Abstract
Candida auris is a recently described rare agent of fungemia. It is notable for its antifungal resistance. A total of 15 C. auris isolates, originating from seven cases of fungemia, three cases of diabetic gangrenous foot, and one case of bronchopneumonia from a tertiary care hospital in south India, were investigated. All of the 15 isolates were identified by sequencing and 14 of these along with 12 C. auris isolates previously reported from two hospitals in Delhi, north India, two each from Japan and Korea were genotyped by amplified fragment length polymorphism (AFLP). In vitro antifungal susceptibility testing (AFST) was done by the Clinical and Laboratory Standards Institute (CLSI) broth microdilution method. Candida auris isolates were misidentified as Candida haemulonii by VITEK. All were resistant to fluconazole [geometric mean minimum inhibitory concentration (MIC) 64 μg/ml] and 11 isolates were resistant to voriconazole (MIC ≥1 μg/ml). Forty-seven percent of the C. auris isolates were resistant to flucytosine (MIC ≥64 μg/ml) and 40 % had high MIC (≥1 μg/ml) of caspofungin. Breakthrough fungemia developed in 28.6 % of patients and therapeutic failure in 4 (66.7 %) patients. Interestingly, the 26 Indian C. auris isolates from north and south India were clonal and phenotypically and genotypically distinct from Korean and Japanese isolates. The present study demonstrates that C. auris is a potential emerging pathogen that can cause a wide spectrum of human mycotic infections. The prevalence of a C. auris endemic clonal strain resistant to azoles and other antifungals in Indian hospitals with high rates of therapeutic failure in cases of fungemia is worrisome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Candida auris was identified as a new species in 2009 from the external ear canal of a Japanese patient [1]. This pathogen cannot be identified by commercial methods used for yeast identification. It has recently been described as an agent of fungemia, so far reported only from Korea and India [2, 3]. Sequence analysis of the D1/D2 domain of 26S rDNA and internal transcribed spacer (ITS) regions of nuclear rRNA genes of C. auris demonstrates a close phylogenetic relationship to Candida haemulonii, with which it is usually misidentified by commercial identification systems such as VITEK 2 and API 20C [2–4]. Moreover, C. auris is reported to be resistant to fluconazole and exhibits reduced susceptibility to voriconazole and itraconazole [2–4]. Further, persistent fungemia occurs in patients despite fluconazole and amphotericin B therapy, resulting in fatal outcome [3]. This pathogen has recently assumed a greater clinical significance due to the local prevalence and transmission of drug-resistant clonal strains in healthcare units [2, 4, 5]. Herein, we report C. auris as an etiologic agent of fungemia and its isolation from deep sterile sites in patients from a tertiary care hospital in south India. Our results indicate that the strains of C. auris exhibiting resistance to azole antifungal agents, from both north and south India, were clonal.
Materials and methods
Fungal isolates and their morphological and physiological characterization
The study analyzed 245 yeast isolates from cases of fungemia and other invasive mycosis cultured during the period November 2011 to June 2013 at a university hospital in Kochi in southern India. Of the 245 yeast isolates, 15 (6.1 %) isolates, initially identified as C. haemulonii by VITEK® 2 Compact (bioMérieux, Marcy I’Etoile, France), were confirmed as C. auris by ITS and D1/D2 sequencing, and were included in the present study. The phenotypic identification was based on the colony color of the isolates on CHROMagar™ Candida medium (Difco, Becton, Dickinson and Company, Baltimore, MD, USA), germ tube test, and morphology on rice Tween 80 agar. Growth of the isolates at 37, 42, and 45 °C was also tested.
Sequencing of ITS and D1/D2 regions
Amplification of the ITS region of the ribosomal subunit and D1/D2 region of the large ribosomal subunit was done, followed by sequencing and alignment of both the strands using the Sequencing Analysis 5.3.1 software (Applied Biosystems, Foster City, CA, USA), as described previously [6–8]. GenBank Basic Local Alignment Search Tool (BLAST) searches (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi) were performed for species identification. For phylogenetic analysis, we included ITS and D1/D2 sequences of the recently reported 12 new clonal strains of C. auris prevalent in two hospitals in northern India (CBS 12766–12777), two Korean isolates, and a solitary Japanese isolate [1–3]. Sequences were aligned using the ClustalW program (v1.82) and the final alignments were edited manually.
