Abstract
Selecting high-risk patients may improve the cost-effectiveness of rapid diagnostics. Our objective was to assess whether model-based selection or clinical selection is better for selecting high-risk patients with a high rate of bacteremia and/or DNAemia. This study involved a model-based, retrospective selection of patients from a cohort from which clinicians selected high-risk patients for rapid direct-from-blood diagnostic testing. Patients were included if they were suspected of sepsis and had blood cultures ordered at the emergency department. Patients were selected by the model by adding those with the highest probability of bacteremia until the number of high-risk patients selected by clinicians was reached. The primary outcome was bacteremia rate. Secondary outcomes were DNAemia rate, and 30-day mortality. Data were collected for 1395 blood cultures. Following exclusion, 1142 patients were included in the analysis. In each high-risk group, 220/1142 were selected, where 55 were selected both by clinicians and the model. For the remaining 165 in each group, the model selected for a higher bacteremia rate (74/165, 44.8% vs. 45/165, 27.3%, p = 0.001), and a higher 30-day mortality (49/165, 29.7% vs. 19/165, 11.5%, p = 0.00004) than the clinically selected group. The model outperformed clinicians in selecting patients with a high rate of bacteremia. Using such a model for risk stratification may contribute towards closing the gap in cost between rapid and culture-based diagnostics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Appropriate empirical antibiotic therapy decreases mortality with an odds ratio of about 1.6, relative to patients where appropriate therapy is initiated when a pathogen has been isolated and tested for its susceptibility to antibiotics [1]. Susceptibility results are typically available 48–72 h after blood is drawn for culture [2].
Rapid molecular diagnostics applied directly to primary blood samples have the potential to significantly shorten the time to pathogen identification, with turnaround times of 6–8 h [3, 4], thus shortening the time to appropriate therapy, which may lead to reduced mortality [1]. These rapid methods are, however, more expensive than the standard diagnostic procedure. A review by the National Institute for Health and Care Excellence (NICE) estimated that the incremental cost of rapid molecular diagnostics, such as those based on the polymerase chain reaction (PCR), range from 127€ to 373€ [5], whereas the price of a blood culture (BC) is approximately 33€ [6].
Selecting patients with higher pre-test probability of positive PCR result and higher mortality could contribute to improving the cost-effectiveness of rapid diagnostic methods. A method for selecting high-risk patients for rapid diagnostics could potentially be implemented as a clinical decision support system, integrated into the hospital’s electronic health record.
SepsisFinder (SF) is a stochastic model implemented as a causal probabilistic network, which predicts the probability of bacteremia based on laboratory variables and vital signs [7, 8]. Assuming that a high probability of bacteremia correlates with a high pre-test probability of a positive PCR, then SF may be able to predict a positive PCR based on a high probability of bacteremia. It has been shown that positive BC and positive PCR can be predicted using the same factors [9]. SF’s estimate of the probability of bacteremia could then be used to select patients for rapid diagnostic testing by stratifying them into low- and high-risk groups. For use in a laboratory setting, we used a modified version of SF: SepsisFinder-Lab (SFL).
The aim of this paper was to compare two strategies for selection of “high-risk” patients from among those who had blood cultures. The rates of bacteremia, DNAemia, and 30-day mortality were compared in groups of high-risk patients selected either by SFL or clinically selected based on Sepsis-3 definitions [10].
Methods
Patients and setting
Data were collected retrospectively for patients in the emergency department or infectious disease ward who had BCs drawn between May 1 and December 31, 2016 at five hospitals in the southern part of the Emilia-Romagna region of Italy. All patients were admitted through the emergency ward, but due to the local procedures at two of the hospitals, some patients had blood cultures drawn at the infectious disease ward. Inclusion criteria were suspicion of infection (drawing of BCs within 2 h of hospital admission, as part of standard clinical care) and age 18 years or older. Infection variables were collected retrospectively from the laboratory information system and included albumin, bilirubin, C-reactive protein (CRP), lactate, neutrophil fraction, and platelets. Values with timestamps closest to the time of BC draw, but within a window of 48 h before to 18 h after, were chosen.
Exclusion criteria
Patients were excluded:
-
If the associated data contained two or fewer infection variables
-
If the infection variables were not available within 1 h after BC draw
-
If the BC test result was not available
-
For patients with multiple visits, only the first encounter was included
Mortality data were collected from the local government population register.
