Abstract
Norway has one of the world’s highest incidences of colorectal cancer (CRC). Accumulating research suggests that the intestinal microbiota may have an important role in initiation and progression of colorectal cancer. In order to evaluate microbiome-based biomarkers for non-invasive detection of CRC, the levels of Fusobacterium nucleatum and selected Escherichia coli toxin genes in stool and mucosa from a small cohort of Norwegian patients were investigated. The study cohort included 72 patients scheduled for colonoscopy. The patients were divided into three groups upon their examinations: cancer, polyp, and control groups. Levels of F. nucleatum in stool samples were significantly higher in the cancer group compared with the control group and the polyp group. High levels of F. nucleatum in stool reflected detection of F. nucleatum in the tumor tissues of colorectal cancer patients. However, no difference in the levels of E. coli toxin genes in neither stool nor biopsy samples between the patient groups was observed. This study suggests that a quantitative PCR assay targeting F. nucleatum in stool samples has the potential to be included in a larger panel of biomarkers for non-invasive testing for colorectal cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Despite developments in diagnostics and treatment, colorectal cancer (CRC) remains the second most common cause of cancer mortality in the western countries. Norway has one of the world’s highest incidences of colon cancer, with 4332 new cases reported in 2017 (Cancer Registry of Norway: www.kreftregisteret.no). Early detection results in significantly improved outcomes for CRC patients. When diagnosis is set at stage I, patients have a 90–95% survival rate with surgical intervention while at stage IV, the survival rate is reduced to just 5–10% (https://seer.cancer.gov/statistics/). Currently, sigmoidoscopy and colonoscopy are used for diagnostics of CRC in Norway. These are invasive methods that require extensive resources and therefore high costs. Alternatively, non-invasive methods such as immunochemical tests for hidden (occult) blood in stool samples (iFOBT) are also in use. The advantages with using these tests lie in the easily accessible sample material and low analytical costs in the laboratory. The drawbacks are, however, poor diagnostic sensitivity and specificity [1]. Therefore, adding new non-invasive biological markers to increase the overall sensitivity and specificity is highly attractive.
Colorectal cancer development is a multifactorial process involving an accumulation of mutations in the genes such as APC, KRAS, BRAF, and TP53, resulting in uncontrolled cell growth and tumor formation. The triggers for initiation of colorectal cancer are not fully characterized, but the risk factors are associated with high intake of animal fat, red meat, processed food, and low consumption of fiber-rich food [2]. We are becoming increasingly aware that the intestinal microbiota may be involved in the initiation and facilitation of CRC [3, 4]. Several models for bacterial involvement in initiation and development of CRC have been presented (reviewed in [5]). The mechanisms include DNA damage from bacterial virulence factors such as genotoxins, activation of inflammatory and oncogenic signaling pathways, and production of tumor-promoting metabolites such as secondary bile acids. A number of microbes have been proposed as candidates for CRC initiation, in particular, Fusobacterium nucleatum and Escherichia coli. An expert review from 2016 summarizes the evidence supporting oncogenic roles of F. nucleatum and E. coli [6]. The authors present supporting evidence from molecular studies illustrating a strong correlation between F. nucleatum and carcinogenesis. F. nucleatum may directly contribute to carcinogenesis through attachment of its FadA adhesion protein to the extracellular domain of E-cadherin on the surface of intestinal epithelial cells. This may result in E-cadherin/β-catenin activation via the WNT signaling pathway [7,8,9,10,11]. β-Catenin accumulates in the cytosol, translocates into the nucleus, and activates its target genes such as NK-κB and the proto-oncogenes C-MYK and C-JUN [7]. The contributory rather than consequential role of F. nucleatum in tumor formation is supported by studies using models with preexisting mutations in the tumor suppressor gene APC. “First-hit” APC mutations appear to be sufficient for F. nucleatum to exert its inflammatory and oncogenic effects [12]. The present hypothesis is therefore that F. nucleatum emerges early in cancer development and is a risk factor for progression from adenoma to cancer [13]. Furthermore, invasive F. nucleatum can also be found in liver metastases, suggesting a persistent association with tumor cells during various stages of colorectal cancer development [14].