Amplified fragment length polymorphism (AFLP)
Of the 15 south Indian C. auris isolates, the genotypic diversity of 14 isolates was determined together with the previously described clonal north Indian (n = 12), Japanese (n = 2), and Korean (n = 2) isolates using amplified fragment length polymorphism (AFLP) fingerprint analysis [9]. Candida auris reference isolates (KCTC 17809, KCTC 17810, JCM 15448, and DSMZ 21092), C. haemulonii (CBS 7801, CBS 7802, CBS 5149, and CBS 5140), C. duobushaemulonii (CBS 7798–7800 and CBS 9754), and C. pseudohaemulonii (KCTC 1787, CBS 10004, and JCM 12453) were used as controls to assign the AFLP genotypes. Briefly, approximately 50 ng of genomic DNA was subjected to a combined restriction–ligation procedure containing EcoRI and MseI restriction enzymes (New England Biolabs, Beverly, MA, USA) and complementary adaptors. Prior to further usage, the restriction–ligation reaction was diluted by adding 80 μl Tris/HCl (pH 8.3) buffer. One microliter of the diluted product was used for amplification in a final volume of 25 μl, for which the selective primers EcoRI (5′-FLU-GACTGCGTACCAATTCAC-3′) and MseI (5′-GATGAGTCCTGACTAAC-3′) were used. One microliter of the 10× diluted amplicon was added to a mixture of 8.9 μl water and 0.1 μl LIZ600 internal size marker (Applied Biosystems), followed by heating the diluted sample for 1 min to 95 °C and subsequent fragment analysis on an ABI 3500xL Genetic Analyzer (Applied Biosystems). Raw data were analyzed using Bionumerics v6.0 (Applied Maths, Sint-Martens-Latem, Belgium) and a dendrogram was generated using standard Pearson and unweighted pair group method with arithmetic mean (UPGMA) settings.
In vitro antifungal susceptibility testing (AFST)
Antifungal susceptibility testing (AFST) was performed using the Clinical and Laboratory Standards Institute (CLSI) broth microdilution method, following the M27-A3 guidelines [10]. The antifungals tested were amphotericin B (Sigma, St. Louis, MO, USA), fluconazole (Pfizer, Groton, CT, USA), itraconazole (Lee Pharma, Hyderabad, India), voriconazole (Pfizer), posaconazole (Merck, Whitehouse Station, NJ, USA), isavuconazole (Basilea Pharmaceutica, Basel, Switzerland), flucytosine (Sigma), caspofungin (Merck), micafungin (Astellas, Toyama, Japan), and anidulafungin (Pfizer). CLSI-recommended Candida krusei ATCC6258 and Candida parapsilosis ATCC22019 were used as quality control strains with each test. The minimum inhibitory concentration (MIC) end points for azoles and echinocandins were defined as the lowest drug concentration that caused a prominent decrease in growth (50 %) vis-à-vis the controls and read visually after 24 h, as validated recently by Pfaller et al. [11, 12]. For amphotericin B, the MIC was defined as the lowest concentration at which there was 100 % inhibition of growth compared with the drug-free control wells. The geometric mean (GM) and MICs for each drug at which 50 % (MIC50) and 90 % (MIC90) of the tested isolates were inhibited were also determined. Newly revised clinical breakpoints (CBPs) defined for C. albicans were used for the interpretation of MIC data. These are described as susceptible (S) ≤2 μg/ml, susceptible dose dependent (SDD) 4 μg/ml, resistant (R) ≥8 μg/ml for fluconazole; S ≤0.12 μg/ml, SDD 0.25–0.5 μg/ml, R ≥1 μg/ml for itraconazole; S ≤0.12 μg/ml, intermediate (I) 0.25–0.5 μg/ml, R ≥1 μg/ml for voriconazole; and S ≤0.25 μg/ml, I 0.5 μg/ml, R ≥1 μg/ml for echinocandins [13]. Epidemiological cutoff values (ECVs) were used for amphotericin B (≤2 μg/ml), flucytosine (≤0.5 μg/ml), and posaconazole (≤0.06 μg/ml) [13].
