Abstract
This study describes, for the first time, the genetic and phenotypic diversity among 93 Streptococcus agalactiae (group B Streptococcus, GBS) isolates collected from Guelma, Algeria and Marseille, France. All strains were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). The molecular support of antibiotic resistance and serotyping were investigated by polymerase chain reaction (PCR). The phylogenetic lineage of each GBS isolate was determined by multilocus sequence typing (MLST) and grouped into clonal complexes (CCs) using eBURST. The isolates represented 37 sequence types (STs), 16 of which were novel, grouped into five CCs, and belonging to seven serotypes. Serotype V was the most prevalent serotype in our collection (44.1 %). GBS isolates of each serotype were distributed among multiple CCs, including cps III/CC19, cps V/CC1, cps Ia/CC23, cps II/CC10, and cps III/CC17. All isolates presented susceptibility to penicillin, whereas resistance to erythromycin was detected in 40 % and tetracycline in 82.2 % of isolates. Of the 37 erythromycin-resistant isolates, 75.7 % showed the macrolide–lincosamide–streptogramin B (MLSB)-resistant phenotype and 24.3 % exhibited the macrolide (M)-resistant phenotype. Constitutive MLSB resistance (46 %) mediated by the ermB gene was significantly associated with the Guelma isolates, whereas the M resistance phenotype (24.3 %) mediated by the mefA/E gene dominated among the Marseille isolates and belonged to ST-23. Tetracycline resistance was predominantly due to tetM, which was detected alone (95.1 %) or associated with tetO (3.7 %). These results provide epidemiological data in these regions that establish a basis for monitoring increased resistance to erythromycin and also provide insight into correlations among clones, serotypes, and resistance genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptococcus agalactiae (group B Streptococcus, GBS) is a Gram-positive species commensal of human gastrointestinal and genitourinary flora and is responsible for severe diseases in susceptible hosts [1]. Moreover, this species can cause life-threatening invasive diseases in pregnant women and newborns [2]. The Centers for Disease Control and Prevention (CDC) recommends a strategy based on intrapartum chemoprophylaxis for pregnant women to decrease GBS infections in neonates [3]. However, in the last several decades, GBS strains have also been associated with invasive disease in non-pregnant adults, the elderly, and patients with underlying medical conditions, such as malignancy, diabetes, or liver disease [4–6]. To prevent S. agalactiae infections in newborns, β-lactams are recommended as a first-line antibiotic prophylaxis in parturient women, and macrolides–lincosamides remain the therapeutic alternative in cases of allergy to β-lactam [7]. However, the use of these antibiotics as alternative agents for prophylaxis is questioned because of increasing trends in the rates of resistance to erythromycin and clindamycin among S. agalactiae [8]. The first erythromycin resistance mechanism reported in GBS was the erm gene-encoded modification of their ribosomal targets via methylation, resulting in cross-resistance to macrolide–lincosamide–streptogramin B (MLSB) antibiotics [9, 10]. Erythromycin resistance can also be due to efflux pumps, mediated by the mefA/E gene, which causes resistance to 14- and 15-membered macrolide compounds and produces the so-called M phenotype [11, 12]. The increasing resistance to macrolides observed worldwide emphasizes the need for more detailed studies on GBS macrolide-resistant populations [13, 14]. Similarly, the resistance to tetracycline of GBS is also common and frequently associated with the tetM gene [15], and tetracycline resistance genes are often found on the same mobile genetic elements that carry macrolide resistance genes [16]. Capsular serotyping is the classical method used in epidemiological studies, which defines ten GBS serotypes (Ia, Ib, II–IX) [17]. One of the most important factors involved in virulence is capsular polysaccharide (CPS) [18], and the multivalent CPS–protein conjugate vaccines have been developed in the last decade, raising the possibility of preventing perinatal GBS disease via maternal vaccination [19]. Among the methods applied for GBS typing, several molecular tools have been developed, especially multilocus typing sequence (MLST), which has a high discriminatory power [20]. Indeed, the MLST approach has contributed to a better characterization of GBS isolates and the classification of bacterial genogroups [21] and has been utilized in epidemiological studies of the population genetics of human pathogenic bacteria [20]. More recently, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) has emerged as a powerful, cost-effective, and rapid tool for bacterial identification in clinical laboratories and biotyping [22]. Other aspects of this technology are potentially interesting, such as its contribution to the field of new species characterization and the detection of certain mechanisms of resistance and simultaneous identification of strains belonging to the highly virulent ST-17 or emerging ST-1 clones [23, 24]. Only one report focused on GBS isolates from Algeria, which investigated risk factors associated with newborn infections by GBS and serotyping using the sero-agglutination method [25]. However, in this study, for the first time, we targeted molecular support of antibiotic resistance, molecular serotyping, and the clonality of a collection of GBS clinical isolates from Guelma, Algeria and compared them to those from Marseille, France.