Despite the accumulating evidence correlating F. nucleatum to colorectal cancer, this has only been shown for a subgroup of colorectal cancer patients. Given the large interindividual differences in intestinal microbiota, it remains unlikely that bacterially induced oncogenic progression is driven by the same bacterial species in all cancer patients. E. coli have repeatedly been associated with colorectal cancer tumor sites, and evidence for a contributory role of E. coli toxins in colorectal cancer is emerging [15, 16]. The virulence factor colibactin, part of the polyketide synthase (pks) island, has been most widely studied. It is suggested to promote tumor progression by miRNA silencing of p53 and hence cellular senescence and hepatocyte growth factor secretion [5]. Other Escherichia virulence factors studied in relation to CRC are cytolethal distending toxin (cdt), toxin coregulated pilus synthesis outer membrane protein C (tcpC), and arginine succinyltransferase (astA) [17, 18].
The majority of studies have investigated F. nucleatum and E. coli toxins in mucosal samples, from tumor and adjacent tissues [15, 19,20,21]. Recent studies also illustrate that F. nucleatum can often be detected in stool samples from CRC patients [22,23,24]. The presence of F. nucleatum and E. coli toxin genes has not previously been studied in samples from Norwegian cancer patients, and their use as marker genes for CRC in the Norwegian population has therefore not been evaluated. The aims of the present study were to estimate the levels of F. nucleatum and selected E. coli toxin genes in stool and mucosa samples from Norwegian colorectal cancer patients and to evaluate the use of F. nucleatum and E. coli quantitative PCRs in a panel of non-invasive biomarkers for detection of colorectal cancer in its early stages.
Materials and methods
Study population and samples
Patients with scheduled colonoscopy at Akershus University Hospital (Ahus) from 2014 to 2017 were included in the study (n = 72). The indications for colonoscopy were gastrointestinal bleeding, weight loss, changes in bowel movements, or detection of polyps/tumors on CT colonography. Prior to the procedure, patients were informed that additional samples would be taken, given their consent, and informed of their rights to withdraw from the study at any time. Written informed consent was obtained from all included participants. The regional committee for medical and health-related research ethics (REK 2012/1944) and the data protection manager at Ahus have approved the study.
The 72 patients were divided into three groups based on findings during colonoscopy: one group with colorectal cancer (n = 25), one group with adenomatous polyps (n = 25), and one control group without pathological findings (n = 22). Median age was 70, 69, and 57 for the cancer, polyp, and control groups, respectively. The proportions of females were 28%, 56%, and 41%, respectively, in the three different groups. Each patient collected a stool sample in RNAlater RNA Stabilization buffer (Qiagen, Hilden, Germany) prior to their bowel preparation or 1 week after colonoscopy. When samples arrived at the laboratory, they were homogenized and stored at − 80 °C. During colonoscopy, biopsies (2 × 2 mm on average) were collected from different positions in the colon: position 1, colon ascendens; position 2, from the polyp or cancerous tissue; position 3, adjacent healthy tissue; and position 4, colon sigmoideum. Immediately after collection, the biopsies were fixed in Allprotect Tissue Reagent (Qiagen, Hilden, Germany) and stored according to the manufacturer’s recommendations.
Classification of the tumors
All tumors were examined by the Dept. of Pathology and classified into stages based on the UICC TNM Classification system. In this anatomically based system, the category T describes the primary tumor size and/or extent, the category N describes the regional lymph node involvement, and the category M describes the presence of distant metastatic spread. According to the National Health Department in Norway, the TNM should be given with the prefix p, indicating assessment done by a pathologist (Table 1).
