 strains had increased mean metronidazole MICs compared to other strains [6, 27, 28]. C. difficile RTs 176, 001, 126, 078 and 198 were also associated with reduced susceptibility to metronidazole [6, 26, 27]. Overall, mechanisms of metronidazole resistance remain unclear but increasingly appear to be multigenic with a role for iron metabolism.
Fidaxomicin
Fidaxomicin, a tiacumicin antimicrobial, inhibits RNA synthesis by interfering with the formation of the DNA–RNA polymerase complex before transcription can be initiated [39]. For C. difficile, it is a bactericidal and narrow spectrum, with minimal side effects, similar rates of initial cure as vancomycin and a significant reduction in recurrence [40]. However, Sears et al. found a mean faecal concentration of ~ 1396 mg/kg, and at such high concentrations, its impact on the gut microbiome may be broader than expected [41]. Similar to vancomycin, this high concentration is far above known MICs, and so future increases in MIC values may not result in reduced susceptibility in clinical settings [27].
Both fidaxomicin and rifamycins inhibit bacterial transcription, binding at adjacent but not overlapping target sites [4, 6]. Mutations in rpoB are responsible for resistance in both, but fortunately, they require separate mutations and are not cross-protective [6]. To examine the effect of rpoB mutations on fidaxomicin MICs, Kuehne et al. generated isogenic mutants, each with a single SNP in rpoB, resulting in the following changes: Val1143Asp, Val1143Gly and Val1143Phe [40]. These mutants had reduced susceptibility to fidaxomicin and reduced virulence and fitness (sporulation, growth and toxin production) in vitro [40]. This indicates that fidaxomicin-resistant mutations may be rarer due to negative effects on fitness, a finding supported by in vitro work demonstrating reduced susceptibility developed easily but was paired with severe fitness defects [42]. This study detected three resistance mutations in rpoB but also identified one isolate that showed a dramatically increased MIC of 64 mg/L with less severe reductions in toxin production and sporulation, and without the expected reduction in replication rate [42]. Mutation in a marR (repressor for a multiple AMR operon) homologue (CD22120) or rpoC (Asp273Tyr change) may also increase fidaxomicin MICs [11, 23].
Despite approval for fidaxomicin use for CDI in 2010, uptake has been limited [21], possibly due to its higher cost [43], and there are currently no breakpoints available [8]. Unlike rifamycins, fidaxomicin resistance is very rare [5, 8, 27, 28, 44].
Rifamycins
Rifamycins inhibit bacterial DNA-dependent RNA polymerase. The bacterial RNA polymerase RpoB, especially the β-subunit [6], is the primary site for resistance-conferring mutations [4]. These mutations may disrupt the direct interaction between target and antimicrobial or modify the rifamycin-binding pocket [11]. Figure 2 depicts the location of resistance-conferring amino acid substitutions in rpoB. Mutations in rpoB resulting in rifamycin resistance in C. difficile do not seem to confer a fitness cost, in contrast to species such as Neisseria meningitidis and M. tuberculosis [40]. Resistant strains may have MICs 1000-fold higher than susceptible strains [45], and there is a high agreement between phenotypic and genotypic resistance in rifaximin [46]. However, a US study found that epidemic C. difficile RT027 strains had unusual intermediate MICs to rifamycin, suggesting the potential for alternate resistance mechanisms [25]. The susceptibility of rifampin correlates with that of rifaximin, and thus, both are discussed in the following section.
Resistance-conferring mutations in RpoB. Known amino acid substitutions in RpoB that confer resistance to rifamycins or fidaxomicin. Mutation-rich regions have been enlarged with line breaks indicated by triple black lines. Substitutions are displayed in standard notation (single-letter amino acid codes and amino acid site; S = serine, T = threonine, D = aspartic acid, N = asparagine, V = valine, P = proline, H = histidine, Y = tyrosine, L = leucine, R = arginine, K = lysine, I = isoleucine, M = methionine, F = phenylalanine) [35, 42, 46, 47, 49, 123,124,125,126,127]
While effective at reducing recurrence, the rapid development of resistance makes rifamycin use risky. In one case, resistance developed in a C. difficile RT056 strain within 3 days of rifaximin therapy, with the MIC increasing from 0.002 to > 32 mg/L [47]. Rifamycins in long-term regimes are commonly used in therapy for tuberculosis, a leading cause of infectious disease-related deaths globally [44, 48]. This combination of long-term, frequent use and the single mutation required leaves rifamycins particularly vulnerable to the development of resistance. In a study of several Asia–Pacific countries, Lew et al. noted that rifaximin resistance was not found in C. difficile strains from countries with low tuberculosis rates (Singapore, Australia and Japan) [44].
A 2020 meta-analysis and a 2018 European study found 42.4% and ~ 15% rifampin resistance, respectively [6, 8]. A separate European study from 2015 found rifampin resistance in 17/22 countries (breakpoint 16 mg/L) [28]. Resistance was not uniform across a location or RT [6], e.g. rifampicin resistance was present in 62.4% of Italian isolates compared to 13.4% of the whole collection [28], and mutations in rpoB were strongly associated with high-risk strains, e.g. multi-locus sequence types (ST) 11 (e.g. RT078) and 37 (e.g. RT017) [49]. While both location and strain appear to be linked to AMR, they do not always agree; e.g. RT001/072 from the Czech Republic demonstrated rifampicin resistance, but RT001/072 from Germany, Latvia or Slovakia did not [28].
Ridinilazole
Ridinilazole is a narrow-spectrum, potent (C. difficile MIC90 0.2 mg/L) antimicrobial currently in phase III trials that has a novel but unclear mechanism of action [16]. High-resolution microscopy suggests an effect on cell division resulting in a filamentous cell [50]. Ridinilazole also reduces in vitro production of toxins A and B in C. difficile RT027 at both sub- and supra-MIC levels. It is poorly absorbed, and high concentrations can be found in stool. In vitro testing suggested that higher concentrations were required to kill gut microbiota than fidaxomicin, thus reducing gut dysbiosis [50]. In both animal models and human trials, ridinilazole demonstrated improved sustained cure rates over vancomycin without significant adverse effects [16, 51]. Sustained cure rates consider both initial cure and recurrence rates, and the superiority of ridinilazole over vancomycin was principally driven by a significant reduction in recurrences [51]. In a phase II trial, those patients who developed a recurrence all had strains that remained susceptible to ridinilazole, suggesting an alternate reason for treatment failure [50].
Treatments under development
Several potential treatments are currently under development but have not yet completed phase III trials, including ramizol, clofazimine, DS-2969b, CRS3123, ibezapolstat, DNV3837/DNV3681, MGB-BP-3, LFF571 and ramoplanin [16]. Some have similar mechanisms or targets to existing antimicrobials, e.g. DS-2969b which targets GyrB and thus may be vulnerable to amino acid substitutions as with the fluoroquinolones. Others, such as ibezapolstat that inhibits DNA polymerase IIIC, have novel mechanisms of action. Surotomycin, a potent bactericidal cyclic lipopeptide, completed phase III trials but did not meet its goals of non-inferiority to vancomycin and is no longer in development [16]. Studies determined resistance was likely to be extremely rare, involving proteins with potential roles in membrane structure or biosynthesis [45, 52]. Similarly, cadazolid, a novel oxazolidinone, did not progress after phase III trials [16].
Resistance to other antimicrobials
Fluoroquinolones
Fluoroquinolones are bactericidal antimicrobials that target the DNA replication enzymes DNA gyrase and type IV topoisomerase, inhibiting their ligase functions while continuing to allow them to generate double-stranded DNA breaks in the chromosome [53]. Newer generation fluoroquinolones such as moxifloxacin show increased activity against Gram-positive and anaerobic bacteria [53, 54]. However, moxifloxacin and levofloxacin may fail to reach inhibitory concentrations in the intestine in early treatment, and resistance develops frequently after exposure [6].
Resistance in C. difficile occurs via mutations that reduce binding affinity in the target site, the quinolone resistance-determining region (QRDR) of DNA gyrase subunits A and B [9, 55]. Figure 3 depicts the major GyrA and GyrB substitutions that confer fluoroquinolone resistance. While there are many amino acid substitutions responsible for resistance, Thr82Ile in GyrA is the most common [56]. In one study, 179/186 (96.2%) strains with reduced susceptibility had this alteration [26]. This mutation lacks a detectable fitness cost [57], and in at least one study, resistant strains exhibited increased fitness [26], suggesting it may be maintained in the population regardless of the presence of antimicrobials [6]. The prevalence of this mutation in particular may be explained by higher fitness costs and increased mutation requirements for other amino acid changes such as Thr82Val in GyrA [57]. Alterations in GyrB are less common than those in GyrA [57]. Different mutations provide different levels of resistance, with Thr82Ile associated with high-level resistance while others lead to lower MICs in comparison [57]. While QRDR mutations are the major mechanism, some resistance-conferring mutations have been found outside the QRDR and may be more common than previously thought [58]. Resistance to newer fluoroquinolones appears to be somewhat cross-protective, with one study finding all strains resistant to moxifloxacin were also resistant to gatifloxacin, levofloxacin and ciprofloxacin [35]. While there is generally high concordance between phenotypic and genotypic (gyrA and gyrB) resistance typing [46], one study has identified ciprofloxacin-resistant strains with no mutations in any part of the gyrA/gyrB regions [58], so other resistance mechanisms may yet be discovered.