Materials and methods
Bacterial strains
Ninety-three GBS isolates were collected using vaginal swabs in Guelma, Algeria (n = 44), between January 2011 and February 2012 and from different samples, including vaginal swabs (n = 30), urine (n = 10), and blood culture (n = 9), from the microbiology laboratory of Timone Hospital at Marseille, France, between October 2013 and January 2014.
Identification of GBS
All isolates were grown on the selective medium Todd–Hewitt broth (bioMérieux, France) and incubated at 37 °C for 24 h. After that, the isolates were cultivated on Columbia agar base enriched with 5 % sheep blood (bioMérieux, France) in a 10 % CO2 atmosphere. The identification was carried out on the basis of the following criteria: hemolysis on blood agar, negative reaction with catalase reagent, a positive CAMP (Christie–Atkins–Munch–Petersen) test, and Lancefield grouping with type B antisera (Pastorex™ Strep B, Bio-Rad, France). The identification of isolates was confirmed using MALDI-TOF MS (Bruker Daltonics, Germany), and their obtained specific spectra were used to build a main spectrum profile (MSP) dendrogram using Biotyper 3.0 software (Bruker Daltonics, Leipzig, Germany), as described previously [26, 27]. We used MALDI-TOF MS to search for GBS biomarkers associated with ST-1 and ST-17 strains at the species identification stage [23].
Antimicrobial susceptibility
The antibiotic susceptibility of our isolates was assessed using the disk diffusion method on Muller–Hinton agar plates supplemented with 5 % blood (bioMérieux, France), according to the recommendation of the French Society for Microbiology standards (CA-SFM). The tested antibiotics were penicillin G, amoxicillin, gentamicin, tetracycline, erythromycin, clindamycin, pristinamycin, rifampicin, and vancomycin (Oxoid, France). The erythromycin (15 μg)–clindamycin (2 μg) double-disk test was used to determine constitutive MLSB (cMLSB) resistance, inducible MLSB (iMLSB) resistance, and the M resistance phenotype, as previously described [10]. Erythromycin and clindamycin minimum inhibitory concentrations (MICs) were determined using the Etest method (bioMérieux, France).
Molecular characterization of antibiotic resistance-encoding genes
DNA extraction of all isolates was performed using EZ1 Advanced XL extractor with EZ1® DNA Tissue Kit (Qiagen, Courtaboeuf, France) and DNA Bacteria Card (Qiagen), according to the manufacturer’s instructions. Antibiotic resistance genes (ermA, ermB, mefA/E, tetM, tetO, tetK, and tetL) were searched by polymerase chain reaction (PCR), as previously described [28, 29].
Sequencing
All obtained PCR products were purified and sequenced using the BigDye Terminator® v1.1 Cycle Sequencing Kit (Applied Biosystems, Courtaboeuf, France). The sequencing products were then processed using an ABI PRISM 3130 automated sequencer (Applied Biosystems, Foster City, CA, USA) [30]. The obtained sequences were aligned and compared with those in GenBank using the BLAST program against the NCBI and ARG-ANNOT databases [31].
Determination of capsular serotypes
Identification of the capsular type (Ia, Ib, II–IX) of all GBS isolates was performed by the multiplex PCR assay according to a previously published procedure [17]. Non-typeable isolates were designated as NT.
Multilocus sequence typing (MLST)
Seven housekeeping genes were used for GBS characterization using the MLST scheme, including adhP (alcohol dehydrogenase), pheS (phenylalanyl transfer RNA synthetase), atr (amino-acid transporter protein), glnA (glutamine synthetase), sdhA (L-serine dehydratase), glcK (glucose kinase), and tkt (transketolase), as described previously [20]. An online database (http://pubmlst.org/sagalactiae/) was used for assigning allele numbers and sequence types (STs). GBS 2603 (serotype V; ST-110) was used as a reference strain. The sequences of the seven loci obtained were concatenated and used to build a phylogenetic tree by the MEGA5 program with the neighbor-joining method [32]. The eBURST program was used to group isolates into clonal complexes (CCs), the members of which shared at least six of the seven MLST loci [33]; otherwise, an ST was considered a singleton.