Patient information
The clinical details of the patients, such as demographic characteristics, clinical diagnosis, risk factors for candidemia, antifungal therapy, and outcome, were collected retrospectively. In cases of candidemia, therapeutic failure was defined either as the persistence of Candida despite the administration of 3 days of antifungal therapy or as the development of breakthrough candidemia while receiving antifungal agents for 3 days.
Results
Microbiology
A total of 15 C. auris isolates originated from 12 patients. Of these, seven isolates were cultured from blood, three each from central venous catheter (CVC) tips and surgically excised tissues, and one each from bronchoalveolar lavage (BAL) fluid and pus (Table 1). All isolates developed a pink color on CHROMagar™ Candida medium and showed ovoid to elongated budding yeast cells occurring singly or in pairs. The isolates grew well at 37 °C and 42 °C, whereas no growth was seen at 45 °C. They were inhibited on 0.01 % cycloheximide-containing medium. In addition, all C. auris isolates assimilated N-acetylglucosamine (NAG), unlike the reference Korean and Japanese C. auris isolates.
Molecular characterization
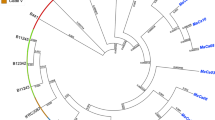
ITS sequences of 15 test isolates (GenBank accession nos. KF689009–KF689023) showed 99 % homology (query coverage ranging from 98 to 100 %) with Korean and the previously described north Indian C. auris isolates (accession nos. EU884184, EU884180, and KC692050) in GenBank. The D1/D2 sequence similarity of our isolates (GenBank accession nos. KF689024–KF689038) was 99 % with the Korean isolates (GenBank accession numbers EU881965, EU881967, and EU881968). Of the 15 south Indian C. auris isolates, 14 have been deposited at the CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands, and the accession numbers assigned are CBS 12874–12887. Candida auris isolates from the present study clustered together with the previously published north Indian C. auris isolates in the ITS phylogenetic tree and were distinct from the Japanese and Korean isolates. AFLP analysis showed separate clades of C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii distinct from C. auris. The 26 C. auris isolates, including 12 from north and 14 from south India, were clonal and clustered together, irrespective of their geographical origin, with an overall similarity of 80.2 % (Fig. 1). They were genotypically distinct from the Japanese and the Korean isolates, which had a similarity of 63.4 % with the Indian C. auris isolates (Fig. 1). In addition, AFLP clearly showed that the most genetically related sibling species of C. auris, i.e., C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii, had a similarity of 9.9 % among these three species versus C. auris (Fig. 1).
Amplified fragment length polymorphism (AFLP) analysis showing fingerprinting of Candida auris isolates from India (north and south), Japan, Korea, and members of the C. haemulonii complex. The dendrogram was constructed using unweighted pair group method with arithmetic mean (UPGMA) analysis in combination with the Pearson correlation coefficient and was restricted to fragments in the range 60–400 bp. The scale bar indicates the percentage similarity
Antifungal susceptibility testing
All 15 C. auris isolates were resistant to fluconazole with GM MICs of 64 μg/ml and 11 (73 %) were resistant to voriconazole (MIC ≥1 μg/ml). Furthermore, 47 % of C. auris isolates were resistant to flucytosine (n = 7; MIC ≥64 μg/ml). In addition, 40 % of C. auris isolates revealed high MICs (≥1 μg/ml) of caspofungin. Posaconazole exhibited the most potent activity, with a GM MIC of 0.03 μg/ml, followed by itraconazole (GM MIC, 0.14 μg/ml) and isavuconazole (GM MIC, 0.23 μg/ml). Amphotericin B showed good activity, with a GM MIC of 0.64 μg/ml. Also, micafungin (GM MIC, 0.10 μg/ml) and anidulafungin (GM MIC, 0.14 μg/ml) were highly active (Table 2).