Fluoroquinolone resistance mutations in GyrA and GyrB. Known amino acid substitutions in GyrA and GyrB conferring resistance to fluoroquinolones. Mutation-rich regions have been enlarged, with line breaks indicated by triple black lines. The quinolone resistance-determining region (QRDR) is identified by a yellow box, with extra-QRDR substitutions noted in blue. Substitutions are displayed in standard notation (single-letter amino acid codes and amino acid site; S = serine, A = alanine, R = arginine, G = glycine, D = aspartic acid, V = valine, N = asparagine, L = leucine, F = phenylalanine, K = lysine, E = glutamic acid, T = threonine, P = proline) [11, 35, 46, 49, 54, 55, 58, 70, 112,113,114,115,116, 128, 129]
Interestingly, antimicrobial adjuncts may improve the efficacy of some antimicrobials for CDI. A recent study by Pellissery et al. found that sub-MIC levels of sodium selenite significantly increased the susceptibility of C. difficile to ciprofloxacin in vitro while not affecting beneficial microbiota [59]. Such findings indicate that AMR may not condemn a drug to permanent disuse; instead, more research is needed on potential combinations.
Fluoroquinolone resistance is widely regarded as having played a significant role in the spread of hypervirulent strain C. difficile RT027 which caused substantial outbreaks across North America and Europe in the 2000s. In the 1990s and 2000s, North America had a heavy reliance on fluoroquinolones [60] such as levofloxacin which was recommended by IDSA/Thoracic Society for therapy for community-acquired pneumonia [61]. In contrast, Australia had strict limitations on fluoroquinolone use, in both humans and livestock, which contributed to the much-delayed importation and limited local spread of RT027 [62,63,64]. C. difficile RT027 was characterised by binary toxin production and fluoroquinolone resistance and responsible for dramatically increased mortality rates [65]. These epidemic strains of C. difficile RT027 had increased resistance to fluoroquinolones compared to pre-epidemic strains [6], with the prevalence of moxifloxacin resistance in Europe rising from 6.6% (1991–1997, before the introduction of moxifloxacin) to 39.9% (2011–2012) [54].
The prevalence of C. difficile RT027 has decreased significantly over the last decade, from 31% in 2011–2012 to 14% in 2015–2017 in the USA, resulting in a drop in moxifloxacin resistance rates [25]. While C. difficile RT027 is the best-known fluoroquinolone-resistant RT, resistance is found in many strains and appears to be becoming more common [6], with C. difficile RT001, RT017, ST35, ST3 and ST54 in particular with a high prevalence of up to 99% [8, 54, 66], although clonal outbreaks may significantly impact resistance rates. There were statistically significant differences in resistance rates across continents, with the highest rates found in South America, followed by Asia and North America [8].
Tetracyclines
Tetracyclines are broad-spectrum antimicrobials that target the 30S ribosome, preventing attachment of the aminoacyl-tRNA to inhibit protein synthesis [11]. Resistance in C. difficile involves protection of the ribosomes via elongation factor homologs encoded on various tet genes (e.g. tetM, tetW, tetA(P) and tetB(P)) carried by transposons (e.g. Tn916, Tn5397 and Tn4453) [9, 11, 54, 56, 67,68,69]. The tetracycline efflux pump gene tet40 has also been detected in C. difficile RT078 [69]. A study of UK, European and North American isolates found that tetM was the most common tetracycline resistance determinant [69]. Some strains have multiple tet genes, but it is unclear whether or not this affects the MIC [11]. Interestingly, isolates that were tetM positive but tetracycline susceptible were discovered in a study of hospital strains in China, although no mechanism was described [70].
Several tetracycline resistance determinants show evidence of movement between species due to their location on MGEs such as plasmids and transposons, and the widespread use of tetracyclines. Selection pressure from extensive tetracycline use in agriculture [8, 69], rather than human medicine, appears to be driving an expansion of tetracycline-resistant clones, particularly in RT078 [69]. C. difficile RT078, commonly found in livestock, has since spread to humans, with increasing numbers of cases (particularly community-acquired cases) first detected in 2005–2008 across Europe and America [69]. It has high rates of tetracycline resistance and frequently possesses tetracycline resistance determinants with high sequence identity to other zoonotic pathogens. Human and animal isolates of RT078 are highly genetically similar [69], and a large-scale phylogenetic study of isolates from 22 countries identified frequent zoonotic transmission of RT078 across geographic borders [71]. Whole-genome sequencing of isolates from humans and pigs has found genetically identical or closely related strains in both [71,72,73,74]. A 2019 study found close genetic links (in both sequence similarity and location) between tetP genes in Paeniclostridium sordellii and C. difficile, suggestive of horizontal gene transfer between clostridial species [75].
As with other antimicrobials, strain differences exist; increasing tetracycline resistance rates were found in C. difficile RT027 clinical strains [25], while C. difficile RT078 isolates contain more of the tetM determinant than other strains [69]. Location plays a significant role in resistance distribution even within single strains; higher rates of tetracycline resistance were detected in C. difficile RT017 isolates from China (82.4–85.7%) than from European countries (27.8%) [54]. The highest rates of tetracycline resistance were found in Oceania, Asia and Europe [8].
Several tetracyclines have been proposed as potential alternative CDI treatments. Tigecycline has been suggested for the treatment of severe CDI [4], as it is efficacious, inhibits spore formation and decreases toxin production; however, it reduces the healthy microbiota of the gut [16] and thus may contribute to CDI recurrence. A pan-European study of 2698 isolates in 2011–2014 found no resistance to tigecycline [27], but a more recent study characterising over 500 isolates from phase III surotomycin clinical trials found 2.5% were resistant [76]. Omadacycline, another tetracycline that has completed phase III clinical trials for various infections, demonstrated in vitro activity against C. difficile (with lower MICs than vancomycin) and does not induce CDI in in vitro models [77]; however, it also has a significant impact on the gut microbiota [16]. Further research in vivo is required, as well as a comprehensive study of the effect tetracycline resistance determinants may have on this drug. Eravacycline also shows activity against C. difficile, but once again is broad-spectrum and thus not ideal for treatment [16].
Macrolide-lincosamide-streptogramin B antimicrobials
Macrolide-lincosamide-streptogramin B (MLSB) antimicrobials (e.g. erythromycin and clindamycin) bind to the 50S subunit of bacterial ribosomes, preventing elongation of peptides and thus inhibiting protein synthesis. MLSB resistance in C. difficile is principally conferred by ribosomal methylation, but other mechanisms have been implicated and much is still unknown. The most common determinants of resistance are 23S ribosomal RNA methyltransferases encoded by erm genes, which are typically carried on transposons. The most prevalent erm gene is ermB, with two copies carried on a 9.6-kb conjugative transposon Tn5398 [6]. In one study in China, 89.3% of MLSB resistant isolates carried ermB [78]. Alternative genetic contexts for ermB have been discovered, including a 12.8-kb plasmid fragment [49], a truncated version of Tn5398 that does not confer resistance (ΔermB isolates) [79] and Tn6215 transduced by phage φC2 [80]. Other erm genes including ermG, ermQ, ermFS and ermC have also been detected on transposons including Tn6398, Tn6194, Tn6149 and Tn6215, some of which are transmissible across species boundaries [4, 9, 46, 55, 68, 81].
An as-yet unexplained but common phenomenon is the existence of C. difficile strains with MLSB resistant phenotype but lacking erm genes of any type, indicating the presence of other resistance mechanisms [6, 9, 11, 46, 67, 78]. Alterations to ribosomal proteins and rDNA are present in non-resistant isolates, and pump inhibitors such as reserpine had no effect despite suggestions of a role for the cme efflux pump [11], providing evidence against target alteration or efflux pump mechanisms [35, 55]. Several other putative mechanisms and genes of interest have been identified; erythromycin-resistant but clindamycin-susceptible strains have been identified with the msrA gene [49] which acts as a protein-oxidation repair enzyme in Escherichia coli, and one strain with high-level clindamycin and erythromycin resistance but lacking any known resistance genes encoded an rRNA methyltransferase enzyme identical to one found in Erysipelotrichaceae (accession AYM48329.1) [46].
A cluster of MLSB resistance-associated genes was found in MDR C. difficile isolates, encoding ermG, mefA and msrD (macrolide efflux) and vat (streptogramin A acetyltransferase) [81]. This cluster appeared to be on a putative mobile genetic element, exhibiting a mosaic structure and including a type 1 restriction-modification (RM) system, an intact prophage and an IS66 family transposase as well as the AMR determinants. The element was later detected in several other C. difficile genomes in GenBank [81].