The eligibility criteria for this study were designed such that the inclusion criteria were the same as, and the inclusion period spanned the period where a prospective trial of a rapid microbiology (RM) assay based on universal PCR amplification coupled with mass spectrometry (PCR/ESI-MS), referred to as the “RM study,” was conducted by AUSL Romagna [11].
Mathematical prediction model: SFL
We modified the original SF, a causal probabilistic network model that included temperature, heart rate, calculated mean arterial pressure, mental status, neutrophil fraction, platelets, CRP, lactate, creatinine, and albumin as input variables for prediction of the probability of bacteremia [7, 8].
The modified SF model was trained using the EM-learning procedure provided in Hugin (Hugin Expert, Aalborg, Denmark) following the same process used in construction of SF [7]. The training dataset was based on 4707 patients with a community acquired infection at Beilinson Hospital, Petah Tikva, Israel. For the training of the bacteremia prediction, the input variables were limited to those available in the data from the Emilia-Romagna hospitals (neutrophil fraction, platelets, bilirubin, CRP, lactate, creatinine, and albumin) (27th European Congress of Clinical Microbiology and Infectious Diseases, abstract P2055). Creatinine was subsequently eliminated from the model because it did not contribute to the predictive performance.
On the training dataset, the model had an area under the receiver operating characteristic curve (AUROC) = 0.717 (0.691–0.744) for prediction of bacteremia (Fig. E1a in Online Resource 1).
Risk assessment
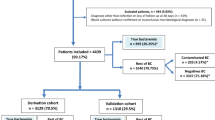
The goal of the present study is to compare the ability of clinicians to select high-risk patients to that of SFL. Two subgroups were defined to make the comparison: Clinical high risk and SFL high risk; see flow diagram (Fig. 1).
Clinical high risk
Physicians were requested to include high-risk patients in the RM study (defined as those with a SOFA score of 2 or more, thereby fulfilling the Sepsis-3 definitions [10]) by ordering concurrent blood samples for RM at the time of BC order. Where SOFA was not available, clinical judgment was used as to whether patients were included or not.
SFL high risk
An alternative selection was performed by SFL. We chose to include a number of SFL high-risk patients equal to the number of clinical high-risk patients by choosing the threshold value for high-risk post hoc in order to achieve the same number of patients in each group.
Microbiology
Positive BCs and positive PCR tests were considered clinically significant when contaminants were excluded. Such positive BCs and PCR tests, including those with fungal isolates, will be referred to as bacteremic (BC+) and DNAemic (DNA+), respectively. Bacillus spp. (except B. anthracis), coagulase-negative staphylococci, Corynebacterium spp., and Micrococcus spp. were considered contaminants, in accordance with local policy at AUSL Romagna. In the absence of clinical data indicating the site of infection, viridans streptococci were also considered contaminants, even though they may occasionally be pathogens.
Blood cultures were performed using the BacT/Alert® system (bioMérieux, Marcy l’Etoile, France), and RM with the Iridica-BAC BSI assay (Ibis Biosciences-Abbott Molecular, Des Plaines, IL). The techniques used are described fully in the RM study [11].
Outcomes and statistical analysis
The primary outcome was the rate of BC+. Secondary outcomes were all-cause 30-day mortality (M30) and the rate of DNA+. Outcomes were compared for the clinically and SFL-selected groups.
Dichotomous categorical variables (including the two outcomes) were compared using the binomial test, while continuous variables were compared using the Mann-Whitney U test or Wilcoxon signed-rank test. Difference in survival was computed using the Mantel-Cox log-rank test. Differences in rates across subgroups were tested using Pearson’s chi-squared test or Fisher’s exact test, as appropriate. SFL probability of BC+ was calculated using Hugin Researcher 7.6 (Hugin Expert A/S). Graphical presentations were constructed in Excel 2017 (Microsoft Corporation) and MATLAB R2016a (MathWorks Inc.) and statistical analyses were performed in SPSS 25 (IBM Corporation) and Excel 2017.
A preliminary version of the results in this manuscript was presented at the 27th European Congress of Clinical Microbiology and Infectious Diseases, Vienna, Austria, April 24, 2017 (OS0558B).