DNA extraction from stool and biopsies
DNA was extracted from all fecal samples with PSP® Spin Stool DNA Kit (Stratec Molecular Gmbh, Berlin, Germany) based on previous results for optimal DNA output and bacterial diversity [25]. Biopsies were extracted with AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) according to the procedure described by Moen et al. [26]. NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was used for measurements of concentration and purity of the extracted DNA. DNA from biopsy and stool samples were diluted 10- and 100-fold, respectively, in PCR-grade water to facilitate downstream analysis.
Quantitative PCR (qPCR)
Fusobacterium nucleatum–specific qPCR
The PCR reactions were performed with primers for F. nucleatum designed by Flanagan et al. [13] but with a slightly modified reverse sequence (Table 2). The primers amplify a portion of the gene encoding the antitermination factor NusG from F. nucleatum. The quantitative PCR (qPCR) reactions were performed on the Applied Biosystems 7900 Real-Time PCR System (Thermo Fisher Scientific) using SYBR® Select Master Mix (Thermo Fisher Scientific) in 20 μl reactions. The cycling conditions were as follows: 50 °C for 2 min (uracil N-glycosylase activation), followed by one hold at 96 °C for 2 min (activation of Taq DNA polymerase), and 40 cycles at 95 °C/15 s, 57 °C/15 s, and 72 °C/20 s.
Escherichia coli toxin–specific qPCR
Four qPCR reactions targeting four different E. coli toxin genes (pks, CNF1, tcpC, and astA) were performed using cycling conditions as previously described [17, 18, 27] (Table 2). The PCR reactions were performed with SYBR® Select Master Mix (Thermo Fisher Scientific) in 20 μl reactions.
Reference gene qPCR
Total bacterial DNA determined by 16S rRNA qPCR was used to normalize the target genes in stool samples. The PCR master mix and cycling conditions were as described for F. nucleatum [28]. A qPCR targeting the human β-globin gene was used for target gene normalization in biopsies [29] (Table 2).
qPCR evaluations
PCR efficiency and limit of detection (LOD) were determined for all qPCR assays using 10-fold dilution series of genomic DNA from control strains of F. nucleatum, extraintestinal E. coli (ExPEC), enteroaggregative E. coli (EAEC), and enterohemorrhagic E. coli (EHEC) (taxonomy confirmed by 16S rRNA gene sequence and/or characterized at the Norwegian Institute of Public Health). Specificity of the PCR primers was determined using a panel of genomic DNA isolated from stool-associated bacterial species (n = 20).
Statistical analysis of qPCR data
Stool samples
Stool samples from the cancer group (n = 23), polyp group (n = 25), and control group (n = 22) were analyzed for quantities of F. nucleatum and the presence of four E. coli toxin genes using qPCR as described above. All qPCR reactions were performed in duplicate. Relative quantities of F. nucleatum and E. coli toxin genes in each stool sample were determined with 2−ΔCt using the 16S rRNA gene as a reference gene: ΔCt = CtFn/E.coli − Ct16S rRNA [13]. The fold difference in F. nucleatum or E. coli toxin gene levels between the patient groups were calculated by the 2−ΔΔCt method. For statistical comparison of means between independent groups, one-way ANOVAs were performed to test differences in both F. nucleatum and E. coli toxin levels between the cancer, polyp, and control groups. Significance was set at α = 0.05; a comparison of means was performed using the post hoc Tukey multiple comparison test. A test of normality was carried out using the Shapiro-Wilk test and also Levene’s test for homogeneity of variance. Due to the non-Gaussian nature of the qPCR data distribution, non-parametric testing was deemed appropriate; consequently, the one-way ANOVA and the Tukey post hoc test were replaced with the Kruskal-Wallis rank sum test and the pairwise Wilcoxon rank sum test, respectively. Statistical comparisons between groups were performed in the open-source software R (version 3.3.2 (2016-10-31)).