The cfr and cfr-like 23S rRNA methyltransferases confer resistance to a large group of antimicrobials (including phenicols, lincosamides, oxazolidinones, pleuromutilins, streptogramin A and large macrolides) in several bacterial species [82, 83]. While several cfr genes have now been detected in C. difficile, often carried on MGEs [84], their contribution to resistance requires further research. Some variants of Tn6218 contain cfrB, suggested to be a mechanism of clindamycin and erythromycin resistance in C. difficile [6, 49]. The cfrC gene has also been found in clindamycin-resistant strains [49], while Stojkovic et al. identified a new cfr-like gene named cfrE in a clinical isolate from South America [84]. In vitro testing showed that both cfrC and cfrE had methylation activity in vitro in E. coli, but poor activity in C. difficile [84]. A study of an MDR (erythromycin, chloramphenicol, clindamycin and florfenicol, linezolid and streptogramin A) C. difficile isolate discovered a constitutively expressed cfrC gene which was later identified in search of several other C. difficile genomes [83]. The genomic context surrounding the gene indicated that C. difficile acquired cfrC some time ago and may be a reservoir for other bacteria [83]. Putative plasmids containing cfr genes have been detected in C. difficile; a putative plasmid containing cfrC was detected in silico (which has not yet been experimentally verified) [30], and a 6961 bp-sized plasmid named pCd13-Lar was identified carrying both the aphA3 aminoglycoside resistance gene and a cfrC gene 99% similar to one found in the pTx40 plasmid from Campylobacter coli [49]. All cfr genes were uncommon in C. difficile, with a ~ 4% (cfrC), 1.3% (cfrB) and 0.09% (cfrE) detection rate according to examination of protein sequences from > 2000 genomes [84].
Clindamycin use poses a high risk for inducing CDI [6], and it was the first antimicrobial to be recognised as such [54]. Indeed, the disease was initially called clindamycin-associated colitis. Resistance is most prevalent in Asia and South America [8] and very common in human isolates in Australia (84%) [85]. A meta-analysis found no significant [79] difference in resistance rates for clindamycin between historic and modern strains [8]; however, a study in Eastern China found a statistically significant increase in MICs for erythromycin and clindamycin between 2015 and 2020 [78], which were the most common resistances found in China along with ciprofloxacin [66].
Oxazolidinones
Oxazolidinones such as linezolid are broad-spectrum antimicrobials active against Gram-positive bacteria. They inhibit protein synthesis by binding to the 50S ribosomal subunit and preventing the formation of the initiation complex [83, 86]. Linezolid inhibits protein synthesis by targeting the 23S rRNA and inhibited C. difficile cytotoxin production in in vitro models [11]; however, it is not recommended for CDI treatment [84]. Linezolid causes gut dysbiosis due to its broad spectrum of activity [87], but this appears short-lived [88]. Reports have emerged of linezolid-induced C. difficile colitis, and there is a suggestion that faecal concentrations may not be sufficient due to renal metabolism [89]. The cfrC gene carried via a transposon and a mutation in the rplC (ribosomal protein 3) gene has been associated with resistance to linezolid [11, 54]. While C. difficile is generally susceptible to linezolid, a small number of resistant strains have been reported in Europe, with estimates of prevalence ranging from 1–5.7% [54].
Chloramphenicol
Chloramphenicol inhibits protein synthesis by binding to the 50S subunit of the ribosome and blocking the action of peptidyl transferase and thus peptide bond formation. It is active against a wide range of bacteria including Gram-positives and anaerobes. Resistance mechanisms focus on protecting the ribosomes via methylation or inactivating the antimicrobial via enzymes such as the chloramphenicol acetyltransferase encoded by the catD gene [55]. In C. difficile, catD has been found on transposons Tn4453a and Tn4453b [6]. Problems with toxicity to human cells limit the use of chloramphenicol for C. difficile, so research is rare. Resistance is uncommon [55] with < 5% of strains from prevalent RTs resistant in in vitro tests according to the ClosER study of 22 European countries [28].
Aminoglycosides
Aminoglycosides target the 16S rRNA aminoacyl-tRNA recognition site and inhibit protein synthesis. Intrinsic resistance in anaerobes means that the presence of aminoglycoside genes is likely a poor indicator of phenotype [81]. Despite intrinsic resistance, some strains of C. difficile also contain acquired aminoglycoside resistance genes, indicating transfer from other bacteria [46]. Resistance mechanisms mainly involve aminoglycoside-inactivating enzymes [90]. Multiple resistance genes have been identified such as npmA (a 16S rRNA methyltransferase), aph(3’)-III (an aminoglycoside phosphotransferase) and ant(6)-Ia (an aminoglycoside nucleotidyltransferase) [91], as well as a range of putative resistance determinants commonly found in other species, such as aadE (an aminoglycoside 6-adenylyltransferase) and aac(6’)-Ie-aph(2″)-Ia [81]. CLSI breakpoints are not defined for aminoglycosides in C. difficile [91].
Newer aminoglycosides such as amikacin, arbekacin and plazomicin are less vulnerable to some resistance mechanisms than the earlier gentamicin and tobramycin but are still affected by some mechanisms such as 16S-rRNA methyltransferases and certain aac and aph genes [90]. In C. difficile, resistance to apramycin, a newer aminoglycoside under development, is rare and associated with the npmA gene [90].
β-Lactams
β-Lactam antimicrobials, including penicillins, carbapenems and cephalosporins, feature a β-lactam ring and inactivate cell wall synthesis enzymes. C. difficile is intrinsically resistant to many β-lactams due to the presence of chromosomal class D β-lactamase genes (blaCDDs) which confer broad-spectrum resistance to β-lactam antimicrobials including penicillins, monobactams and all generations of cephalosporins [92, 93]. Unlike many β-lactamases, the blaCDD genes appear not to be mobile elements, as they are not surrounded by known MGE genes and are only distantly related to other β-lactamases [46] while maintaining high amino acid sequence identity with blaCDDs from other C. difficile strains [92]. They remain associated with the cell surface and are only active under reducing conditions [93]. The regulation of blaCDDs is debated, with Toth et al. finding blaCDD enzymes were poorly expressed [92], while Sandhu et al. found a high expression that was inducible and both dosage- and antimicrobial-dependent [93]. Regulation is controlled by blaR (sensor) and blaI (repressor), with disruption of blaI resulting in improved growth in ampicillin [93]. A co-transcribed lipoprotein, blaX, enhances blaCDD activity but is not essential [93].
β-Lactamase inhibitors in combination with antimicrobials (e.g. amoxicillin-clavulanate) were effective against C. difficile in at least one study, although these agents are not often included in AMR surveillance [54]. Carbapenems possess a unique structure that confers resistance to most β-lactamases [94]; they bind to PBPs 1, 2 and 3 [94]. C. difficile has generally been considered susceptible to carbapenems, but resistance has begun to be reported [54] primarily in RT017 [95], which have been found with imipenem-resistance mutations in the active site of penicillin-binding protein genes pbp1 and pbp3 [54, 81]. A fifth pbp5 on a mobile element was also found in resistant strains, but its effects are unclear [95].
Resistance to cephalosporins is high in C. difficile, and their widespread use is recognised as a high risk for inducing CDI [6], while penicillins and carbapenems have significantly lower MICs [92]. According to Toth et al., testing of several β-lactam antimicrobials (ampicillin, penicillin G, oxacillin, cephalothin, cefotaxime, ceftriaxone, cefoxitin, ceftazidime, cefepime, aztreonam, imipenem and meropenem) revealed that the highest MICs were to aztreonam, followed by ceftazidime, cefotaxime and cefoxitin, with the lowest MICs to meropenem, followed by ampicillin, penicillin G and imipenem [92].
Efflux mechanisms
Efflux pumps are classified into ATP-binding cassette (ABC) transporters or secondary multidrug transporters, which are further divided into resistance-nodulation-cell division (RND), small multidrug resistance, major facilitator superfamily (MFS) and multidrug and toxic compound extrusion (MATE) groups [23, 96]. Their role in C. difficile AMR has not been clearly described; however, C. difficile does possess efflux pump genes known to confer resistance when tested in other species; the cdeA gene encodes an MATE efflux mechanism that confers resistance to norfloxacin and ciprofloxacin in E. coli, while the cme MFS mechanism confers erythromycin resistance in E. faecalis [23, 97]. The cdeA transporter conferred resistance only under the control of the Plac promoter, not its natural promoter [98].
ABC transporters conferring AMR have been found in other Clostridium spp. as well, including cmpA in C. hathewayi and CPE1506 in C. perfringens [99]. A putative ABC transporter is also present in C. difficile; cd2068 expression is induced by antimicrobials (cloxacillin, ampicillin, cefoxitin, ciprofloxacin, gentamicin and levofloxacin) and confers increased resistance to these antimicrobials [99]. Interestingly, C. difficile has an efflux pump complex homologous to the TolC pump found in E. coli, in which it locates in the outer membrane and thus has not otherwise been found in Gram-positive species [96]. Mutants lacking this complex had their growth inhibited by tetracyclines and penicillins as well as acidic pH, fungicides and some inorganic and organic compounds not used against C. difficile [96].