Results
The eligible study population included 1395 patients. Following exclusion, BCs were analyzed for a total of 1142 patients (Fig. 1). Of 1142 patients, 384 (33.6%) had a positive BC, and after excluding contaminants 296 (25.9%) were considered BC+. Of 296 BC+, 186 (62.8%) had a single Gram-negative isolate, 84 (28.4%) had a single Gram-positive isolate, and 2 (0.7%) had a fungal isolate, while 24 (8.1%) were polymicrobial (9 with multiple Gram-negatives, 1 with multiple Gram-positives and 14 mixed Gram-negative and Gram-positive). Table 1 presents a breakdown of the BC isolates.
The infection variables CRP, neutrophil fraction, and platelets were available for > 98% of patients. Bilirubin was recorded for 53% of patients. Albumin and lactate were only available for 0.9% and 0.7% of patients, respectively. Descriptive statistics are presented in Table 2.
Clinical high risk
Two hundred twenty patients were in the clinical high-risk group. According to the original RM study design, these patients should have had SOFA > = 2 on admission. However, calculating a SOFA score for these patients prior to inclusion was impractical. Retrospectively, the SOFA score was available for 180/220 patients, with 45 patients having SOFA < 2. Among the patients for whom SOFA was available, there was no difference in BC+ (33.3% vs. 31.1%, p = 0.78) or 30-day mortality (13.3% vs. 14.8%, p = 0.81) for patients with SOFA < 2 and with SOFA > = 2. There was a significant difference between the rate of BC+ in the clinical high-risk group compared to the remaining low-risk patients (32.3% vs. 24.4%, p = 0.02) (Fig. 2a). The sensitivity of the clinical high-risk for BC+ was 24.0% (71/296), while the specificity was 82.4% (697/846). The predictive power of inclusion in the clinical and SFL high-risk groups for BC+ prediction is detailed in Table 3.
SFL high risk
The SFL high-risk group had significantly higher BC+ rate than the SFL low-risk group (45.5% vs. 21.3%, p < 10−12, Fig. 2b). The sensitivity of SFL high risk for BC+ was 33.8% (100/296) and the specificity was 85.8% (726/846). The area under the ROC curve for prediction of BC+ was 0.70 for the study data (Fig. E3a in Online Resource 1). Calibration plots for training and study data can be found in Fig. E2 and E4 of Online Resource 1, respectively.
Figure 2c compares the clinical and SFL high-risk patients. Fifty-five patients were in both the clinical and the SFL high-risk groups (intersecting selections). The clinical and SFL high-risk groups are therefore not independent. To form independent groups, allowing the use of the binomial test for differences between clinical and SFL high-risk groups, we defined three groups which were then compared pairwise. The three groups defined by the overlapping sets are referred to as clinical only, both, and SFL only. The rate of BC+ was higher in the SFL-only group than in the clinical-only group (44.8% vs. 27.3%, p = 0.001), and patients had higher mortality than those in the clinical-only group (29.7% vs. 11.5%, p = 0.00004). There were no significant differences, neither in mortality nor in BC+ rate for SFL-only group compared to the both groups (p = 0.26, p = 0.75, respectively). However, in both groups, the rate of BC+ was significantly higher compared to the clinical-only selection (47.3% vs. 27.3%, p = 0.006), while the difference in 30-day mortality (21.8% vs. 11.5%, p = 0.06) did not reach significance. The DNA+ rate was also significantly higher (52.7% vs. 32.1%, p = 0.006) in both group compared to the clinical-only group.
Survival analysis
Figure 3 shows log-survival curves for patients included in the study. The upper panel (a) presents curves for SFL high-risk and low-risk patients, and the lower panel (b) presents curves for clinical high-risk and low-risk patients. SFL high-risk patients had lower survival than low-risk patients (p < 0.001). There was no significant difference for clinical high-risk vs. low-risk patients (p = 0.98). The patterns in survival remained when stratifying by age; the separation between survival curves for SFL high-risk and low-risk patients was still present (Fig. E5a in Online Resource 1), as was the lack of separation for the clinical high-risk and low-risk groups (Fig. E5b in Online Resource 1).
Discussion
The primary finding was that risk assessment provided by SFL significantly outperformed the clinical risk stratification in terms of selecting patients with higher bacteremia rates. These patients may also have a higher DNAemia rate, if it is accepted that the DNAemia rate is linked to the bacteremia rate by a fixed odds ratio. In addition, the clinical selection appeared to fail: although there was a difference in BC+ rate between clinical high risk and clinical low risk, there was no difference between the Clinical only group and the Clinical low-risk. In addition, there was no difference in 30-day mortality for clinical high-risk and low-risk patients.
