Abstract
We explore the genetic diversity of class D oxacillinases, including OXA-23, -24 (-40), -58 and, particularly, the intrinsic OXA-51-like genes, among multidrug-resistant (MDR) Acinetobacter baumannii strains from inpatients at a tertiary care hospital in Riyadh, Saudi Arabia. Sequence-based typing (SBT) of the OXA-51-like gene was carried out on 253 isolates. Selected isolates (n = 66) were subjected to multilocus sequence typing (MLST). The polymerase chain reaction (PCR) typing results showed that all isolates (n = 253) contained the OXA-51-like and OXA-23 genes. However, the OXA-58 gene was detected in five isolates. Further, none of the isolates had the OXA-40 (identical to the OXA-24) gene. SBT revealed a high OXA-51-like genotypic diversity and showed that all isolates were clustered into four main groups: OXA-66 (62.3 %), followed by OXA-69 (19.1 %), OXA-132 (7.6 %) and other OXA-51-like genes (10.3 %), including OXA-79, -82, -92, -131 and -197. MLST revealed four main sequence types (STs), 2, 19, 20 and 25, among the isolates, in addition to six isolates with newly designated ST194–ST197 singletons. Further, a high prevalence (81.4 %) of OXA-66 and OXA-69-like genes in A. baumannii was identified. More studies are essential in order to explore the molecular mechanisms that confer carbapenem-resistant phenotypes for A. baumannii isolates and to investigate the genetic diversity of other OXA-D genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Healthcare-associated infections (HAIs) due to Acinetobacter baumannii are an increasing problem globally, including countries of the Gulf Cooperation Council (GCC) [1, 2]. A. baumannii is widespread in hospitals and the healthcare environment, and plays an important role as a causative agent for HAIs [3]. OXA-51-like genes are genetically diverse and have been identified worldwide, including Europe, North and South Americas, and the Far East [4]. Recent publications from neighbouring countries have identified the prevalence of A. baumannii in HAIs and have further identified the presence of OXA-51 and OXA-23 among their isolates [5, 6]. However, the prevalence and genetic diversity of OXA-51-like oxacillinases in the Middle East in general and Saudi Arabia in particular have not been explicitly described [7]. The purpose of the current study was to identify the genetic diversity of OXA-51-like genes using sequence-based typing (SBT) and multilocus sequence typing (MLST) among multidrug-resistant (MDR) A. baumannii in Saudi Arabia [8].
Methods
Bacterial identification and susceptibility testing
The clinical isolates used in this study were collected at the microbiology laboratory of King Abdulaziz Medical City (KAMC) hospital in Riyadh between 2006 and 2008. Isolates that exhibited aerobic growth, and were catalase-positive and oxidase-negative, were further identified as Acinetobacter by commercial biochemical system MicroScan® WalkAway (Siemens Healthcare). Isolates were initially identified at the species level using MicroScan and were then confirmed by the RNA polymerase β-subunit (rpoB) gene polymerase chain reaction (PCR) and sequencing [2]. Isolates were tested for their minimum inhibitory concentrations (MICs) using the Clinical and Laboratory Standards Institute (CLSI) guidelines [9]. All results that exhibited resistance were confirmed by the Etest® (bioMérieux) to detect the exact MIC value. Only one isolate per patient per year was included in the current analysis. Quality control was performed by testing these same antimicrobials against Escherichia coli ATCC 25922, E. coli ATCC 35218 and Pseudomonas aeruginosa ATCC 27853. The isolates were defined as MDR if they were found to be resistant to three or more classes of antibiotics, including: β-lactam/β-lactamase inhibitors, combinations of extended-spectrum cephalosporins, aminoglycosides, fluoroquinolones and carbapenems [10].
DNA extraction and detection of resistance genes
Genomic DNA was extracted from 100 μL of fresh tryptic soy broth subcultured samples using the DNA extraction kit MagNA Pure Compact (Roche), according to the manufacturer’s instructions. Isolate species were confirmed to be A. baumannii using rpoB PCR primers Ac696F (5′-TAYCGYAAAGAYTTGAAAGAAG-3′) and Ac1093R (5′-CMACACCYTTGTTMCCRTGA-3′) [2]. Samples were stored at −20 °C until further analysis. Eleven pairs of primers were used for A. baumannii resistance genotyping (Eurofins, Germany), including OXA A, B, C and 58, as previously explained by Hujer et al. [11]. Some modifications to the annealing temperature, number of cycles etc. were made to optimise the amplification for certain genes. The PCR products were analysed by electrophoresis on a 1.5 % (wt/vol) agarose gel containing 0.125 μg/mL ethidium bromide.