Multidrug resistance
MDR in C. difficile, defined as resistance to three or more antimicrobial agents, is common and usually involves clindamycin, erythromycin, fluoroquinolones and cephalosporins [6, 44]. Like AMR rates, MDR rates vary by location and strain type. The majority of epidemic and emerging strains such as C. difficile RTs 176 and 356/607 are MDR [6]. In Europe, MDR estimates varied between 28 and 77%, with RTs 027, 017 and 198 resistant to five or more antimicrobials and some strains even resistant to nine antimicrobials [6, 27, 35]. Oddly, rifampicin resistance was always associated with moxifloxacin resistance in one study [35]. In the Asia–Pacific region, multidrug resistance was present in 100% of RT369, 92.7% of RT018 and 66.1% of RT017 [44]. Li et al. found MDR varied from 5.8% in Southwest China to 43.75% in South China and suggested increasing wealth correlated to increasing MDR [66], possibly due to the availability of more drug classes. MDR also remained common in US RT027 strains, with a significant increase in co-resistance to clindamycin, moxifloxacin, rifamycins and tetracyclines over time [25].
Difficulties in determining AMR in C. difficile
Characterising AMR in a strain of C. difficile can be surprisingly difficult. Heteroresistance (small numbers of resistant cells within a susceptible population) and reduced susceptibility can prevent accurate determination by standard susceptibility tests [4]. Treatment failure unrelated to known resistance mechanisms also occurs. While known mutations in gyrA, gyrB and rpoB were highly concordant with their respective resistance phenotypes [46], the same cannot be said for other resistance mechanisms, highlighting a key issue with modern methods of AMR determination in silico: While in silico methods can provide rapid, high-volume detection of many antimicrobial resistance determinants at once, genotype does not always correlate with phenotype. In a recent study of Australian C. difficile ST11 strains, in silico AMR screening matched well (100%) with agar dilution MIC results for fluoroquinolones and tetracycline, but only poorly for MLSBs (36%, screened using ermB), further indicating alternate mechanisms exist [100].
In silico methods also depend on comprehensive knowledge of genetic features to search for, and in many cases, this information is not yet known, or there is a delay between discovery and implementation in databases. Figure 4 provides an overview of potential resistance-conferring loci. While large AMR gene databases and search tools exist, there are several, each with differences, and they require a separate set of skills from in vitro testing. In vitro testing is not immune to these challenges either, with a variety of methods and standards in use making comparisons difficult. Antimicrobial susceptibility tests include the agar dilution assay (gold standard), E-tests (most commonly used [6, 8]) and broth microdilution [4]. The E-test is less laborious than agar dilution but can have discrepancies in MICs which are generally lower than those determined by agar dilution [4, 67]. Many methods are based on set ‘checkpoints’ of concentration or dilution, thus limiting exact measurements, and chemical properties, e.g. solubility, may limit testing of concentrations outside a set range [11]. Even only using the E-test, one study found clindamycin MICs changed around the resistance breakpoint depending on how MIC endpoints were read, either via the product insert or using methods described in the literature [25]. When MICs cluster around breakpoints (as is common with cephalosporins), small changes (e.g. changes in media or testing method) can have large consequences for perceived resistance rates [101].
Genomic loci suggested to play a role in AMR in C. difficile. Graphical representation of the C. difficile genome with loci that have been implicated as potentially playing a role in AMR indicated by coloured lines (red = metronidazole, blue = vancomycin, purple = fidaxomicin, teal = efflux, yellow = MLSB). These genes are listed beside with gene names and the suggested antimicrobial affected. GC content is indicated on the inner circle, with above average in yellow and below average in purple
Similar strains may also compromise testing; in one study, a newly discovered clindamycin-resistant strain RT591 was misidentified via Xpert C. difficile PCR (Cepheid) as RT027 due to similar genetic composition of the PaLoc and presence of cdtB [102]. C. difficile RT244 in an Australian outbreak was also misidentified as RT027 by the same test [103].
One of the most common issues for in vitro testing is the lack of a single standard, making comparisons between studies difficult. Various measures are reported such as weighted pooled resistance (WPR), MIC50 and MIC90, S/I/R classifications, cumulative mean resistance, high- or low-level resistance, or resistance prevalence. Clinical breakpoints characterise microbes as susceptible, intermediate or resistant depending on whether a level of antimicrobial activity is associated with a likelihood of treatment success or failure [104]. Breakpoints from CLSI, EUCAST and prior literature are used, with some taken from their use in other bacteria as breakpoints are not available [5]. Epidemiological cut-off values may also be used, characterising microbes as wild type (absence of acquired or mutational resistance mechanism) or non-wild type (presence of acquired or mutational resistance mechanism) [104].
EUCAST and CLSI guidelines are the most commonly used in determining the classification of bacteria as susceptible/intermediate/resistant [67]; however, the breakpoints of each are not equivalent. For metronidazole, for example, the CLSI breakpoint is ≥ 32 mg/L [105], while the EUCAST breakpoint is ≥ 2 mg/L [106]. One study comparing AMR in several Gram-negative bacteria found that resistance rates were dramatically different with different breakpoints, as much as 52.3% (EUCAST) vs 19.9% (CLSI) for amoxicillin-clavulanic acid [107]. Also, the CLSI breakpoint is based on serum rather than intestinal concentration, but ESCMID guidelines are based on intestinal activity [67]. Due to differences in classification schemes and disagreement between methods, some groups have suggested that the use of direct MIC data would better enable comparisons between studies [5].
On a broader scale, it can be difficult to definitively state levels of resistance of certain C. difficile populations. Saha et al. analysed 60 studies between 1982 and 2017, including data from 27 countries and over 8000 isolates [5], and found the majority of reports were from Europe or North America. Unfortunately, this is a consistent issue with limited publications from many regions, e.g. Africa and many parts of Asia. Antimicrobial susceptibility testing is not routinely performed for C. difficile in most places [30], and diagnostic assays for CDI differ between locations, affecting the recruitment of cases/strains for study [44]. The quality of studies also has a statistically significant impact on AMR, with vancomycin resistance threefold higher in studies rated low quality compared to those rated high quality [8].
Adding to these challenges is the sheer diversity in this species. C. difficile has one of the lowest gene conservation rates of all bacterial species, with only an estimated 16–32% of genes conserved [108, 109] out of a large, open pangenome of over 10,000 genes [100, 108, 109]. Moreover, C. difficile is taxonomically incoherent, displaying genomic characteristics representative of distinct species divided along with the major evolutionary clades [110]. It also has a highly mobile genome with a demonstrated capacity to exchange genes with other species [46, 108], enhanced by the ability of C. difficile to persist as spores, and its niche within environments considered hotspots for genetic exchange (such as the human gut, soil and sewage) [49, 62]. In Southeast Asia, more than half the non-toxigenic strains studied carried mobile elements featuring AMR genes, many of which were similar to those found in other bacterial species of the porcine gut, such as E. rhusiopathiae and S. suis [46]. AMR genes included tetM, ermB, tetO, tetB, catP, the ant(6)-iae aph(30)-IIIl sat4A cluster and aac(60)-Ie-aph(200)-Ia [46]. While non-toxigenic strains have been suggested as an option for reducing CDI due to the protective effect they have against toxigenic strains [111], their ability to gain or pass on resistance genes to other bacteria remains a risk.
Epidemic strains are typically more resistant than non-epidemic counterparts [62, 67], with a lower diversity of RTs also correlated with increased AMR [28]. C. difficile RT017 (common in Asia) was reported to have greater resistance to several antimicrobials [44] as well as higher rates of MDR. Analysis of isolates from the Asia–Pacific region found that RT014/020 and RT002 (both common in Australia) to have lower rates of resistance [44, 54]. C. difficile RTs 027, 001/072, 017, 018 and 356 were associated with resistance in Europe [28]. Surviving antimicrobial treatment may not be the only benefit to resistant strains, with Baines and Wilcox suggesting strains with reduced susceptibility hold a competitive advantage as they would be able to recolonise the gut sooner after antimicrobial treatment than other strains [11].
Several analyses showed that rates of AMR varied significantly not just with strain but with location (country and continent), wealth and patient age (with AMR increasing to a peak in the 65–85 year age group) [8, 44, 66, 67]. As the evolution of resistance is largely driven by selective pressure, differences in population dynamics and antimicrobial use policies explain much of this finding [9, 28]. An excellent example of this is the rapid expansion of C. difficile RT027 across North America where fluoroquinolone usage was high [9], compared to the scarcity and much-delayed development of the same strain in Australia, where fluoroquinolones were strictly limited [62], although the lack of a local reservoir likely contributed to the failure of RT027 to establish in Australia.
Economic factors may also play a part. The association with the gross domestic product (inversely correlated to combined AMR) [44] may be due to a host of factors such as less stringent antimicrobial stewardship, poorer healthcare infrastructure and increased rates of other pathogens requiring antimicrobial treatment. Many Asian countries have some of the world’s highest antimicrobial consumption rates, with availability often not requiring prescriptions nor appropriate diagnosis [44]. Australia, in contrast, with higher wealth and strict antimicrobial stewardship policies, had the lowest mean cumulative resistance score (2.58) of 12 Asia–Pacific countries [44].