Statistical analysis
Fisher’s exact test was used to evaluate differences in the distributions of isolates using Epi Info software version 7, according to CDC recommendations (http://www.openepi.com/Menu/OE_Menu.htm). A p-value ≤ 0.05 was considered significant.
Results
Bacterial isolation and identification
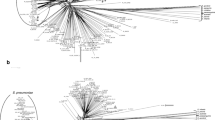
The study collection consisted of 93 isolates of S. agalactiae, 44 from Guelma, Algeria and 49 from Marseille, France. The most prevalent source of isolates from Marseille was vaginal samples (30/49; 61.2 %), followed by urine (20.4 %) and blood cultures (18.4 %) (Figs. 1 and 2). Using MALDI-TOF MS, all GBS isolates were correctly identified at the species level, with score values >2.1 using Bruker Biotyper 3.0 software. The MSP dendrogram of our isolates revealed three clusters according to the geographical origin to an arbitrary cut-off at a distance level of 700, as shown in Fig. 1. The 49 isolates from Marseille contained 36 S. agalactiae grouped into two clusters as follows: cluster C1 (vaginal; n = 8 samples) and cluster C2 (vaginal; n = 9, blood; n = 9, and urine; n = 10 samples). Conversely, cluster C3 contained 42 isolates of S. agalactiae from Guelma (vaginal samples). Clusters C1 and C2 were significantly associated with Marseille isolates, whereas cluster C3 was associated with Algerian isolates (p < 10−6) (Fig. 1). Additionally, MALDI-TOF MS identified a 6250-Da protein specific to sequence type ST-1 strains (and no mass peak at 6888 Da) (n = 15) and a 7625-Da protein specific to ST-17 strains (n = 3). However, these two peaks were also present in other STs, including a peak at 6250 in ST-460 (n = 1) and ST-693 (n = 2), and a peak at 7625 in ST-106 (n = 1), as shown in Fig. 2.
Neighbor-joining tree of 37 STs constructed from multilocus sequence typing (MLST) analysis found using the MEGA5 program among 93 S. agalactiae isolates. The dendrogram shows genetic diversity and phenotypic characterization of macrolide and tetracycline S. agalactiae resistant isolates, genetic relationships among the different serotypes, resistance genes, sequences type, and clonal complexes. CC, clonal complexes; ST, sequence type; *, new ST; NT, not typeable; c/iMLSB, constitutive/induced macrolides, lincosamides, and streptogramins B-type resistance; ermA/B, erythromycin ribosome methylase A/B; M, macrolides resistance phenotype; Mef, macrolide efflux; RF, reference strain
Antimicrobial susceptibility testing
Among the 93 clinical isolates, we found that they were all susceptible to penicillin. However, we also found that 34 out of 74 (46 %) vaginal isolates (74/93) were resistant to erythromycin (20 out of 44 isolates in Guelma versus 14 out of 30 in Marseille, p = 0.91). Resistance to clindamycin was found in 37.8 %. All the resistance phenotypes detected are provided in Table 1. For tetracycline, 100 % of the isolates from Guelma were resistant versus 86.6 % from Marseille (Table 1).
Macrolide resistance genotypes and tetracycline resistance determinants
Among 34 out of 74 isolates resistant to erythromycin, the most prevalent determinant of resistance was the ermB gene (73.5 %), and significantly more isolates carrying the ermB gene were isolated in Guelma (n = 18) compared to Marseille (n = 7; p = 0.009). Conversely, the mefA/E gene (14.7 %) was significantly more frequently detected in the isolates from Marseille (n = 4) compared to those from Guelma (n = 1; p = 0.05). An exception occurred for four strains that exhibited a combination of ermA/ermB genes (8.8 %), represented by one isolate in Guelma and two isolates in Marseille, and ermA/mefA/E genes (3 %), represented only by one isolate in Marseille. All ermA, ermB, and mefA/E gene-positive isolates expressed iMLSB, cMLSB, and the M resistance phenotypes, respectively, as shown in Table 1. The tetM gene was detected in 100 % of the isolates from Guelma and 84.6 % of the isolates from Marseille; tetO was found in only 3.8 % of the isolates, and a co-occurrence of both tetM and tetO was only found in 11.6 % of the Marseille isolates. In contrast, tetK and tetL were not detected in our isolates (Tables 1 and 2).