SBT and sequencing
The PCR amplicons were purified by using the QIAquick PCR extraction kit (Qiagen). Both strands of the amplicons were sequenced by the same gene amplification primers using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems™, Austin, TX, USA). Similarity searches were done using the BLAST program of PubMed National Centre for Biotechnology Information.
MLST and lineage tree
We performed MLST of 66 randomly selected isolates representing all four clusters from the SBT phylogenies. MLST was carried out using seven housekeeping genes as described by Diancourt et al. [12] and the PCR conditions were as previously described elsewhere [12, 13]. Data were collected and analysed by Excel, SPSS and START2 software. A lineage tree was generated using the unweighted pair group method with arithmetic mean.
Results
MDR phenotypes and OXA genotyping
A total of 253 isolates were collected from the clinical microbiology laboratory between 2006 and 2008. The sources of the clinical isolates were as follows: respiratory 117/253 (46.24 %), wound and burn 61/253 (24.11 %), blood 34/253 (13.43 %), urine 8/253 (3.16 %) and others 33/253 (13.04 %). Susceptibility testing showed that all isolates were resistant to four or more antimicrobial groups. Antibiotic susceptibility and MIC values are presented in Table 1. Carbapenem resistance, where the MIC was ≥8 μg/ml for meropenem and imipenem, was found in 96.44 % (244/253) and 88.53 % (224/253) of the isolates, respectively. OXA-D PCR typing showed that all 253 isolates carried OXA-51-like and OXA-23 genes. The OXA-58 gene was detected in only five isolates, while none of the isolates carried the OXA-40 (identical to the OXA-24) gene.
SBT
The SBT OXA data showed A. baumannii isolates carrying a wide range of OXA-51-like genes, including OXA-66, -69, -79, -82, -98, -94, -95, -131, -132 and -197. These were clustered into four main groups, including OXA-66 (62.3 %), OXA-69 (19.1 %), OXA-132 (7.6 %) and OXA-51-like genes (11 %), such as OXA-79, -82, -98, -94, -95, -131 and -197. Moreover, OXA-66 gene sequencing showed the partial insertion sequence of ISAba1 in the upstream position, while the insertion sequence ISAba3-like sequence transposase (tnpA) was identified with the carbapenem-hydrolysing oxacillinase OXA-58 (blaOXA-58)
STs of A. baumannii isolates and identification of regional clones
The sequence types (STs) of 66 isolates typed by MLST were grouped into four clusters. The most common ST was ST2 (n = 26), which is part of clonal complex 2 (CC2) and represents the European clone 2 (EU2) or worldwide lineage 2 (WW2). The next most common type was ST20 (CC1) (n = 15), followed by other STs (ST19, ST49, ST141, ST154). Eleven isolates were not designated with STs due to missing one or more MLST allele. Nevertheless, they were identified with mixed OXA-51-like genotypes, such as OXA-66, -131 and -197. MLST revealed four new isolates (194–197) with novel allelic profiles (Table 2). The MLST allelic profiles of the four novel ST isolates were entered into the Institut Pasteur MLST database (http://www.pasteur.fr/mlst) on September 2012 with the following Pasteur website IDs: 602, 612, 617, 620, 633 and 640.
Association of OXA-51 diversity and MLST typing
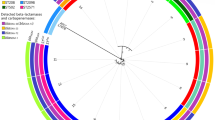
A lineage tree was generated from the MLST allelic profile and SBT OXA using the unweighted pair group method with arithmetic mean (Fig. 1). MDR A. baumannii isolates were grouped into four clusters, where the first cluster consisted of isolates that carried the OXA-66 gene and belonged to ST2 (CC2). On the contrary, isolates typed as ST20 (CC1) and possessed mostly the OXA-69 gene were the main contributors to the second cluster. However, the third and fourth clusters were more intermixed, where the OXA-51-like genes and MLST included: 65 (ST197), 69 (ST1), 98 (ST49), 132 (ST25) and 197 (ST154, ST196).