A solution to these issues is unlikely to be easily found. In silico testing appears promising with its ability to avoid much of the methodological variance of in vitro testing and the increasing use of genome sequencing. Further research to identify currently unknown genetic determinants, as well as a greater scope of gene search databases to include potential AMR acquisition from other species, would greatly improve the predictive value of in silico testing.
Barring the development of a quick, easy, cheap and effective testing method suitable for both clinical use and the research lab, there are simply too many factors at play to easily resolve the variance in physical testing around the world. Perhaps a more achievable solution would be to improve data sharing and understanding of these factors so that they may be taken into account when determining resistance. Large databases are already widely used for a variety of purposes, such as the ENA, GenBank or CARD, and enable large-scale analyses by pooling research from around the globe. Integration of susceptibility testing data, along with methods used, into a centralised database may enable a greater understanding of the factors influencing the determination of resistance. Such a database would ideally allow for filtering and comparison of methodology factors (such as test used, media, storage and breakpoints) to account for their effects. This would enable comparisons of resistance between datasets using different methods without requiring global standardisation of laboratories with different needs and resources.
Concluding remarks
AMR in C. difficile remains a serious threat and burden to healthcare systems, with many mechanisms of resistance still poorly understood and MDR widespread. Treatment options are narrowing, with both metronidazole and vancomycin seeing increasing treatment failures with no clear answer, while rifaximin is especially vulnerable to the rapid development of resistance. Evidence is increasingly pointing towards a diverse array of environmental and animal reservoirs of C. difficile with the transfer of AMR genes to and from other species. Resistance-conferring plasmids have now been identified, while previously, they were assumed to play no role in C. difficile AMR.
With high diversity, a mobile resistome and new resistance mechanisms being discovered, ongoing genomic-focused public health surveillance of C. difficile is critical for understanding and combating CDI. Yet, determining AMR in C. difficile remains challenging for a variety of genetic, epidemiological and practical reasons. Continuing AMR research with a One Health perspective, as well as improvements to collaborative data collection, distribution and use, will be key to combating this pathogen in the future.
Data availability
Not applicable.
Code availability
Not applicable.
References
O’Neill J (2016) Tackling drug-resistant infections globally: final report and recommendations. The review on antimicrobial resistance. Web citation: https://amr-review.org/. Accessed 20 Apr 2021
Centers for Disease Control and Prevention (2013) CDC antibiotic resistance threats in the United States, 2013. Web citation: http://www.cdc.gov/drugresistance/threat-report-2013/. Accessed 20 Apr 2021
Centers for Disease Control and Prevention (2019) CDC antibiotic resistance threats in the United States, 2019. Web citation: https://www.cdc.gov/drugresistance/biggest-threats.html. Accessed 20 Apr 2021
Peng Z, Jin D, Kim HB, Stratton CW, Wu B, Tang YW, Sun X (2017) Update on antimicrobial resistance in Clostridium difficile: resistance mechanisms and antimicrobial susceptibility testing. J Clin Microbiol 55(7):1998–2008
Saha S, Kapoor S, Tariq R, Schuetz AN, Tosh PK, Pardi DS, S K, (2019) Increasing antibiotic resistance in Clostridioides difficile: a systematic review and meta-analysis. Anaerobe 58:35–46
Spigaglia P, Mastrantonio P, Barbanti F (2018) Antibiotic resistances of Clostridium difficile. Springer International Publishing, Cham, pp 137–159
Slimings C, Riley TV (2014) Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother 69(4):881–891
Sholeh M, Krutova M, Forouzesh M, Mironov S, Sadeghifard N, Molaeipour L, Maleki A, Kouhsari E (2020) Antimicrobial resistance in Clostridioides (Clostridium) difficile derived from humans: a systematic review and meta-analysis. Antimicrob Resist Infect Control 9(1):158
Hong S, Knight DR, Riley TV (2019) The impact of antimicrobial resistance on induction, transmission and treatment of Clostridium difficile infection. Microbiol Aust 40(2):77–81
Deshpande A, Pasupuleti V, Thota P, Pant C, Rolston DDK, Sferra TJ, Hernandez AV, Donskey CJ (2013) Community-associated Clostridium difficile infection and antibiotics: a meta-analysis. J Antimicrob Chemother 68(9):1951–1961
Baines SD, Wilcox MH (2015) Antimicrobial resistance and reduced susceptibility in Clostridium difficile: potential consequences for induction, treatment, and recurrence of C. difficile infection. Antibiotics (Basel) 4(3):267–298
Rao K, Malani PN (2020) Diagnosis and treatment of Clostridioides (Clostridium) difficile infection in adults in 2020. J Am Med Assoc 323(14):1403–1404
McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH (2018) Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66(7):e1–e48
Trubiano JA, Cheng AC, Korman TM, Roder C, Campbell A, May MLA, Blyth CC, Ferguson JK, Blackmore TK, Riley TV, Athan E (2016) Australasian Society of Infectious Diseases updated guidelines for the management of Clostridium difficile infection in adults and children in Australia and New Zealand. J Intern Med 46(4):479–493
Debast SB, Bauer MP, Kuijper EJ (2014) European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect 20:1–26
Carlson TJ, Gonzales-Luna AJ (2020) Antibiotic treatment pipeline for Clostridioides difficile infection (CDI): a wide array of narrow-spectrum agents. Curr Infect Dis Rep 22(8):20
Pu M, Cho JM, Cunningham SA, Behera G, Becker S, Amjad T, Greenwood-Quaintance KE, Mendes-Soares H, Jones-Hall Y, Jeraldo PR, Chen J, Dunny G, Patel R, Kashyap PC (2020) Plasmid acquisition alters vancomycin susceptibility in Clostridioides difficile. Gastroenterology 160(3):941–945
The European Committee on Antimicrobial Susceptibility Testing (2021) Breakpoint tables for interpretation of MICs and zone diameters. Web citation: https://eucast.org/clinical_breakpoints/?debug=1. Accessed 02 Mar 2021
Woods E, Wetzel D, Mukerjee M, McBride S (2018) Examination of the Clostridioides (Clostridium) difficile VanZ ortholog, CD1240. Anaerobe 53:108–115
Ammam F, Marvaud J, Lambert T (2012) Distribution of the vanG-like gene cluster in Clostridium difficile clinical isolates. Can J Microbiol 58:547–551
Shen WJ, Deshpande A, Hevener KE, Endres BT, Garey KW, Palmer KL, Hurdle JG (2020) Constitutive expression of the cryptic vanGCd operon promotes vancomycin resistance in Clostridioides difficile clinical isolates. J Antimicrob Chemother 75(4):859–867
Knight DR, Androga GO, Ballard SA, Howden BP, Riley TV (2016) A phenotypically silent vanB2 operon carried on a Tn1549-like element in Clostridium difficile. mSphere 1(4):e00177-00116
Harnvoravongchai P, Pipatthana M, Chankhamhaengdecha S, Janvilisri T (2017) Insights into drug resistance mechanisms in Clostridium difficile. Essays Biochem 61(1):81–88
Jarrad AM, Blaskovich MAT, Prasetyoputri A, Karoli T, Hansford KA, Cooper MA (2018) Detection and investigation of eagle effect resistance to vancomycin in Clostridium difficile with an ATP-bioluminescence assay. Front Microbiol 9:1420
Tickler IA, Obradovich AE, Goering RV, Fang FC, Tenover FC, Consortium TH, Consortium HAI (2019) Changes in molecular epidemiology and antimicrobial resistance profiles of Clostridioides (Clostridium) difficile strains in the United States between 2011 and 2017. Anaerobe 60:102050
Krutova M, Capek V, Nycova E, Vojackova S, Balejova M, Geigerova L, Tejkalova R, Havlinova L, Vagnerova I, Cermak P, Ryskova L, Jezek P, Zamazalova D, Vesela D, Kucharova A, Nemcova D, Curdova M, Nyc O, Drevinek P (2020) The association of a reduced susceptibility to moxifloxacin in causative Clostridium (Clostridioides) difficile strain with the clinical outcome of patients. Antimicrob Resist Infect Control 9(1):98