Clinical evaluation of the patients
The clinical features, risk factors, and response to antifungal therapy of the 12 patients with C. auris-associated infections are summarized in Table 1. Seven patients were diagnosed to have candidemia and three had an infection of a gangrenous diabetic foot. Of the fungemic patients, two patients also had CVC tip cultures positive for C. auris, while another patient had post-operative wound sepsis due to C. auris. In one patient, only a CVC tip culture was positive and in a solitary patient, BAL fluid cultures yielded C. auris. The age group of patients ranged from 2 to 87 years (mean age 56.2 years). A mean of 7 of the 10 risk factors for candidiasis were present in each patient. The most common risk factor, present in all the patients, was the concomitant use of broad-spectrum antibiotics and the least was neutropenia, present in two (16.7 %) patients. Nine (75 %) patients had immunosuppressive conditions, such as diabetes mellitus in 5 patients (55.5 %), chronic kidney disease in 4 (44.4 %), and hematologic malignancies or history of cancer chemotherapy in 3 (33.3 %) patients. Among the other important risk factors, admission to the intensive care unit and the presence of an indwelling urinary catheter were noted in 11 (91.6 %) patients each, followed by CVC placement in 10 (83.3 %). Other risk factors included parenteral nutrition in 8 (66.6 %), concomitant bacteremia and a recent history of surgery in 7 (58.3 %) each, and the use of antifungals in 5 (41.6 %) patients. Breakthrough fungemia was observed in two (28.6 %) patients. Details of therapy were available for 11 patients. Of these, 9 (81.8 %) received antifungal therapy, the details of which are given in Table 1. Persistent fungemia was noted in 4 of 6 patients whose records were available. Of these six patients, cultures of CVC could be done in only two patients. Overall, 4 (36.4 %) patients expired, all of whom had fungemia. Of the remaining seven cases that survived, three underwent surgical excision of the gangrenous tissue; the solitary case of bronchopneumonia was treated with fluconazole and caspofungin, while another case (case 12) was managed by removal of the central line.
Discussion
Although C. auris has been previously reported as a rare agent of fungemia, this report highlights the isolation of C. auris from diverse clinical samples not reported so far [2, 3]. The isolation of C. auris from pus, surgically resected tissues, and BAL fluid suggests its potential role in varied diseases. In addition, the study documents seven well-characterized cases of fungemia due to this rare pathogen. In the present study, C. auris represented 8.6 % of annual candidemia cases in a tertiary care hospital. Previously, one-third (30 %) cases of candidemia have been attributed to this pathogen in a single tertiary care hospital of north India [2]. To date, there is a paucity of information regarding the clinical characteristics and antifungal susceptibility profiles of C. auris. The major risk factors associated with C. auris infections in the present study were similar to those of invasive candidiasis, which were broad-spectrum antibiotics, indwelling catheters, and immunosuppressive conditions [14]. Breakthrough fungemia developed in 28.6 % patients and therapeutic failure was observed in 66.7 % patients with fungemia. Also, previously high rates of therapeutic failures in patients with fungemia have been reported with this pathogen [2, 3]. In the present study, therapeutic failure was observed in patients with fungemia who received fluconazole, to which the isolate was highly resistant. The successful treatment outcome in the remaining two patients could be attributed to the timely institution of caspofungin and amphotericin B. It is noteworthy that low MICs of amphotericin B have been reported previously [2–4], as well as in the present study. In contrast, C. haemulonii, with which C. auris is commonly misidentified by routine identification systems, is notably resistant to amphotericin B and azoles [4, 15]. Therefore, the correct identification of C. auris by sequencing and the need for undertaking antifungal susceptibility testing for this pathogen can hardly be overemphasized. It is difficult to ascertain the clinical significance of C. auris isolates recovered from patients with diabetic foot and BAL. Although the histopathologic evidence was lacking in aforementioned cases, the favorable response to surgical excision of the gangrenous tissue supports a causative role of this agent. Also, the case with bronchopneumonia from which no obvious etiological agent except C. auris was isolated had underlying risk factors and responded well to caspofungin. The patient had indwelling catheters, which could be the source of infection. However, blood and catheter tips were not cultured. The south Indian C. auris isolates also assimilated NAG, just like the previous isolates from north India [2]. Thus, the emerging Indian C. auris isolates were phenotypically and genotypically distinct from the Japanese and Korean strains. Additionally, AFLP analysis of 26 Indian C. auris strains from three geographically distant hospitals located at a distance of 2,081 km, one in Kochi, south India, and two in Delhi, north India, revealed the presence of an endemic clonal strain, suggesting widespread transmission. The fact that not only geographical clusters of C. auris could be segregated, as well as that the sibling species of C. auris, i.e., C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii, were revealed to be only distantly related by AFLP analysis, indicated that this genotyping method is a good discriminatory tool to study the epidemiology of this emerging yeast pathogen. To conclude, C. auris is an emerging pathogen which shows reduced susceptibility to antifungal agents and demonstrates a potential to cause a wide spectrum of human mycotic infections.