Lineage tree generated from the multilocus sequence typing (MLST) allelic profile showing the MLST alleles, sequence type, corresponding OXA-D genotypes, number of resistant antibiotics and patient demographic data (age, gender, isolate source) of MDR A. baumannii isolates form Saudi Arabia. The new sequence type isolates are shown by the arrows
Discussion
We identified OXA-66 as the major OXA-51-like gene, which is consistent with several other reports from Europe, the United States, South Americas, Turkey and China [14–16]. The sequences showed that our isolates carried the OXA-66 gene, along with the upstream partial insertion sequence of ISAba1, which was previously described as an OXA-66 promotor associated with carbapenem resistance among A. baumannii isolates [15, 17]. Another gene identified, OXA-69, known to confer carbapenem resistance, was found to a lesser extent among our clinical isolates, 19.1 % [18]. Both OXA-66 and OXA-69 were associated to a particular epidemic lineage of A. baumannii, which is consistent with the findings of other research groups [18, 19]. The OXA-132 gene was present in 7.6 % of our isolates, a gene which has been previously reported among clinical isolates from Saudi Arabia and Portugal [7, 20]. Five of our isolates showed that OXA-58 coexisted with the partial insertion sequence ISAba3 and tnpA gene, which may contribute to the resistance phenotype by integrating the insertion sequences into host genomes to confer drug resistance [21–24]. Three OXA-58 isolates contained new MLST alleles which could not be categorised into the major worldwide clonal lineage; hence, they are noted as new singletons (two ST194 and one ST196). However, this is the first report of the molecular typing of OXA-23 and OXA-58 in our region.
Conclusion
Our findings noticeably emphasise that the rate of carbapenem resistance is astounding, where 81 % of our multidrug-resistant (MDR) Acinetobacter baumannii clinical isolates carry OXA-66 and OXA-69 genes that belong to the worldwide lineage 2 (WW2, clonal complex CC2) and WW1 (CC1), respectively. Moreover, the identification of the novel sequence types ST194–ST197 isolates represents a local genetic diversity that needs further investigation, as well as the molecular epidemiology of carbapenem-resistant A. baumannii, which has become an endemic pathogen within the Middle Eastern countries.
References
Aly M, Balkhy HH (2012) The prevalence of antimicrobial resistance in clinical isolates from Gulf Corporation Council countries. Antimicrob Resist Infect Control 1(1):26
Bonomo RA, Szabo D (2006) Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis 43(Suppl 2):S49–S56. doi:10.1086/504477
Munoz-Price LS, Weinstein RA (2008) Acinetobacter infection. N Engl J Med 358(12):1271–1281. doi:10.1056/NEJMra070741
Brown S, Amyes S (2006) OXA (beta)-lactamases in Acinetobacter: the story so far. J Antimicrob Chemother 57(1):1–3. doi:10.1093/jac/dki425
Fouad M, Attia AS, Tawakkol WM, Hashem AM (2013) Emergence of carbapenem-resistant Acinetobacter baumannii harboring the OXA-23 carbapenemase in intensive care units of Egyptian hospitals. Int J Infect Dis 17(12):e1252–e1254. doi:10.1016/j.ijid.2013.07.012
Hasan B, Perveen K, Olsen B, Zahra R (2014) Emergence of carbapenem-resistant Acinetobacter baumannii in hospitals in Pakistan. J Med Microbiol 63(Pt 1):50–55. doi:10.1099/jmm.0.063925-0
Alsultan AA, Hamouda A, Evans BA, Amyes SG (2009) Acinetobacter baumannii: emergence of four strains with novel bla(OXA-51-like) genes in patients with diabetes mellitus. J Chemother 21(3):290–295
Giannouli M, Cuccurullo S, Crivaro V, Di Popolo A, Bernardo M, Tomasone F, Amato G, Brisse S, Triassi M, Utili R, Zarrilli R (2010) Molecular epidemiology of multidrug-resistant Acinetobacter baumannii in a tertiary care hospital in Naples, Italy, shows the emergence of a novel epidemic clone. J Clin Microbiol 48(4):1223–1230. doi:10.1128/jcm.02263-09