Freeman J, Vernon J, Pilling S, Morris K, Nicholson S, Shearman S, Longshaw C, Wilcox MH (2018) The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011–2014. Clin Microbiol Infect 24(7):724–731
Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, Wilcox MH (2015) Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect 21(3):248.e249-248.e216
Dingsdag SA, Hunter N (2017) Metronidazole: an update on metabolism, structure–cytotoxicity and resistance mechanisms. J Antimicrob Chemother 73(2):265–279
Boekhoud IM, Hornung BVH, Sevilla E, Harmanus C, Bos-Sanders I, Terveer EM, Bolea R, Corver J, Kuijper EJ, Smits WK (2020) Plasmid-mediated metronidazole resistance in Clostridioides difficile. Nat Commun 11(1):598
Moura I, Monot M, Tani C, Spigaglia P, Barbanti F, Norais N, Dupuy B, Bouza E, Mastrantonio P (2014) Multidisciplinary analysis of a nontoxigenic Clostridium difficile strain with stable resistance to metronidazole. Antimicrob Agents Chemother 58(8):4957–4960
Chong PM, Lynch T, McCorrister S, Kibsey P, Miller M, Gravel D, Westmacott GR, Mulvey MR, the Canadian Nosocomial Infection Surveillance P (2014) Proteomic analysis of a NAP1 Clostridium difficile clinical isolate resistant to metronidazole. Plos One 9(1):e82622
Deshpande A, Wu X, Huo W, Palmer KL, Hurdle JG (2020) Chromosomal resistance to metronidazole in Clostridioides difficile can be mediated by epistasis between iron homeostasis and oxidoreductases. Antimicrob Agents Chemother 64(8):e00415-00420
Boekhoud IM, Sidorov I, Nooij S, Harmanus C, Bos-Sanders IMJG, Viprey V, Spittal W, Clark E, Davies K, Freeman J, Kuijper EJ, Smits WK, Consortium C-C (2021) Haem is crucial for medium-dependent metronidazole resistance in clinical isolates of Clostridioides difficile. J Antimicrob Chemother:dkab097 76(7):1731–1740. https://doi.org/10.1093/jac/dkab097
Spigaglia P, Barbanti F, Mastrantonio P, on behalf of the European Study Group on Clostridium difficile, Ackermann G, Balmelli C, Barbut F, Bouza E, Brazier J, Delmée M, Drudy D, Kuijper E, Ladas H, Mastrantonio P, Nagy E, Pituch H, Poxton I, Rupnik M, Wullt M, Yücesoy M (2011) Multidrug resistance in European Clostridium difficile clinical isolates. J Antimicrob Chemother 66(10):2227–2234
Lynch T, Chong P, Zhang J, Hizon R, Du T, Graham MR, Beniac DR, Booth TF, Kibsey P, Miller M, Gravel D, Mulvey MR, Canadian Nosocomial Infection Surveillance Program (2013) Characterization of a stable, metronidazole-resistant Clostridium difficile clinical isolate. PLoS One 8(1):e53757
Peláez T, Cercenado E, Alcalá L, Marín M, Martín-López A, Martínez-Alarcón J, Catalán P, Sánchez-Somolinos M, Bouza E (2008) Metronidazole resistance in Clostridium difficile is heterogeneous. J Clin Microbiol 46(9):3028–3032
Krutova A, Kinross P, Barbut F, Hajdu A, Wilcox MH, Kuijper EJ (2017) How to: surveillance of Clostridium difficile infections. Clin Microbiol Infect 24:469–475
Artsimovitch I, Seddon J, Sears P (2012) Fidaxomicin is an inhibitor of the initiation of bacterial RNA synthesis. Clin Infect Dis 55(Suppl 2):S127–S131
Kuehne SA, Dempster AW, Collery MM, Joshi N, Jowett J, Kelly ML, Cave R, Longshaw CM, Minton NP (2018) Characterization of the impact of rpoB mutations on the in vitro and in vivo competitive fitness of Clostridium difficile and susceptibility to fidaxomicin. J Antimicrob Chemother 73(4):973–980
Sears P, Crook DW, Louie TJ, Miller MA, Weiss K (2012) Fidaxomicin attains high fecal concentrations with minimal plasma concentrations following oral administration in patients with Clostridium difficile infection. Clin Infect Dis 55:S116–S120
Schwanbeck J, Riedel T, Laukien F, Schober I, Oehmig I, Zimmermann O, Overmann J, Groß U, Zautner AE, Bohne W (2019) Characterization of a clinical Clostridioides difficile isolate with markedly reduced fidaxomicin susceptibility and a V1143D mutation in rpoB. J Antimicrob Chemother 74(1):6–10
Nelson RL, Suda KJ, Evans CT (2017) Antibiotic treatment for Clostridium difficile‐associated diarrhoea in adults. Cochrane Database Syst Rev 3(3):CD004610
Lew T, Putsathit P, Sohn KM, Wu Y, Ouchi K, Ishii Y, Tateda K, Riley TV, Collins DA (2020) Antimicrobial susceptibilities of Clostridium difficile isolates from 12 Asia-Pacific countries in 2014 and 2015. Antimicrob Agents Chemother 64(7):e00296-e220
Mascio CT, Chesnel L, Thorne G, Silverman JA (2014) Surotomycin demonstrates low in vitro frequency of resistance and rapid bactericidal activity in Clostridium difficile, Enterococcus faecalis, and Enterococcus faecium. Antimicrob Agents Chemother 58(7):3976–3982
Imwattana K, Kiratisin P, Riley TV, Knight DR (2020) Genomic basis of antimicrobial resistance in non-toxigenic Clostridium difficile in Southeast Asia. Anaerobe 66:102290
Carman RJ, Boone JH, Grover H, Wickham KN, Chen L (2012) In vivo selection of rifamycin-resistant Clostridium difficile during rifaximin therapy. Antimicrob Agents Chemother 56(11):6019–6020
World Health Organisation (2020) Global Tuberculosis Report 2020. Web citation: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosisreport-2020. Accessed 10 May 2021
Chatedaki C, Voulgaridi I, Kachrimanidou M, Hrabak J, Papagiannitsis CC, Petinaki E (2019) Antimicrobial susceptibility and mechanisms of resistance of Greek Clostridium difficile clinical isolates. J Glob Antimicrob Resist 16:53–58
Carlson TJ, Endres BT, Bassères E, Gonzales-Luna AJ, Garey KW (2019) Ridinilazole for the treatment of Clostridioides difficile infection. Expert Opin Investig Drugs 28(4):303–310
Vickers RJ, Tillotson GS, Nathan R, Hazan S, Pullman J, Lucasti C, Deck K, Yacyshyn B, Maliakkal B, Pesant Y, Tejura B, Roblin D, Gerding DN, Wilcox MH, Dsg Co (2017) Efficacy and safety of ridinilazole compared with vancomycin for the treatment of Clostridium difficile infection: a phase 2, randomised, double-blind, active-controlled, non-inferiority study. Lancet Infect Dis 17(7):735–744
Adams HM, Li X, Mascio C, Chesnel L, Palmer KL (2015) Mutations associated with reduced surotomycin susceptibility in Clostridium difficile and Enterococcus species. Antimicrob Agents Chemother 59(7):4139–4147
Oliphant CM, Green GM (2002) Quinolones: a comprehensive review. Am Fam Physician 65(3):455–464
Imwattana K, Knight DR, Kullin B, Collins DA, Putsathit P, Kiratisin P, Riley TV (2020) Antimicrobial resistance in Clostridium difficile ribotype 017. Expert Rev Anti Infect Ther 18(1):17–25
Spigaglia P (2016) Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis 3(1):23–42
Mackin KE, Elliott B, Kotsanas D, Howden BP, Carter GP, Korman TM, Riley TV, Rood JI, Jenkin GA, Lyras D (2015) Molecular characterization and antimicrobial susceptibilities of Clostridium difficile clinical isolates from Victoria, Australia. Anaerobe 34:80–83
Wasels F, Kuehne S, Cartman S, Spigaglia P, Barbanti F, Minton N, Mastrantonio P (2014) Fluoroquinolone resistance does not impose a cost on the fitness of Clostridium difficile in vitro. Antimicrob Agents Chemother 59(3):1794–1796
Mac Aogáin M, Kilkenny S, Walsh C, Lindsay S, Moloney G, Morris T, Jones S, Rogers TR (2015) Identification of a novel mutation at the primary dimer interface of GyrA conferring fluoroquinolone resistance in Clostridium difficile. J Glob Antimicrob Resist 3(4):295–299
Pellissery AJ, Vinayamohan PG, Yin H-B, Mooyottu S, Venkitanarayanan K (2019) In vitro efficacy of sodium selenite in reducing toxin production, spore outgrowth and antibiotic resistance in hypervirulent Clostridium difficile. J Med Microbiol 68(7):1118–1128
Linder JA, Huang ES, Steinman MA, Gonzales R, Stafford RS (2005) Fluoroquinolone prescribing in the United States: 1995 to 2002. Am J Med 118(3):259–268
Niederman MS, Mandell LA, Anzueto A, Bass JB, Broughton WA, Campbell GD, Dean N, File T, Fine MJ, Gross PA, Martinez F, Marrie TJ, Plouffe JF, Ramirez J, Sarosi GA, Torres A, Wilson R, Yu VL (2001) Guidelines for the management of adults with community-acquired pneumonia. Am J Respir Crit Care Med 163(7):1730–1754