References
Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H (2009) Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53(11):41–44
Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF (2013) New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19(10):1670–1673
Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC (2011) First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49(9):3139–3142
Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, Lee JS, Jung SI, Park KH, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW (2009) Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48(6):e57–e61
Oh BJ, Shin JH, Kim MN, Sung H, Lee K, Joo MY, Shin MG, Suh SP, Ryang DW (2011) Biofilm formation and genotyping of Candida haemulonii, Candida pseudohaemulonii, and a proposed new species (Candida auris) isolates from Korea. Med Mycol 49(1):98–102
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Kurtzman CP, Robnett CJ (1997) Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol 35(5):1216–1223
Chowdhary A, Agarwal K, Kathuria S, Singh PK, Roy P, Gaur SN, de Hoog GS, Meis JF (2013) Clinical significance of filamentous basidiomycetes illustrated by isolates of the novel opportunist Ceriporia lacerata from the human respiratory tract. J Clin Microbiol 51(2):585–90
Illnait-Zaragozí MT, Martínez-Machín GF, Fernández-Andreu CM, Perurena-Lancha MR, Theelen B, Boekhout T, Meis JF, Klaassen CH (2012) Environmental isolation and characterisation of Cryptococcus species from living trees in Havana city, Cuba. Mycoses 55(3):e138–144
Clinical and Laboratory Standards Institute (CLSI) (2008) Reference method for broth dilution antifungal susceptibility testing of yeasts; Approved standard—third edition. CLSI document M27-A3. CLSI, Wayne, PA
Pfaller MA, Boyken LB, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Diekema DJ (2008) Validation of 24-hour fluconazole MIC readings versus the CLSI 48-hour broth microdilution reference method: results from a global Candida antifungal surveillance program. J Clin Microbiol 46(11):3585–3590
Pfaller MA, Boyken LB, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Diekema DJ (2011) Validation of 24-hour posaconazole and voriconazole MIC readings versus the CLSI 48-hour broth microdilution reference method: application of epidemiological cutoff values to results from a global Candida antifungal surveillance program. J Clin Microbiol 49(4):1274–1279
Pfaller MA, Diekema DJ (2012) Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J Clin Microbiol 50(9):2846–2856
Cornely OA, Bassetti M, Calandra T, Garbino J, Kullberg BJ, Lortholary O, Meersseman W, Akova M, Arendrup MC, Arikan-Akdagli S, Bille J, Castagnola E, Cuenca-Estrella M, Donnelly JP, Groll AH, Herbrecht R, Hope WW, Jensen HE, Lass-Flörl C, Petrikkos G, Richardson MD, Roilides E, Verweij PE, Viscoli C, Ullmann AJ; ESCMID Fungal Infection Study Group (2012) ESCMID* guideline for the diagnosis and management of Candida diseases 2012: non-neutropenic adult patients. Clin Microbiol Infect 18(Suppl 7):19–37
Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gómez-López A, Boekhout T (2012) Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol 50(11):3641–3651
Acknowledgments
CS is supported by a University Grants Commission Research Fellowship (F.2-15/2003 SA-I). JFM has been supported by Qatar National Research Fund grant NPRP 5-298-3-086.
Conflict of interest
JFM received grants from Astellas, Merck, and Schering-Plough. He has been a consultant to Astellas, Basilea, and Merck, and received speaker’s fees from Merck. All other authors: no potential conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chowdhary, A., Anil Kumar, V., Sharma, C. et al. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33, 919–926 (2014). https://doi.org/10.1007/s10096-013-2027-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-013-2027-1