A strength of SFL is that the prediction of bacteremia seems robust with consistent areas under the ROC curve of 0.72 for the training set (Israel) and 0.70 for the validation set (Italy). SFL also provided a “fair” area under the ROC curve, even though the number of available variables was limited: none of the vital parameters and only three of the six lab values in the original version of SF [7, 8] were available. The absence of lactate in the present dataset was mainly logistical. Clinical practice at the time meant that lactate was measured by blood gas analysis in the hospital and available in the hospital’s patient record, but not in the laboratory information system. It is expected that the addition of both lactate and albumin measurements (particularly for ER patients) would improve predictive performance. Albumin is linked to mortality [12], while lactate is also used as a severity marker or a marker for septic shock [10, 13]. Bilirubin was also added in SFL due to data availability and lack of a measure of liver dysfunction in the model. In practice, the ability to stratify the patients based on lab values alone may be logistically important, since lab values in most hospitals/microbiology laboratories can be drawn automatically from the laboratory information system.
The clinical selection of high-risk patients performed poorly in identifying patients with a higher rate of bacteremia and higher mortality. We believe there are three possible explanations why clinicians were outperformed by SFL; the first explanation is that the mortality-based SOFA score, used according to the Sepsis-3 definitions [10], is unsuitable, or it was not used as intended. The unsuitability of SOFA is supported by the finding that there were no significant differences in mortality and bacteremia in patients with SOFA < 2 and SOFA ≥ 2. This is supported by other studies showing that while the Sepsis-3 criteria provide a good prognostic indication in the ICU [14], they are less effective in the emergency department [15]. However, it is a limitation of the study that clinical data, and thus also SOFA scores, were only collected for the clinically selected patients.
The second explanation is that the SOFA score has not been applied appropriately. It is possible that some patients who were critically ill were not clinically selected due to the severity of their illness, with the clinicians perhaps focused on fluid resuscitation and timely antimicrobial treatment [16] instead of enrolment in the RM study. This would create a selection bias disfavoring the clinical selection, but the study is limited in the sense that it does not provide the clinical data required to confirm or reject this explanation. It is also possible that clinicians may err on the side of caution, partly due to incomplete history and clinical data [17], resulting in an overestimation of the risk of bacteremia [18]. This is backed by the fact that 49 patients with SOFA < 2 were included in the clinical high-risk group.
The third explanation is that the comparison between SFL and the SOFA score is not fair: SFL selects for high bacteremia rates, while the SOFA score selects for high mortality. However, this explanation is incompatible with the finding that SFL selection outperforms the clinical selection with a significantly higher mortality.
It remains unexplained why the rate of positive BCs (33.9%) as well as of bacteremia (26.3%) was high compared to rates reported in the literature (5–15%) [19,20,21] for patients suspected of sepsis. However, the bacteremia rate in this study is consistent with historical averages for AUSL Romagna. Yearly bacteremia rates for cultures drawn in the emergency department ranged from 23 to 25% for 2013–2016 (Report di Sorveglianza Microbiologica AUSL della Romagna 2013–2016, available on request). One possible explanation of the high rate of bacteremia is that blood may not have been drawn for culture in all low-risk patients. However, this does not explain that the observed rate of bacteremia is twice as high as that predicted by SFL (see Fig. E4 in Online Resource 1). Other speculative reasons for the high rate of bacteremia include differences in workflow and/or culture-related processes such as temperature control of the sample or the rate of antibiotic therapy preceding blood sampling, but the exact reasons have not been determined. Improved culture technique would be consistent with the observation that the contamination rate (6.8%) is also about twice as high as those reported in the literature [21,22,23].
The lack of access to clinical data for the patients included in the study also made it difficult to provide a true determination of the contamination rate. It is possible, for example, that a number of the patients, whose blood cultures grew CoNS or viridans streptococci, had an infection where one of these organisms was the causative pathogen.
Since the completion of the study, the RM product has been withdrawn from the market. However, the concept of risk-based stratification remains valid as a selection method for improving the utilization of rapid microbiology. The methods presented here are test agnostic.