Biopsy samples
Biopsies from 21 cancer patients, 11 polyp patients, and 11 control patients were analyzed for quantities of F. nucleatum and the presence of four E. coli toxin genes using qPCR as described above. Levels of F. nucleatum and E. coli toxin genes were related to human β-globin: ΔCt = CtFn/E.coli − Ctβ-globin. Repeated measures ANOVAs were performed to test differences in both F. nucleatum and E. coli toxin levels between the colon ascendens, cancerous tissue, adjacent healthy tissue, and colon sigmoideum biopsies in colon cancer patients. Post hoc Tukey multiple comparisons of means were again implemented to identify which pair(s) of biopsy position means differed significantly. We used the Shapiro-Wilk normality test and Mauchly’s test of sphericity to formally test these assumptions. Non-parametric testing was performed when appropriate, replacing the repeated measures ANOVA and the Tukey post hoc test with the Friedman rank sum test and the pairwise Friedman-Nemenyi multiple comparison test, respectively. All significance levels were set at α = 0.05.
16S rRNA massive parallel sequencing
For verification of qPCR results, stool samples from the cancer and control groups (n = 27), as well as two negative processing controls (included in the whole sample processing procedure from DNA extraction), were sent for 16S rRNA massive parallel sequencing by a commercial laboratory, Omega Bioservices (Atlanta, GA, USA). 12.5 ng DNA was used as input and the V3-V4 region of the bacterial 16S rRNA gene sequences was amplified using the primer pair 341F-805R (Table 2) and KAPA HiFi HotStart ReadyMix (Kapa Biosystems, Wilmington, MA). PCR products were purified with Mag-Bind RxnPure Plus magnetic beads (Omega Bio-tek, Norcross, GA) before performing a second index PCR amplification with the same master mix. Libraries (~ 600 bp) were normalized, pooled, and sequenced (2 × 300 bp paired-end read setting) using the MiSeq platform (Illumina, San Diego, CA, USA).
Sequences were preprocessed, quality filtered, and analyzed using QIIME2 1.9. Further analysis was performed in the Illumina BaceSpace 16S metagenomics application. The read classifier was Illumina-modified RDP, and taxonomy was assigned using the Illumina-curated version of the May 2013 release of the Greengenes Consortium Database 13.5 (performed by Omega Bioservices). Taxonomic IDs with only one aligned sequence read were discarded. The microbial classifications were compared at different taxonomic levels (genus and species) between the groups.
Results
Quantitative PCR results
Levels of Fusobacterium nucleatum in stool samples are associated with colorectal cancer and reflect the tumor environment
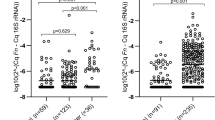
Significantly higher levels of Fusobacterium nucleatum were observed in stool samples from the cancer group compared with the polyp group (Tukey p = 0.00074 and Wilcoxon p = 0.0028) and the cancer group compared with the control group (Tukey p = 0.00014 and Wilcoxon p = 0.0073) using qPCR (Fig. 1). Comparison of F. nucleatum levels between the cancer and control groups with the 2−ΔΔCt method illustrated a fold difference of 66, in favor of the cancer group. The difference between the cancer and polyp groups was 97-fold. No significant difference was detected between the polyp and control groups (Tukey p = 0.928 and Wilcoxon p = 0.495).
Figure 2 illustrates that 35% of cancer patients and none of the control or polyp patients in this study would be identified with a ∆Ct (CtFn − Ct16S rRNA) cutoff of 12. High levels of F. nucleatum in stool samples correlated with detection of F. nucleatum in tumor tissue. In 69% (9/13) of cancer patients that tested positive for F. nucleatum in their cancerous tumor biopsies, high levels were identified in stool.