Clinical and Laboratory Standards Institute (CLSI) (2011) Performance standards for antimicrobial susceptibility testing; Twenty-first informational supplement. CLSI, Wayne, PA
Peleg AY, Seifert H, Paterson DL (2008) Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21(3):538–582. doi:10.1128/cmr.00058-07
Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA (2006) Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50(12):4114–4123. doi:10.1128/aac.00778-06
Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S (2010) The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5(4):e10034. doi:10.1371/journal.pone.0010034
Bacila I, Jakab E, Ferencz B, Popescu O (2008) MLST method (Multilocus Sequence Typing). Bacteriol Virusol Parazitol Epidemiol 53(1):13–17
Culebras E, González-Romo F, Head J, Gómez M, Morales G, Picazo JJ (2010) Outbreak of Acinetobacter baumannii producing OXA-66 in a Spanish hospital: epidemiology and study of patient movements. Microb Drug Resist 16(4):309–315. doi:10.1089/mdr.2009.0113
Figueiredo S, Poirel L, Croize J, Recule C, Nordmann P (2009) In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural bla(OXA-66) oxacillinase gene. Antimicrob Agents Chemother 53(6):2657–2659. doi:10.1128/AAC.01663-08
Hu WS, Yao SM, Fung CP, Hsieh YP, Liu CP, Lin JF (2007) An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob Agents Chemother 51(11):3844–3852. doi:10.1128/AAC.01512-06
Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL (2006) The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 258(1):72–77. doi:10.1111/j.1574-6968.2006.00195.x
Hamouda A, Evans BA, Towner KJ, Amyes SG (2010) Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of bla(OXA-51-like) genes. J Clin Microbiol 48(7):2476–2483. doi:10.1128/jcm.02431-09
Evans BA, Hamouda A, Towner KJ, Amyes SG (2008) OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin Microbiol Infect 14(3):268–275. doi:10.1111/j.1469-0691.2007.01919.x
Grosso F, Carvalho KR, Quinteira S, Ramos A, Carvalho-Assef APDA, Asensi MD, Peixe L (2011) OXA-23-producing Acinetobacter baumannii: a new hotspot of diversity in Rio de Janeiro? J Antimicrob Chemother 66(1):62–65. doi:10.1093/jac/dkq406
Evans BA, Hamouda A, Towner KJ, Amyes SG (2010) Novel genetic context of multiple bla OXA-58 genes in Acinetobacter genospecies 3. J Antimicrob Chemother 65(8):1586–1588. doi:10.1093/jac/dkq180
Merkier AK, Catalano M, Ramírez MS, Quiroga C, Orman B, Ratier L, Famiglietti A, Vay C, Di Martino A, Kaufman S, Centrón D (2008) Polyclonal spread of bla(OXA-23) and bla(OXA-58) in Acinetobacter baumannii isolates from Argentina. J Infect Dev Ctries 2(3):235–240
Poirel L, Marqué S, Héritier C, Segonds C, Chabanon G, Nordmann P (2005) OXA-58, a novel class D {beta}-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother 49(1):202–208. doi:10.1128/AAC.49.1.202-208.2005
Chen TL, Chang WC, Kuo SC, Lee YT, Chen CP, Siu LK, Cho WL, Fung CP (2010) Contribution of a plasmid-borne blaOXA-58 gene with its hybrid promoter provided by IS1006 and an ISAba3-like element to beta-lactam resistance in acinetobacter genomic species 13TU. Antimicrob Agents Chemother 54(8):3107–3112. doi:10.1128/AAC.00128-10
Acknowledgement
This work has been supported by a grant from King Abdulaziz City for Science and Technology (KACST), Riyadh, Saudi Arabia, grant number: ARP-28-112.
Conflict of interest
All authors confirm that there is no financial relationship with the organisation that sponsored the research. EJA however, is an employee of KACST.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aly, M., Tayeb, H.T., Al Johani, S.M. et al. Genetic diversity of OXA-51-like genes among multidrug-resistant Acinetobacter baumannii in Riyadh, Saudi Arabia. Eur J Clin Microbiol Infect Dis 33, 1223–1228 (2014). https://doi.org/10.1007/s10096-014-2068-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-014-2068-0