A recent review [24] of predictive models for bacteremia included two computerized models. One model was TREAT [20]. SF was originally a component of the decision support system TREAT [25,26,27], which advises on optimal antimicrobial therapy. TREAT requires more data including background information and the physical examination of the patient. The other predictive model was another Bayesian model [28] which focused on hospitalized patients, where many of the input variables would not typically be available in the emergency department. SFL had similar performance characteristics in terms of the AUROC reported for these other models, but with the practical advantage that it only uses data typically available in the emergency department, and that these data are often available in a structured electronic format.
SFL was able to select a group with a higher rate of bacteremia and higher mortality than in the clinically selected high-risk group. Presumably, early appropriate antibiotic treatment is more important in a severely ill group and thus the SFL high-risk group would benefit more from rapid diagnostics than the clinical high-risk patients. A practical implementation of SFL as part of a clinical decision support system for risk stratification may contribute towards improving the selection of emergency department patients with suspected sepsis for rapid diagnostics. Such an implementation should be assessed in a prospective study, where the true benefits can be assessed.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request with permission from AUSL Romagna.
References
Paul M, Shani V, Muchtar E, Kariv G, Robenshtok E, Leibovici L (2010) Systematic review and meta-analysis of the efficacy of appropriate empiric antibiotic therapy for sepsis. Antimicrob Agents Chemother 54:4851–4863. https://doi.org/10.1128/AAC.00627-10
Kirn TJ, Weinstein MP, Grumaz C, Decker SO, Weigand MA, Hofer S et al (2013) Update on blood cultures: how to obtain, process, report, and interpret. Clin Microbiol Infect 1(19):513–520. https://doi.org/10.1111/1469-0691.12180
Liesenfeld O, Lehman L, Hunfeld K-P, Kost G (2014) Molecular diagnosis of sepsis: new aspects and recent developments. Eur J Microbiol Immunol 4:1–25
Bloos F, Reinhart K (2014) Rapid diagnosis of sepsis. Virulence 11(5):154–160. https://doi.org/10.4161/viru.27393
National Institute for Health and Care Excellence (2016) Tests for rapidly identifying bloodstream bacteria and fungi (LightCycler SeptiFast Test MGRADE, SepsiTest and IRIDICA BAC BSI assay) [Internet]. [cited 2017 28]; https://www.nice.org.uk/guidance/dg20/chapter/4-Outcomes
Perl B, Gottehrer NP, Raveh D, Schlesinger Y, Rudensky B, Yinnon AM (1999) Cost-effectiveness of blood cultures for adult patients with cellulitis. Clin Infect Dis 1(29):1483–1488. https://doi.org/10.1086/313525
Ward LM (2016) Gradation of the severity of sepsis - learning in a causal probabilistic network. https://doi.org/10.5278/VBN.PHD.MED.00064
Ward L, Møller JK, Eliakim-Raz N, Andreassen S (2018) Prediction of bacteraemia and of 30-day mortality among patients with suspected infection using a CPN model of systemic inflammation. IFAC-PapersOnLine 51:116–121. https://doi.org/10.1016/j.ifacol.2018.11.657
Leli C, Cardaccia A, D’Alo F, Ferri C, Bistoni F, Mencacci A et al (2014) A prediction model for real-time PCR results in blood samples from febrile patients with suspected sepsis. J Med Microbiol 63:649–658. https://doi.org/10.1099/jmm.0.064097-0
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M et al (2016) The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 23(315):801. https://doi.org/10.1001/jama.2016.0287
Tassinari M, Zannoli S, Farabegoli P, Pedna MF, Pierro A, Mastroianni A et al (2018) Rapid diagnosis of bloodstream infections in the critically ill: evaluation of the broad-range PCR/ESI-MS technology. PLoS One 13:e0197436. https://doi.org/10.1371/journal.pone.0197436
Vincent J-L, Dubois M-J, Navickis RJ, Wilkes MM (2003) Hypoalbuminemia in acute illness: is there a rationale for intervention?: a meta-analysis of cohort studies and controlled trials. Ann Surg 237:319
Howell MD, Donnino M, Clardy P, Talmor D, Shapiro NI (2007) Occult hypoperfusion and mortality in patients with suspected infection. Intensive Care Med 33:1892–1899