Serotype identification
The isolates studied represented seven capsular types. The most represented serotypes isolated in Guelma and Marseille were type V (44.6 %), followed by serotypes II and III (19 % both) and serotype Ia (8 %). Moreover, serotypes IV, VII, and VIII (1.3 % each) were found only in Marseille isolates. Finally, 5.4 % of the isolates from Marseille were non-typeable (p = 0.005) compared to Guelma, as shown in Table 2 and Fig. 2.
Multilocus sequence typing (MLST)
The MLST results of the S. agalactiae isolates were analyzed and presented in a phylogenetic tree; isolates demonstrated the existence of different genetic lineages (Fig. 2). A total of 37 individual STs were identified (Fig. 2). Moreover, 16 novel STs were detected: seven in Guelma and nine in Marseille. These new STs were entered in the S. agalactiae MLST database (STs 685 to 700). Thirty-one of the STs were clustered into five CCs, and six were singleton STs (Fig. 2). Among these, 93.5 % (87/93) of the isolates were found within five CCs; CC1, CC10, CC17, CC19, and CC23; 6.5 % (6/93) of the isolates identified were not part of a cluster. The most prevalent of these complexes was CC19 (including STs: 19, 28, 106, 158, 190, 241, 233, 521, 677, 685*, 687*, 691*; 35.5 %), followed by CC1 (including STs: 1, 321, 460, 689*, 693*, 694*, 697*, 700*; 24.7 %), which regrouped all vaginal isolates from Guelma and Marseille, then CC10 (18.3 %), CC23 (11.8 %), and CC17 (3.2 %), which were more common among the Marseille isolates (Fig. 2).
Correlation between phenotype, genotype, serotype, and MLST analysis
Our results showed that the cMLSB phenotype was significantly more common among the Guelma isolates as compared to the Marseille isolates (p = 0.03) and carried the ermB gene (p = 0.009), whereas the M phenotype was associated with the Marseille isolates (p = 0.02), expressed the mefA/E gene, and belonged to ST-23 (p = 10−7) (Fig. 2). The MLST analysis showed that 82.6 % of serotype V GBS clustered into CC1 (ST-1/V, p = 0.012), 72.7 % of serotype Ia GBS clustered into CC23 (ST-23/Ia, p = 10−7), 100 % of serotype III GBS clustered into CC17 (ST-17, p = 10−4), and 70.6 % of serotype II and the non-typeable capsular serotype clustered into CC10 (ST-10/II, p = 2.10−5 and ST-8/NT, p = 7.10−6). In contrast, 45.4 and 42.4 %, respectively, of serotype V and serotype III isolates clustered into CC19 (ST-19/V, p = 0.7312 and ST-19/III, p = 4.10−7) (Table 2 and Fig. 2).
Discussion
This report presents, for the first time, a comprehensive molecular analysis of GBS isolates circulating in Guelma, Algeria and Marseille, France. The MSP dendrogram clustering of isolates using MALDI-TOF MS [27] showed significant clusters according to the geographical source. Such grouping of isolates has recently been reported for Klebsiella pneumoniae isolates [22]. Moreover, Lartigue et al. report that MALDI-TOF MS analysis was also able to identify strains belonging to the highly virulent ST-17 clone or to the emerging ST-1 clone [23]. However, this was not true in our hands, as four isolates outside these two STs harbored these peaks.
One of the main objectives of this investigation was to determine the genetic basis of antibiotic resistance. In this study, all strains remained uniformly susceptible to penicillin [5, 15, 34]. However, the overall rate of erythromycin resistance among our isolates analyzed was 40 % (45.4 and 34.7 % in Guelma and Marseille, respectively). Such a level of resistance has been reported in Taiwan (44 %), Tunisia (40 %), Morocco (38.5 %), Switzerland (30 %), France (20.2–35.3 %), the USA (32–54 %) [12, 14, 16, 35–38], and extremely high in China (85.7 %) [39]. Due to this high level of resistance, the CDC guidelines no longer recommend erythromycin [40]. The increasing emergence of resistance to macrolides among GBS is a therapeutic problem among patients allergic to β-lactams. This observation emphasizes the need for the continuous monitoring of antimicrobial susceptibility profiles.
In our study, there was a predominance of the cMLSB phenotype in Guelma isolates mediated by the ermB gene, whereas the M phenotype was more common in Marseille isolates, which carried the mefA/E genes. A predominance of the MLSB phenotype has been reported in Australia, Switzerland, and Tunisia [8, 14, 16], whereas in Brazil and Italy, the cMLSB and M phenotypes were detected with equal frequencies [13, 19]. In France (Paris), the iMLSB phenotype was more common in 2001, yet cMLSB was more dominant from 2007 to 2010 [38, 41]. Interestingly, we detected a coexistence of ermA/ermB and ermA/mefA/E genes in iMLSB and M phenotypes, respectively. The co-occurrence of both genes has been documented previously [10, 42].