Knight DR, Riley TV (2016) Clostridium difficile clade 5 in Australia: antimicrobial susceptibility profiling of PCR ribotypes of human and animal origin. J Antimicrob Chemother 71(8):2213–2217
Knight DR, Giglio S, Huntington PG, Korman TM, Kotsanas D, Moore CV, Paterson DL, Prendergast L, Huber CA, Robson J, Waring L, Wehrhahn MC, Weldhagen GF, Wilson RM, Riley TV (2015) Surveillance for antimicrobial resistance in Australian isolates of Clostridium difficile, 2013–14. J Antimicrob Chemother 70(11):2992–2999
Australian Commission on Safety and Quality in Health Care (2018) Clostridium difficile infection. Monitoring the national burden of Clostridium difficile. Sydney. Web citation: https://www.safetyandquality.gov.au/publications-and-resources/resource-library/monitoringnational-burden-clostridium-difficileinfection. Accessed 21 Oct 2021
Alyousef AA (2018) Clostridium difficile: epidemiology, pathogenicity, and an update on the limitations of and challenges in its diagnosis. J AOAC Int 101(4):1119–1126
Li H, Li W-G, Zhang W-Z, Yu S-B, Liu Z-J, Zhang X, Wu Y, Lu J-X (2019) Antibiotic resistance of clinical isolates of Clostridioides difficile in China and its association with geographical regions and patient age. Anaerobe 60:102094
Banawas SS (2018) Clostridium difficile infections: a global overview of drug sensitivity and resistance mechanisms. Biomed Res Int 2018:8414257
Schmidt C, Loffler B, Ackermann G (2007) Antimicrobial phenotypes and molecular basis in clinical strains of Clostridium difficile. Diagn Microbiol Infect Dis 59(1):1–5
Dingle KE, Didelot X, Quan TP, Eyre DW, Stoesser N, Marwick CA, Coia J, Brown D, Buchanan S, Ijaz UZ, Goswami C, Douce G, Fawley WN, Wilcox MH, Peto TEA, Walker AS, Crook DW (2019) A role for tetracycline selection in recent evolution of agriculture-associated Clostridium difficile PCR ribotype 078. mBio 10(2):e02790-02718
Dong D, Zhang L, Chen X, Jiang C, Yu B, Wang X, Peng Y (2013) Antimicrobial susceptibility and resistance mechanisms of clinical Clostridium difficile from a Chinese tertiary hospital. Int J Antimicrob Agents 41(1):80–84
Knetsch CW, Kumar N, Forster SC, Connor TR, Browne HP, Harmanus C, Sanders IM, Harris SR, Turner L, Morris T, Perry M, Miyajima F, Roberts P, Pirmohamed M, Songer JG, Weese JS, Indra A, Corver J, Rupnik M, Wren BW, Riley TV, Kuijper EJ, Lawley TD (2018) Zoonotic transfer of Clostridium difficile harboring antimicrobial resistance between farm animals and humans. J Clin Micro 56(3):e01384-e1317
Knetsch CW, Connor TR, Mutreja A, van Dorp SM, Sanders IM, Browne HP, Harris D, Lipman L, Keessen EC, Corver J, Kuijper EJ, Lawley TD (2014) Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill 19(45):20954
Werner A, Mölling P, Fagerström A, Dyrkell F, Arnellos D, Johansson K, Sundqvist M, Norén T (2020) Whole genome sequencing of Clostridioides difficile PCR ribotype 046 suggests transmission between pigs and humans. PLoS One 15(12):e0244227
Knight DR, Squire MM, Collins DA, Riley TV (2017) Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front Microbiol 7:2138
Vidor CJ, Bulach D, Awad M, D L, (2019) Paeniclostridium sordellii and Clostridioides difficile encode similar and clinically relevant tetracycline resistance loci in diverse genomic locations. BMC Microbiol 19(1):53
Cheknis A, Devaris D, Chesnel L, Dale SE, Nary J, Sambol SP, Citron DM, Goering RV, Johnson S (2020) Characterization of Clostridioides difficile isolates recovered from two phase 3 surotomycin treatment trials by restriction endonuclease analysis, PCR ribotyping and antimicrobial susceptibilities. J Antimicrob Chemother 75(11):3120–3125
Begum K, Bassères E, Miranda J, Lancaster C, Gonzales-Luna AJ, Carlson TJ, Rashid T, Eyre DW, Wilcox MH, Alam MJ, Garey KW (2020) In vitro activity of omadacycline, a new tetracycline analog, and comparators against Clostridioides difficile. Antimicrob Agents Chemother 64(8):e00522-e520
Zhao L, Luo Y, Bian Q, Wang L, Ye J, Song X, Jiang J, Tang Y-W, Wang X, Jin D (2020) High-level resistance of toxigenic Clostridioides difficile genotype to macrolide-lincosamide-streptogramin B in community acquired patients in Eastern China. Infect Drug Resist 13:171–181
Ramírez-Hernández V, Ramírez-Vargas G (2020) A putative erm gene is present in ΔermB Clostridioides difficile isolates showing high levels of resistance to clindamycin. bioRxiv:2020.2011.2020.391128. https://doi.org/10.1101/2020.11.20.391128
Goh S, Hussain H, Chang BJ, Emmett W, Riley TV, Mullany P (2013) Phage ϕC2 mediates transduction of Tn6215, encoding erythromycin resistance, between strains. mBio 4(6):e00840-00813
Isidro J, Menezes J, Serrano M, Borges V, Paixão P, Mimoso M, Martins F, Toscano C, Santos A, Henriques AO, Oleastro M (2018) Genomic study of a Clostridium difficile multidrug resistant outbreak-related clone reveals novel determinants of resistance. Front Microbiol 9(2994):2994
Vester B (2018) The cfr and cfr-like multiple resistance genes. Res Microbiol 169(2):61–66
Candela T, Marvaud JC, Nguyen TK, Lambert T (2017) A cfr-like gene cfr(C) conferring linezolid resistance is common in Clostridium difficile. Int J Antimicrob Agents 50(3):496–500
Stojković V, Ulate MF, Hidalgo-Villeda F, Aguilar E, Monge-Cascante C, Pizarro-Guajardo M, Tsai K, Tzoc E, Camorlinga M, Paredes-Sabja D, Quesada-Gómez C, Fujimori DG, Rodríguez C (2019) cfr(B), cfr(C), and a new cfr-like gene, cfr(E), in Clostridium difficile strains recovered across Latin America. Antimicrob Agents Chemother 64(1):e01074-e1019
Lim S-C, Androga GO, Knight DR, Moono P, Foster NF, Riley TV (2018) Antimicrobial susceptibility of Clostridium difficile isolated from food and environmental sources in Western Australia. Int J Antimicrob Agents 52(3):411–415
Bozdogan B, Appelbaum PC (2004) Oxazolidinones: activity, mode of action, and mechanism of resistance. Int J Antimicrob Agents 23(2):113–119
von der Hartmut Lode N, Höh SZ, KB, Carl Erik Nord, (2001) Ecological effects of linezolid versus amoxicillin/clavulanic acid on the normal intestinal microflora. Scand J Infect Dis 33(12):899–903
Baines SD, Noel AR, Huscroft GS, Todhunter SL, O’Connor R, Hobbs JK, Freeman J, Lovering AM, Wilcox MH (2011) Evaluation of linezolid for the treatment of Clostridium difficile infection caused by epidemic strains using an in vitro human gut model. J Antimicrob Chemother 66(7):1537–1546
Zabel LT, Worm S (2005) Linezolid contributed to Clostridium difficile colitis with fatal outcome. Infection 33(3):155–157
Plattner M, Gysin M, Haldimann K, Becker K, Hobbie SN (2020) Epidemiologic, phenotypic, and structural characterization of aminoglycoside-resistance gene aac(3)-IV. Int J Mol Sci 21(17):6133
Marsh JW, Pacey MP, Ezeonwuka C, Ohm SL, Snyder D, Cooper VS, Harrison LH, Doi Y, Mustapha MM (2019) Clostridioides difficile: a potential source of npmA in the clinical environment. J Antimicrob Chemother 74 (2):521-523
Toth M, Stewart NK, Smith C, Vakulenko SB (2018) Intrinsic class D β-lactamases of Clostridium difficile. mBio 9(6):e01803-01818
Sandhu BK, Edwards AN, Anderson SE, Woods EC, McBride SM (2019) Regulation and anaerobic function of the Clostridioides difficile β-lactamase. Antimicrob Agents Chemother 64(1):e01496-e1419
Codjoe FS, Donkor ES (2017) Carbapenem resistance: a review. Med Sci (Basel, Switzerland) 6(1):1
Isidro J, Santos A, Nunes A, Borges V, Silva C, Vieira L, Mendes AL, Serrano M, Henriques AO, Gomes JP, Oleastro M (2018) Imipenem resistance in Clostridium difficile ribotype 017. Portugal Emerging infectious diseases 24(4):741–745
Lopez JME (2017) Characterization of an efflux pump system, the CD_AcrA-AcrB-TolC complex, in Clostridium difficile. Kansas State University, Department of Plant Pathology, p 66
Lebel S, Bouttier S, Lambert T (2004) The cme gene of Clostridium difficile confers multidrug resistance in Enterococcus faecalis. FEMS Microbiol Lett 238(1):93–100
Dridi L, Tankovic J, Petit JC (2004) CdeA of Clostridium difficile, a new multidrug efflux transporter of the MATE family. Microb Drug Resist 10(3):191–196