The figure illustrates that 35% of stool samples from cancer patients and none of the stool samples from polyp or control patients were detected with ∆Ct (CtF.nucleatum − Ct16S rRNA) values below 12. These samples were all from cancer patients in early stages and represented 69% of cancer patients with F. nucleatum in their superfluous tumor biopsies. Cancer patients with distant metastatic spread had higher ∆Ct values or F. nucleatum was undetected in stool
Fusobacterium nucleatum is associated with tumor tissues
The relative abundance of F. nucleatum in different positions along the colon was determined using qPCR. Irrespective of sample position, F. nucleatum was detected in samples from 60%, 18%, and 18% of patients in the cancer, polyp, and control groups, respectively. Further analyses of samples from the cancer group (n = 21) illustrated that F. nucleatum was detected in 52% (n = 13) of samples from tumor tissue, 24% (n = 6) of healthy tissue sample from the ascending colon, 36% (n = 9) of healthy tissue samples adjacent to the tumor, and 36% (n = 9) of healthy tissue samples from the colon sigmoideum. Comparison of F. nucleatum relative quantities suggested higher levels of F. nucleatum in the tumor tissue compared with adjacent healthy tissue (6.3-fold), colon ascendens (13-fold), and colon sigmoideum (11-fold) (Fig. 3). The non-parametric Friedman test showed significant differences only between cancerous tissue and colon ascendens (p = 0.036).
Relative quantities of F. nucleatum in different positions of the colon in cancer patients. Statistical analysis (Friedman test) showed significant differences only between cancerous tissue (TU) and colon ascendens (CA) (p = 0.036). The boxplot shows the median and interquartile range of the relative F. nucleatum quantifications. CS colon sigmoideum, TI healthy tissue adjacent to the tumor
Association between tumor position, tumor stage, and Fusobacterium nucleatum
Tumors located in the proximal part of the colon were more frequently associated with F. nucleatum. F. nucleatum was detected in 72% (8/11) of tumors in the proximal part of the colon, compared with 35% (5/14) of tumors in the distal part of the colon and rectum. A trend of wider distribution of F. nucleatum in the colorectum, illustrated by detection in multiple positions, was observed when the tumor was located in the proximal part of the colon. Seven of the 13 tumors with F. nucleatum were positive also in adjacent tissues, notably six of these seven tumors were located in the proximal part of the colon (Fig. 4). The qPCR data did not reveal any differences in quantities of F. nucleatum in tumors located in the proximal versus distal part of the colon, nor did they detect any differences in F. nucleatum quantities between different TNM classifications. These numbers are too small for statistical comparison.
Levels of E. coli toxin genes in stool samples and colonic mucosa are not associated with colorectal cancer
No statistical significant differences in E. coli toxin gene levels in stool were observed between the patient groups (one-way ANOVA p = 0.61, Tukey p = 0.41, and Wilcoxon p = 0.59). Irrespective of sample position, E. coli toxin genes were detected in biopsies from 52%, 27%, and 45% of patients in the cancer, polyp, and control groups, respectively. Further analyses of samples from the cancer group did not result in any significant differences between the different positions (repeated measures ANOVA p = 0.36). The E. coli toxin biopsy data met the assumptions of both normality and sphericity. The same toxin gene was detected in two or more positions of the colon in 90% of the patients with E. coli toxins in stool and in three or four positions in 70% of the patients with E. coli toxins in stool, illustrating that the toxin-producing E. coli were distributed in the entire colorectum. The results from biopsies correlated with results from stool samples. In 95% of patients that tested positive for an E. coli toxin gene in a biopsy, the same gene was also detected in the stool sample.
Evaluation of qPCR results by massive parallel sequencing of the bacterial 16S rRNA gene
Illumina 300-bp paired-end sequencing generated, after quality filtering and processing, between 109,348 to 196,219 sequence reads per sample. Between 80 and 95% of the sequences were taxonomically assigned to genus level. The Shannon diversity indexes ranged from 2.273 to 2.296 and between 346 and 535 species were identified per sample. Results from 16S rRNA sequencing confirmed the results from F. nucleatum qPCR. Sequence data on species and genus level illustrated higher relative abundance of F. nucleatum and Fusobacterium spp. in the cancer group compared with the control group (Fig. 5). 16S rRNA sequencing data could not be used to verify the presence or quantities of E. coli toxin genes, but the data illustrated similar quantities of Escherichia sp. between the cancer and control groups (Fig. 5).