We also report a high rate of tetracycline resistance in our study (82.2 %), as already described in Tunisia (97.3 %), France (94 %), Malaysia (71.8 %), and Italy (69.9 %) [5, 7, 16, 43]. Moreover, according to our results, the tetM gene has spread throughout all strains. Thus, we also observed that the majority of isolates carrying the ermB gene also harbored the tetM gene (96.4 %). The contemporary presence of both genes was previously described by Gherardi et al. [15]. Seroprevalence studies are an important measure for determining the incidence and proportion of serotypes that are circulating in a given population [44].
Among the 93 GBS strains studied, all capsular serotypes except VI and IX were found. Our data show that serotype V was the predominant one among isolates (44.1 %), as also reported in Kuwait [45] and Japan [46]. However, other studies showed a predominance of other serotypes, such as serotype III in Morocco and France, serotype IV in the United Arab Emirates, serotype Ia in Brazil, and serotypes VI–VIII in Japan [2, 19, 34, 47]. Furthermore, global serotyping distribution studies have shown that the serotype distribution of GBS varies both geographically and over time [15, 44]. All serotypes (I, III, and V) are frequently associated worldwide with GBS infections [44, 48–50]. The proportion of NT strains showed higher percentages (5.4 %) among Marseille isolates, which could be the result of acquisition of an uncharacterized capsule gene cluster or mutations in capsule genes [44, 51]. Usually, erythromycin-resistant isolates were more frequently found in serotype V [2, 52]. However, in our study, we did not find this association [13], though an association between ST-23 and the M phenotype was established [47].
The population structures of GBS exhibit a remarkable clonal population, with large differences within groups of clones. This is the first report of MLST analysis in GBS strains circulating in Algeria. In this study, 37 STs were identified. The main STs identified in this study have also been observed as major STs for strains isolated among large collections during infectious diseases [19, 47, 53]. Despite this high genetic diversity, all STs found were grouped into five CCs, i.e., CC1, CC10, CC23, CC17, and CC19, which have been previously identified worldwide [54, 55]. The most prevalent of these CCs was CC19, followed by CC1. In addition, the main STs included in these two CCs, such as ST-19 and ST-1, were over-represented among carriage isolates of GBS [20]. Such diverse clonal populations have also been found in other countries, including Italy, Poland, France, the USA, and Senegal [15, 47, 54–56]. Therefore, GBS from these CCs have been shown to cause the majority of both neonatal and adult GBS infections [54, 55]. The diversity of the genetic lineages between countries suggests that most diseases combined with GBS are caused by certain clonal lineages [6].
In conclusion, the data obtained in this study shed new light on the need for a more rigorous characterization and detection of correlations among serotypes, resistance genes, and clonal clusters of GBS isolates circulating in the study areas. Comparative genetic studies of S. agalactiae will be essential to perform epidemiological comparisons between countries and the evolution of isolates, as well as for vaccine development. Finally, as erythromycin resistance rates in GBS have increased, local antibiotic resistance surveillance is advisable in guiding empirical antibiotic therapy to prevent the development of such infections. Further epidemiological studies in other cities in France and in other Algerian cities are needed to support our findings.