Ngernsombat C, Sreesai S, Harnvoravongchai P, Chankhamhaengdecha S, Janvilisri T (2017) CD2068 potentially mediates multidrug efflux in Clostridium difficile. Sci Rep 7(1):9982–9982
Knight DR, Kullin B, Androga GO, Barbut F, Eckert C, Johnson S, Spigaglia P, Tateda K, Tsai P-J, Riley TV (2019) Evolutionary and genomic insights into Clostridioides difficile sequence type 11: a diverse zoonotic and antimicrobial-resistant lineage of global one health importance. mBio 10(2):e00446-00419
Brook I, Wexler HM, Goldstein EJC (2013) Antianaerobic antimicrobials: spectrum and susceptibility testing. Clin Microbiol Rev 26(3):526–546
Skinner AM, Petrella L, Siddiqui F, Sambol SP, Gulvik CA, Gerding DN, Donskey CJ, Johnson S (2020) Unique clindamycin-resistant Clostridioides difficile strain related to fluoroquinolone-resistant epidemic BI/RT027 strain. Emerg Infect Dis 26(2):247–254
Lim SK, Stuart RL, Mackin KE, Carter GP, Kotsanas D, Francis MJ, Easton M, Dimovski K, Elliott B, Riley TV, Hogg G, Paul E, Korman TM, Seemann T, Stinear TP, Lyras D, Jenkin GA (2014) Emergence of a ribotype 244 strain of Clostridium difficile associated with severe disease and related to the epidemic ribotype 027 strain. Clin Infect Dis 58(12):1723–1730
The European Committee on Antimicrobial Susceptibility Testing (2021) EUCAST definitions of clinical breakpoints and epidemiological cut-off values, https://eucast.org/clinical_breakpoints/?debug=1. Accessed 18 Mar 2021
Clinical and Laboratory Standards Institute (CLSI) (2020) Performance standards for antimicrobial susceptibility testing. CLSI supplement M100. Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania 19087 USA (30th ed). https://standards.globalspec.com/std/14371884/CLSI%20M100
The European Committee on Antimicrobial Susceptibility Testing (2021) Breakpoint tables for interpretation of MICs and zone diameters Version: 11.0. http://www.eucast.org
Cusack TP, Ashley EA, Ling CL, Rattanavong S, Roberts T, Turner P, Wangrangsimakul T, Dance DAB (2019) Impact of CLSI and EUCAST breakpoint discrepancies on reporting of antimicrobial susceptibility and AMR surveillance. Clin Microbiol Infect 25(7):910–911
Elliott B, Androga GO, Knight DR, Riley TV (2017) Clostridium difficile infection: evolution, phylogeny and molecular epidemiology. Infect Genet Evol 49:1–11
Ferguson JK, Cheng AC, Gilbert GL, Gottlieb T, Korman T, McGregor A, Richards M, Roberts S, Robson J, Van Gessel H, Riley TV (2011) Clostridium difficile laboratory testing in Australia and New Zealand: national survey results and Australasian Society for Infectious Diseases recommendations for best practice. Pathology 43(5):482–487
Knight DR, Imwattana K, Kullin B, Guerrero-Araya E, Paredes-Sabja D, Didelot X, Dingle KE, Eyre DW, Rodríguez C, Riley TV (2020) The Clostridioides difficile species problem: global phylogenomic analysis uncovers three ancient, toxigenic, genomospecies. bioRxiv:2020.2009.2021.307223
Gerding DN, Sambol SP, Johnson S (2018) Non-toxigenic Clostridioides (formerly Clostridium) difficile for prevention of C. difficile infection: from bench to bedside back to bench and back to bedside. Front Microbiol 9:1700
Spigaglia P, Barbanti F, Mastrantonio P, Brazier JS, Barbut F, Delmée M, Kuijper E, I. RP, on behalf of the European Study Group on Clostridium difficile (2008) Fluoroquinolone resistance in Clostridium difficile isolates from a prospective study of C. difficile infections in Europe. J Med Microbiol 57 (6):784-789
Kuwata Y, Tanimoto S, Sawabe E, Shima M, Takahashi Y, Ushizawa H, Fujie T, Koike R, Tojo N, Kubota T, Saito R (2015) Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from a university teaching hospital in Japan. Eur J Clin Microbiol Infect Dis 34(4):763–772
Liao CH, Ko WC, Lu JJ, Hsueh PR (2012) Characterizations of clinical isolates of Clostridium difficile by toxin genotypes and by susceptibility to 12 antimicrobial agents, including fidaxomicin (OPT-80) and rifaximin: a multicenter study in Taiwan. Antimicrob Agents Chemother 56(7):3943–3949
Walkty A, Boyd DA, Gravel D, Hutchinson J, McGeer A, Moore D, Simor A, Suh K, Taylor G, Miller M, Mulvey MR (2010) Molecular characterization of moxifloxacin resistance from Canadian Clostridium difficile clinical isolates. Diagn Microbiol Infect Dis 66(4):419–424
Huang H, Weintraub A, Fang H, Nord CE (2009) Antimicrobial resistance in Clostridium difficile. Int J Antimicrob Agents 34(6):516–522
He M, Sebaihia M, Lawley TD, Stabler RA, Dawson LF, Martin MJ, Holt KE, Seth-Smith HM, Quail MA, Rance R, Brooks K, Churcher C, Harris D, Bentley SD, Burrows C, Clark L, Corton C, Murray V, Rose G, Thurston S, van Tonder A, Walker D, Wren BW, Dougan G, Parkhill J (2010) Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A 107(16):7527–7532
Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH, Peto TE, Walker AS, Riley TV, Crook DW, Didelot X (2014) Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol 6(1):36–52
Wang H, Roberts AP, Lyras D, Rood JI, Wilks M, Mullany P (2000) Characterization of the ends and target sites of the novel conjugative transposon Tn5397 from Clostridium difficile: excision and circularization is mediated by the large resolvase. TndX J Bacteriol 182(13):3775–3783
Roberts AP, Mullany P (2011) Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol Rev 35(5):856–871
Corver J, Bakker D, Brouwer MSM, Harmanus C, Hensgens MP, Roberts AP, Lipman LJA, Kuijper EJ, van Leeuwen HC (2012) Analysis of a Clostridium difficile PCR ribotype 078 100 kilobase island reveals the presence of a novel transposon, Tn6164. BMC Microbiol 12:130–130
Lyras D, Storie C, Huggins AS, Crellin PK, Bannam TL, Rood JI (1998) Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob Agents Chemother 42(7):1563–1567
O’Connor JR, Galang MA, Sambol SP, Hecht DW, Vedantam G, Gerding DN, Johnson S (2008) Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother 52(8):2813–2817
Pecavar V, Blaschitz M, Hufnagl P, Zeinzinger J, Fiedler A, Allerberger F, Maass M, Indra A (2012) High-resolution melting analysis of the single nucleotide polymorphism hot-spot region in the rpoB gene as an indicator of reduced susceptibility to rifaximin in Clostridium difficile. J Med Microbiol 61(Pt 6):780–785
Miller MA, Blanchette R, Spigaglia P, Barbanti F, Mastrantonio P (2011) Divergent rifamycin susceptibilities of Clostridium difficile strains in Canada and Italy and predictive accuracy of rifampin Etest for rifamycin resistance. J Clin Microbiol 49(12):4319–4321
Carman RJ, Genheimer CW, Rafii F, Park M, Hiltonsmith MF, Lyerly DM (2009) Diversity of moxifloxacin resistance during a nosocomial outbreak of a predominantly ribotype ARU 027 Clostridium difficile diarrhea. Anaerobe 15(6):244–248
Curry SR, Marsh JW, Shutt KA, Muto CA, O’Leary MM, Saul MI, Pasculle AW, Harrison LH (2009) High frequency of rifampin resistance identified in an epidemic Clostridium difficile clone from a large teaching hospital. Clin Infect Dis 48(4):425–429
Ackermann G, Tang YJ, Kueper R, Heisig P, Rodloff AC, Silva J Jr, Cohen SH (2001) Resistance to moxifloxacin in toxigenic Clostridium difficile isolates is associated with mutations in gyrA. Antimicrob Agents Chemother 45(8):2348–2353
Dridi L, Tankovic J, Burghoffer B, Barbut F, Petit JC (2002) gyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob Agents Chemother 46(11):3418–3421
Funding
K.O. is funded by an Australian Government Research Training Program Scholarship. D.R.K. is funded by a Fellowship from the National Health and Medical Research Council (APP1138257) and a grant from the Raine Medical Research Foundation (RPG002-19).
Author information
Authors and Affiliations
Contributions
Not applicable.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
O’Grady, K., Knight, D.R. & Riley, T.V. Antimicrobial resistance in Clostridioides difficile. Eur J Clin Microbiol Infect Dis 40, 2459–2478 (2021). https://doi.org/10.1007/s10096-021-04311-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-021-04311-5