Discussion
The aims of the present study were to determine the levels of F. nucleatum and selected E. coli toxin genes in stool and mucosa samples from Norwegian colon cancer patients and to evaluate the use of F. nucleatum and E. coli quantitative PCRs as potential microbiome-based biomarkers for detection of colorectal cancer in its early stages.
We identified significantly higher levels of F. nucleatum and Fusobacterium spp. in stool samples from the cancer group compared with the control group and the polyp group. Furthermore, F. nucleatum was more frequently detected in biopsies from the cancer patients. This is in line with previous studies from the USA, Canada, China, and Japan [22,23,24, 31], and it illustrates that F. nucleatum is a microbe associated with a subgroup of colorectal cancer patients also in Norway.
A number of previous studies have dismissed stool as sample material since this is not reflective of the microenvironment in colonic mucosa. A study from Mira-Pascual et al. illustrated more similar microbial communities in stool samples between control, adenoma, and CRC patients than within biopsy samples from the same patient [32]. Although stool samples do not represent the same microbiota as tumor tissues in neither richness nor diversity, there is increasing evidence that important microbial signatures associated with CRC are reflected in stool [13, 22,23,24]. To identify cancer patients with high levels of F. nucleatum in stool, a ∆Ct (CtFn − Ct16S rRNA) cutoff of 12 was set. Using this approach, high levels of F. nucleatum in stools were detected in 35% of cancer patients in the present study. These patients were diagnosed in early cancer stages. Three patients with distant metastatic spread had lower levels of F. nucleatum in stool (Fig. 2). No difference in F. nucleatum abundance was identified in stool samples between the polyp group and the control group, and F. nucleatum was identified in the mucosa from only one patient from the polyp group; hence, we find no evidence for F. nucleatum correlation with polyp development. High levels of F. nucleatum in stool may therefore have potential as a non-invasive biomarker for detection of colorectal cancer in its early stages. However, our results indicated that a quantitative PCR targeting fecal amounts of F. nucleatum was capable to identify just a subset of colorectal cancer patients, and therefore it can be used in combination with other biomarkers. The subset of F. nucleatum–high patients may represent a distinct CRC group and will be the focus of further research.
Colorectal cancer is typically classified into rectal, distal colon, and proximal colon cancers, which are known to have different clinical, pathological, and epidemiological features [33]. Several studies have shown that F. nucleatum is frequently associated with CpG island methylator phenotype–high lesions, which typically occur in the proximal colon (reviewed in [34]). Results from the present study suggested that F. nucleatum was detected more frequently and in several positions of the colon when the tumor was located in the proximal part of the colon compared with the distal part and rectum. Comparison of F. nucleatum quantities in tumors versus the other sample sites indicated, but did not confirm statistically, a higher F. nucleatum abundance in the tumor site compared with the other sample positions. Due to the bias of more frequent detection of F. nucleatum in the proximal part of the colon, the comparisons should be performed separately for groups with different tumor locations. This will be the focus in ongoing studies in our group. Our results are in line with Mima et al. who illustrated that the proportion of F. nucleatum–high colorectal cancers gradually increased from the rectum to the cecum [35]. The authors argue that pathogenic influence of the microbiota on neoplastic and immune cells varies along the proximal to distal axis of the colorectum and challenges the prevailing two-colon (proximal vs. distal) dichotomy paradigm [35].
Although E. coli has been suggested to play a role in the pathogenesis of colorectal cancer, the precise role of this diverse species and its toxin genes are not clearly defined. This study found no significant difference in the number or the relative quantities of E. coli toxin genes in stool or biopsy samples between the patient groups, neither did we identify differences between tumor tissue and healthy tissue in CRC patients, as previously suggested [15, 36]. Dutilh et al. found that enterobacterial toxins were among the most highly expressed in CRC tissues while Enterobacteriaceae were not among the most abundant species [37]. This suggests that extended metagenomic and metatranscriptomic studies are needed to identify the exact role of Enterobacteriaceae and toxin contribution to colorectal cancer.