References
Wang NY, Patras KA, Seo HS, Cavaco CK, Rösler B, Neely MN, Sullam PM, Doran KS (2014) Group B streptococcal serine-rich repeat proteins promote interaction with fibrinogen and vaginal colonization. J Infect Dis 210(6):982–991
Otaguiri ES, Morguette AE, Tavares ER, dos Santos PM, Morey AT, Cardoso JD, Perugini MR, Yamauchi LM, Yamada-Ogatta SF (2013) Commensal Streptococcus agalactiae isolated from patients seen at University Hospital of Londrina, Paraná, Brazil: capsular types, genotyping, antimicrobial susceptibility and virulence determinants. BMC Microbiol 13:297
Mukhopadhyay S, Eichenwald EC, Puopolo KM (2013) Neonatal early-onset sepsis evaluations among well-appearing infants: projected impact of changes in CDC GBS guidelines. J Perinatol 33(3):198–205
Skoff TH, Farley MM, Petit S, Craig AS, Schaffner W, Gershman K, Harrison LH, Lynfield R, Mohle-Boetani J, Zansky S, Albanese BA, Stefonek K, Zell ER, Jackson D, Thompson T, Schrag SJ (2009) Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990–2007. Clin Infect Dis 49(1):85–92
Eskandarian N, Ismail Z, Neela V, van Belkum A, Desa MN, Amin Nordin S (2015) Antimicrobial susceptibility profiles, serotype distribution and virulence determinants among invasive, non-invasive and colonizing Streptococcus agalactiae (group B streptococcus) from Malaysian patients. Eur J Clin Microbiol Infect Dis 34(3):579–584
Tien N, Ho CM, Lin HJ, Shih MC, Ho MW, Lin HC, Lin HS, Chang CC, Lu JJ (2011) Multilocus sequence typing of invasive group B Streptococcus in central area of Taiwan. J Microbiol Immunol Infect 44(6):430–434
Poyart C, Jardy L, Quesne G, Berche P, Trieu-Cuot P (2003) Genetic basis of antibiotic resistance in Streptococcus agalactiae strains isolated in a French hospital. Antimicrob Agents Chemother 47(2):794–797
Garland SM, Cottrill E, Markowski L, Pearce C, Clifford V, Ndisang D, Kelly N, Daley AJ; Australasian Group for Antimicrobial Resistance-GBS Resistance Study Group (2011) Antimicrobial resistance in group B streptococcus: the Australian experience. J Med Microbiol 60(Pt 2):230–235
Buter CC, Mouton JW, Klaassen CH, Handgraaf CM, Sunnen S, Melchers WJ, Sturm PD (2010) Prevalence and molecular mechanism of macrolide resistance in beta-haemolytic streptococci in The Netherlands. Int J Antimicrob Agents 35(6):590–592
Culebras E, Rodriguez-Avial I, Betriu C, Redondo M, Picazo JJ (2002) Macrolide and tetracycline resistance and molecular relationships of clinical strains of Streptococcus agalactiae. Antimicrob Agents Chemother 46(5):1574–1576
DiPersio LP, DiPersio JR, Beach JA, Loudon AM, Fuchs AM (2011) Identification and characterization of plasmid-borne erm(T) macrolide resistance in group B and group A Streptococcus. Diagn Microbiol Infect Dis 71(3):217–223
DiPersio LP, DiPersio JR (2006) High rates of erythromycin and clindamycin resistance among OBGYN isolates of group B Streptococcus. Diagn Microbiol Infect Dis 54(1):79–82
Pinto TC, Costa NS, Vianna Souza AR, Silva LG, Corrêa AB, Fernandes FG, Oliveira IC, Mattos MC, Rosado AS, Benchetrit LC (2013) Distribution of serotypes and evaluation of antimicrobial susceptibility among human and bovine Streptococcus agalactiae strains isolated in Brazil between 1980 and 2006. Braz J Infect Dis 17(2):131–136
Capanna F, Emonet SP, Cherkaoui A, Irion O, Schrenzel J, Martinez de Tejada B (2013) Antibiotic resistance patterns among group B Streptococcus isolates: implications for antibiotic prophylaxis for early-onset neonatal sepsis. Swiss Med Wkly 143:w13778
Gherardi G, Imperi M, Baldassarri L, Pataracchia M, Alfarone G, Recchia S, Orefici G, Dicuonzo G, Creti R (2007) Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among group B streptococci in Italy. J Clin Microbiol 45(9):2909–2916
Hraoui M, Boutiba-Ben Boubaker I, Rachdi M, Slim A, Ben Redjeb S (2012) Macrolide and tetracycline resistance in clinical strains of Streptococcus agalactiae isolated in Tunisia. J Med Microbiol 61(Pt 8):1109–1113
Imperi M, Pataracchia M, Alfarone G, Baldassarri L, Orefici G, Creti R (2010) A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J Microbiol Methods 80(2):212–214
Lemire P, Houde M, Lecours MP, Fittipaldi N, Segura M (2012) Role of capsular polysaccharide in Group B Streptococccus interactions with dendritic cells. Microbes Infect 14(12):1064–1076