Seymour CW, Liu VX, Iwashyna TJ, Brunkhorst FM, Rea TD, Scherag A et al (2016) Assessment of clinical criteria for sepsis. JAMA 315:762. https://doi.org/10.1001/jama.2016.0288
Calle P, Cerro L, Valencia J, Jaimes F (2012) Usefulness of severity scores in patients with suspected infection in the emergency department: a systematic review. J Emerg Med 42:379–391. https://doi.org/10.1016/j.jemermed.2011.03.033
Pulia MS, Redwood R, Sharp B (2017) Antimicrobial stewardship in the management of sepsis. Emerg Med Clin North Am 35:199–217. https://doi.org/10.1016/j.emc.2016.09.007
Peterson L-KN, Chase K (2017) Pitfalls in the treatment of sepsis. Emerg Med Clin North Am 35:185–198. https://doi.org/10.1016/j.emc.2016.09.008
Poses RM, Anthony M (1991) Availability, wishful thinking, and physicians’ diagnostic judgments for patients with suspected bacteremia. Med Decis Mak 2(11):159–168. https://doi.org/10.1177/0272989X9101100303
Shapiro NI, Wolfe RE, Wright SB, Moore R, Bates DW (2008) Who needs a blood culture? A prospectively derived and validated prediction rule. J Emerg Med 35:255–264. https://doi.org/10.1016/j.jemermed.2008.04.001
Paul M, Andreassen S, Nielsen AD, Tacconelli E, Almanasreh N, Fraser A et al (2006) Prediction of bacteremia using TREAT, a computerized decision-support system. Clin Infect Dis 1(42):1274–1282. https://doi.org/10.1086/503034
Jessen MK, Mackenhauer J, Hvass AMSW, Ellermann-Eriksen S, Skibsted S, Kirkegaard H et al (2014) Prediction of bacteremia in the emergency department. Eur J Emerg Med 23:44–49. https://doi.org/10.1097/MEJ.0000000000000203
Hall KK, Lyman JA (2006) Updated review of blood culture contamination. Clin Microbiol Rev 1(19):788–802. https://doi.org/10.1128/CMR.00062-05
Weinstein MP (2003) Blood culture contamination: persisting problems and partial progress. J Clin Microbiol 1(41):2275–2278. https://doi.org/10.1128/JCM.41.6.2275-2278.2003
Eliakim-Raz N, Bates DW, Leibovici L (2015) Predicting bacteraemia in validated models—a systematic review. Clin Microbiol Infect 1(21):295–301. https://doi.org/10.1016/j.cmi.2015.01.023
Paul M, Andreassen S, Tacconelli E, Nielsen AD, Almanasreh N, Frank U et al (2006) Improving empirical antibiotic treatment using TREAT, a computerized decision support system: cluster randomized trial. J Antimicrob Chemother 58:1238–1245. https://doi.org/10.1093/jac/dkl372
Leibovici L, Fishman M, Schonheyder HC, Riekehr C, Kristensen B, Shraga I et al (2000) A causal probabilistic network for optimal treatment of bacterial infections. Knowl Data Eng IEEE Trans 12:517–528
Andreassen S, Leibovici L, Paul M, Nielsen AD, Zalounina A, Kristensen LE et al (2005) A probabilistic network for fusion of data and knowledge in clinical microbiology. In: Probabilistic modeling in bioinformatics and medical informatics. Springer, pp 451–472
Jin SJ, Kim M, Yoon JH, Song YG (2013) A new statistical approach to predict bacteremia using electronic medical records. Scand J Infect Dis 45:672–680
Funding
This work was supported by a grant from Innovation Fund Denmark (grant number 5190-00011B) to L.W. and S.A.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
S.A. is a member of the board and shareholder in Treat Systems ApS. L.W. is an employee of Treat Systems ApS. J.A., Z.R., M.F., and V.S. declare no conflict.
Ethical considerations
The patients evaluated in this study were anonymized following a procedure (AVR PPC-09 Rev.2) that completely blinds the researchers to the demographic details of each single patient. This procedure has been reviewed and accepted by the AUSL della Romagna Local Ethical Committee. The research described in this paper is a retrospective analysis of anonymous patient data and did not require informed consent nor separate ethical approval.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 297 kb)
Rights and permissions
About this article
Cite this article
Ward, L., Andreassen, S., Astrup, J.J. et al. Clinical- vs. model-based selection of patients suspected of sepsis for direct-from-blood rapid diagnostics in the emergency department: a retrospective study. Eur J Clin Microbiol Infect Dis 38, 1515–1522 (2019). https://doi.org/10.1007/s10096-019-03581-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03581-4