To our knowledge, this is the first study to evaluate the levels of F. nucleatum and E. coli toxins in Norwegian patients. In the present study, we have included two different control groups, one group with adenomatous polyps and one control group without any pathological findings, although scheduled for colonoscopy based on symptoms. These are patients that need to be differentiated from colorectal cancer patients and are therefore considered representative controls. A healthy control group without any symptoms would possibly reflect larger differences between the groups but would leave a gap of knowledge relating to patients suffering from similar symptoms as colorectal cancer patients.
There are some limitations of the present study. The number of patients included in each group (n = 22–25) is low, and the approach therefore needs to be tested on a larger sample material. Furthermore, F. nucleatum is a heterogeneous species with five proposed subspecies: ss animalis, ss fusiforme, ss nucleatum, ss polymorphum, and ss vincentii. The primers used to detect F. nucleatum do not differentiate between the five subspecies, which could have different contributory effects to colorectal cancer. There could also potentially be cross-reactions between the different species of the Fusobacterium genus.
Conclusion
Approximately 35% of the cancer patients and none of the control patients in the present study were identified as F. nucleatum-high using qPCR on stool samples with a ΔCt cutoff of 12. F. nucleatum qPCR could potentially be included in a larger panel of non-invasive stool biomarkers for detection of colorectal cancer, but further studies are necessary to confirm this hypothesis.
Change history
28 November 2019
Two affiliations of author John Christopher Noone were not included in the original article and have been added here. Also, Acknowledgments of the originally published article is not complete. Please see the corrected section below.
28 November 2019
Two affiliations of author John Christopher Noone were not included in the original article and have been added here. Also, Acknowledgments of the originally published article is not complete. Please see the corrected section below.
References
Ahlquist DA (2015) Multi-target stool DNA test: a new high bar for noninvasive screening. Dig Dis Sci 60(3):623–633
Niederreiter L, Adolph TE, Tilg H (2018) Food, microbiome and colorectal cancer. Dig Liver Dis 50(7):647–652
Tilg H, Adolph TE, Gerner RR, Moschen AR (2018) The intestinal microbiota in colorectal cancer. Cancer Cell 33(6):954–964
Louis P, Hold GL, Flint HJ (2014) The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12(10):661–672
Mima K, Ogino S, Nakagawa S, Sawayama H, Kinoshita K, Krashima R et al (2017) The role of intestinal bacteria in the development and progression of gastrointestinal tract neoplasms. Surg Oncol 26(4):368–376
Leung A, Tsoi H, Yu J (2015) Fusobacterium and Escherichia: models of colorectal cancer driven by microbiota and the utility of microbiota in colorectal cancer screening. Expert Rev Gastroenterol Hepatol 9(5):651–657
Bashir A, Miskeen AY, Bhat A, Fazili KM, Ganai BA (2015) Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. Eur J Cancer Prev 24(5):373–385
Bashir A, Miskeen AY, Hazari YM, Asrafuzzaman S, Fazili KM (2016) Fusobacterium nucleatum, inflammation, and immunity: the fire within human gut. Tumour Biol 37(3):2805–2810
Ma CT, Luo HS, Gao F, Tang QC, Chen W (2018) Fusobacterium nucleatum promotes the progression of colorectal cancer by interacting with E-cadherin. Oncol Lett 16(2):2606–2612
Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H et al (2016) Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol 22(2):557–566
Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW (2013) Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe 14(2):195–206
Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M et al (2013) Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14(2):207–215
Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V et al (2014) Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 33(8):1381–1390
Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D et al (2017) Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358(6369):1443–1448
Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A et al (2013) High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 8(2):56964
Raisch J, Buc E, Bonnet M, Sauvanet P, Vazeille E, de Vallee A et al (2014) Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J Gastroenterol 20(21):6560–6572