De Francesco MA, Caracciolo S, Gargiulo F, Manca N (2012) Phenotypes, genotypes, serotypes and molecular epidemiology of erythromycin-resistant Streptococcus agalactiae in Italy. Eur J Clin Microbiol Infect Dis 31(8):1741–1747
Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS, Kunst F, Glaser P, Rusniok C, Crook DW, Harding RM, Bisharat N, Spratt BG (2003) Multilocus sequence typing system for group B streptococcus. J Clin Microbiol 41(6):2530–2536
Lanotte P, Perivier M, Haguenoer E, Mereghetti L, Burucoa C, Claverol S, Atanassov C (2013) Proteomic biomarkers associated with Streptococcus agalactiae invasive genogroups. PLoS One 8(1):e54393
Berrazeg M, Diene SM, Drissi M, Kempf M, Richet H, Landraud L, Rolain JM (2013) Biotyping of multidrug-resistant Klebsiella pneumoniae clinical isolates from France and Algeria using MALDI-TOF MS. PLoS One 8(4):e61428
Lartigue MF, Kostrzewa M, Salloum M, Haguenoer E, Héry-Arnaud G, Domelier AS, Stumpf S, Quentin R (2011) Rapid detection of “highly virulent” Group B Streptococcus ST-17 and emerging ST-1 clones by MALDI-TOF mass spectrometry. J Microbiol Methods 86(2):262–265
Kempf M, Bakour S, Flaudrops C, Berrazeg M, Brunel JM, Drissi M, Mesli E, Touati A, Rolain JM (2012) Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7(2):e31676
Ait AA, Hamidechi AM (2003) Incidence of group b streptococcus (GBS) in neonates born in Constantine (Algeria) and two of its suburbs usefullness of serotyping. Med Mal Infect 33(8):417–421
Seng P, Abat C, Rolain JM, Colson P, Lagier JC, Gouriet F, Fournier PE, Drancourt M, La Scola B, Raoult D (2013) Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 51(7):2182–2194
Lartigue MF, Héry-Arnaud G, Haguenoer E, Domelier AS, Schmit PO, van der Mee-Marquet N, Lanotte P, Mereghetti L, Kostrzewa M, Quentin R (2009) Identification of Streptococcus agalactiae isolates from various phylogenetic lineages by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 47(7):2284–2287
Malhotra-Kumar S, Lammens C, Piessens J, Goossens H (2005) Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob Agents Chemother 49(11):4798–4800
Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L (1996) Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother 40(11):2562–2566
Belbel Z, Chettibi H, Dekhil M, Ladjama A, Nedjai S, Rolain JM (2014) Outbreak of an armA methyltransferase-producing ST39 Klebsiella pneumoniae clone in a pediatric Algerian Hospital. Microb Drug Resist 20(4):310–315
Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM (2014) ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58(1):212–220
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG (2004) eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186(5):1518–1530
Aitmhand R, Moustaoui N, Belabbes H, Elmdaghri N, Benbachir M (2000) Serotypes and antimicrobial susceptibility of group B streptococcus isolated from neonates in Casablanca. Scand J Infect Dis 32(3):339–340
Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ; Active Bacterial Core surveillance/Emerging Infections Program Network (2008) Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA 299(17):2056–2065
Sahnoun O, Ben Abdallah H, Noomen S, Ben Elhadj Khélifa A, Mastouri M (2007) Antimicrobial susceptibility of Streptococcus agalactiae strains in Monastir. Med Mal Infect 37(11):734–737
Janapatla RP, Ho YR, Yan JJ, Wu HM, Wu JJ (2008) The prevalence of erythromycin resistance in group B streptococcal isolates at a University Hospital in Taiwan. Microb Drug Resist 14(4):293–297
Tazi A, Morand PC, Réglier-Poupet H, Dmytruk N, Billoët A, Antona D, Trieu-Cuot P, Poyart C (2011) Invasive group B streptococcal infections in adults, France (2007–2010). Clin Microbiol Infect 17(10):1587–1589
Wang P, Tong JJ, Ma XH, Song FL, Fan L, Guo CM, Shi W, Yu SJ, Yao KH, Yang YH (2015) Serotypes, antibiotic susceptibilities, and multi-locus sequence type profiles of Streptococcus agalactiae isolates circulating in Beijing, China. PLoS One 10(3):e0120035
Verani JR, McGee L, Schrag SJ; Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention (CDC) (2010) Prevention of perinatal group B streptococcal disease—revised guidelines from CDC, 2010. MMWR Recomm Rep 59(RR-10):1–36
Fitoussi F, Loukil C, Gros I, Clermont O, Mariani P, Bonacorsi S, Le Thomas I, Deforche D, Bingen E (2001) Mechanisms of macrolide resistance in clinical group B streptococci isolated in France. Antimicrob Agents Chemother 45(6):1889–1891
Betriu C, Culebras E, Gómez M, Rodríguez-Avial I, Sánchez BA, Agreda MC, Picazo JJ (2003) Erythromycin and clindamycin resistance and telithromycin susceptibility in Streptococcus agalactiae. Antimicrob Agents Chemother 47(3):1112–1114
Lambiase A, Agangi A, Del Pezzo M, Quaglia F, Testa A, Rossano F, Martinelli P, Catania MR (2012) In vitro resistance to macrolides and clindamycin by Group B Streptococcus isolated from pregnant and nonpregnant women. Infect Dis Obstet Gynecol 2012:913603
Ippolito DL, James WA, Tinnemore D, Huang RR, Dehart MJ, Williams J, Wingerd MA, Demons ST (2010) Group B streptococcus serotype prevalence in reproductive-age women at a tertiary care military medical center relative to global serotype distribution. BMC Infect Dis 10:336
Boswihi SS, Udo EE, Al-Sweih N (2012) Serotypes and antibiotic resistance in Group B streptococcus isolated from patients at the Maternity Hospital, Kuwait. J Med Microbiol 61(Pt 1):126–131
Ueno H, Yamamoto Y, Yamamichi A, Kikuchi K, Kobori S, Miyazaki M (2012) Characterization of group B streptococcus isolated from women in Saitama city, Japan. Jpn J Infect Dis 65(6):516–521
van der Mee-Marquet N, Fourny L, Arnault L, Domelier AS, Salloum M, Lartigue MF, Quentin R (2008) Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J Clin Microbiol 46(9):2906–2911
Abat C, Chaudet H, Raoult D, Colson P (2014) Increasing trend of invasive group B streptococcal infections, Marseille, France. Clin Infect Dis 58(5):750–751
Martins ER, Andreu A, Correia P, Juncosa T, Bosch J, Ramirez M, Melo-Cristino J; Microbiologist Group for the Study of Vertical Transmission Infections from the Catalan Society for Clinical Microbiology and Infectious Diseases (2011) Group B streptococci causing neonatal infections in barcelona are a stable clonal population: 18-year surveillance. J Clin Microbiol 49(8):2911–2918
Martins ER, Melo-Cristino J, Ramirez M; Portuguese Group for the Study of Streptococcal Infections (2012) Dominance of serotype Ia among group B Streptococci causing invasive infections in nonpregnant adults in Portugal. J Clin Microbiol 50(4):1219–1227
Ramaswamy SV, Ferrieri P, Flores AE, Paoletti LC (2006) Molecular characterization of nontypeable group B streptococcus. J Clin Microbiol 44(7):2398–2403
von Both U, Ruess M, Mueller U, Fluegge K, Sander A, Berner R (2003) A serotype V clone is predominant among erythromycin-resistant Streptococcus agalactiae isolates in a southwestern region of Germany. J Clin Microbiol 41(5):2166–2169
Manning SD, Springman AC, Lehotzky E, Lewis MA, Whittam TS, Davies HD (2009) Multilocus sequence types associated with neonatal group B streptococcal sepsis and meningitis in Canada. J Clin Microbiol 47(4):1143–1148
Sadowy E, Matynia B, Hryniewicz W (2010) Population structure, virulence factors and resistance determinants of invasive, non-invasive and colonizing Streptococcus agalactiae in Poland. J Antimicrob Chemother 65(9):1907–1914
Bohnsack JF, Whiting A, Gottschalk M, Dunn DM, Weiss R, Azimi PH, Philips JB 3rd, Weisman LE, Rhoads GG, Lin FY (2008) Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. Academic Centers from 1995 to 1999. J Clin Microbiol 46(4):1285–1291
Brochet M, Couvé E, Bercion R, Sire JM, Glaser P (2009) Population structure of human isolates of Streptococcus agalactiae from Dakar and Bangui. J Clin Microbiol 47(3):800–803
Acknowledgments
We are very grateful to Linda Hadjadj for the technical assistance. We thank American Journal Experts (AJE) for the English corrections.
Funding source
This work was partly funded by CNRS and IHU Mediterranean Infection.
Conflict of interest
The authors declare no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bergal, A., Loucif, L., Benouareth, D.E. et al. Molecular epidemiology and distribution of serotypes, genotypes, and antibiotic resistance genes of Streptococcus agalactiae clinical isolates from Guelma, Algeria and Marseille, France. Eur J Clin Microbiol Infect Dis 34, 2339–2348 (2015). https://doi.org/10.1007/s10096-015-2487-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-015-2487-6