Arthur JC, Gharaibeh RZ, Muhlbauer M, Perez-Chanona E, Uronis JM, McCafferty J et al (2014) Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun 5:4724
Gomez-Moreno R, Robledo IE, Baerga-Ortiz A (2014) Direct detection and quantification of bacterial genes associated with inflammation in DNA isolated from stool. Adv Microbiol 4(15):1065–1075
Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM et al (2012) Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22(2):292–298
Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J et al (2012) Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22(2):299–306
McCoy AN, Araujo-Perez F, Azcarate-Peril A, Yeh JJ, Sandler RS, Keku TO (2013) Fusobacterium is associated with colorectal adenomas. PLoS One 8(1):53653
Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y et al (2013) Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol 66(2):462–470
Baxter NT, MTt R, Rogers MA, Schloss PD (2016) Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med 8(1):37
Suehiro Y, Sakai K, Nishioka M, Hashimoto S, Takami T, Higaki S et al (2017) Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann Clin Biochem 54(1):86–91
Noone JC, Bemanian V, Smith Tunsjø H (2017) DNA-ekstraksjon: Mengde er ikke synonymt med mangfold. Bioingeniøren 6:20–26
Moen AE, Tannaes TM, Vatn S, Ricanek P, Vatn MH, Jahnsen J et al (2016) Simultaneous purification of DNA and RNA from microbiota in a single colonic mucosal biopsy. BMC research notes 9:328
Muller D, Greune L, Heusipp G, Karch H, Fruth A, Tschape H et al (2007) Identification of unconventional intestinal pathogenic Escherichia coli isolates expressing intermediate virulence factor profiles by using a novel single-step multiplex PCR. Appl Environ Microbiol 73(10):3380–3390
Brukner I, Longtin Y, Oughton M, Forgetta V, Dascal A (2015) Assay for estimating total bacterial load: relative qPCR normalisation of bacterial load with associated clinical implications. Diagn Microbiol Infect Dis 83(1):1–6
Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA et al (1985) Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230(4732):1350–1354
Herlemann DP, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME journal 5(10):1571–1579
Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G et al (2017) Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 66(8):1441–1448
Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A et al (2015) Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol 50(2):167–179
Iacopetta B (2001) Are there two sides to colorectal cancer? Int J Cancer 101(5):403–408
Koi M, Okita Y, Carethers JM (2018) Fusobacterium nucleatum infection in colorectal cancer: linking inflammation, DNA mismatch repair and genetic and epigenetic alterations. J anus, rectum and colon 2(2):37–46
Mima K, Cao Y, Chan AT, Qian ZR, Nowak JA, Masugi Y et al (2016) Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin Transl Gastroenterol 7(11):200
Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ et al (2012) Intestinal inflammation targets cancer-inducing activity of the microbiota (2012). Science 338(6103):120–123
Dutilh BE, Backus L, van Hijum SA, Tjalsma H (2013) Screening metatranscriptomes for toxin genes as functional drivers of human colorectal cancer. Best Pract Res Cl Ga 27(1):85–99
Acknowledgments
Students at Oslo Metropolitan University (Mia Sandgren et al., Sammy Ramstock et al., and Silje Mathiassen et al.) have contributed to parts of the contents in this manuscript. We thank the Pathology Dept. at Akershus University Hospital for characterization of tumors. We also express our gratitude to biomedical laboratory scientist Eva Smedsrud, Dept. of Multidisciplinary Laboratory Science and Medical Biochemistry, Genetic Unit, for all help with patient inclusion and sample processing.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical considerations
The regional committee for medical and health-related research ethics (REK 2012/1944) and the data protection manager at Ahus have approved the study. Written informed consent was obtained from all participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tunsjø, H.S., Gundersen, G., Rangnes, F. et al. Detection of Fusobacterium nucleatum in stool and colonic tissues from Norwegian colorectal cancer patients. Eur J Clin Microbiol Infect Dis 38, 1367–1376 (2019). https://doi.org/10.1007/s10096-019-03562-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-019-03562-7