Abstract
Blastocystis is a genetically diverse and widespread intestinal parasite of animals and humans with controversial pathogenic potential. At least nine subtypes of Blastocystis have been found in humans. The genetic diversity of Blastocystis was examined in stool samples from 68 patients from the Stockholm area, Sweden. Blastocystis was identified by light microscopy, and subtyped by sequencing the 5′-end of the small subunit ribosomal RNA gene. Five Blastocystis subtypes were identified in the 63 patients whose samples were successfully subtyped: ST1 (15.9%), ST2 (14.3%), ST3 (47.6%), ST4 (20.6%), and ST7 (1.6%). ST3 was more common in males compared to females (P = 0.049). Comparative molecular analysis of Blastocystis sequences revealed intra-subtype variations within the identified subtypes with the exception of ST4. Among ST4 sequences in this study, as well as in the majority of human GenBank sequences, a limited genetic diversity was found compared to what was found among the other common subtypes (ST1, ST2 and ST3). The relative prevalence of ST4 in this study was comparable to the overall distribution of ST4 in European cohorts (16.5%). This contrasts with the sparse reports of ST4 in studies from other continents, which may indicate that the distribution of this subtype is geographically heterogeneous.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Blastocystis is an anaerobic unicellular eukaryote that can inhabit the intestinal tract of humans and many animals. It is the most commonly found non-fungal eukaryotic organism in human faecal samples [1–3]. The distribution of the parasite is worldwide and the prevalence in human faecal samples ranges from 7 to 20% in developed countries to 30–60% in rural areas in developing countries [4–8]. Much is still unknown about Blastocystis. For instance, its life-cycle and pathogenic potential are still under debate [1–3, 9, 10]. Blastocystis is found in individuals both with and without symptoms. However, reports have been published of patients with gastrointestinal symptoms and Blastocystis as the only detected possible pathogen, and whose symptoms were relieved after successful treatment of Blastocystis [11–13]. Various gastrointestinal symptoms such as diarrhoea, abdominal pain, vomiting, constipation and flatulence have been linked to Blastocystis infection. As with other intestinal parasite infections, diarrhoeal episodes can alternate with normal defecation patterns or even constipation, conditions similar to chronic gastrointestinal illnesses such as irritable bowel syndrome (IBS). In fact, an increased prevalence of Blastocystis has been found in IBS patients [14–17]. Moreover, symptomatic Blastocystis infection has been reported to occur more often in immunocompromised adult patients than in controls [18] and has also been reported to be common in HIV-infected and immunocompromised children with gastrointestinal symptoms [11, 19].
Molecular methods have shown extensive genetic variation among Blastocystis isolates [20, 21]. A consensus approach has assigned Blastocystis isolates from humans, mammals and birds to one of nine subtypes, with sufficient genetic divergence between many of them to be classified as separate species [22]. Recently, four additional subtypes have been identified but only in non-human hosts [23, 24]. This genetic diversity has supported the hypothesis that the variability in symptoms in patients positive for Blastocystis could be due to different pathogenic potential among the subtypes [25–31].
In a previous study investigating bacterial, viral and protozoan enteropathogens in Swedish patients with diarrhoea compared to healthy controls, Blastocystis was found in 4% of the patients and 9% of the controls [32] but no subtyping of Blastocystis was performed. We here present the first investigation of Blastocystis subtypes present in a Swedish collection of faecal samples from a diagnostic laboratory.
Materials and methods
Patient samples
Stool samples from patients in the Stockholm area were sent to the Laboratory of Diagnostic Parasitology of Karolinska University Hospital, Solna, Sweden, for routine ova and (oo)cyst examination. Notifications from clinicians on travel history and type of symptoms were only available in a minority of cases. Samples were sent fresh and Blastocystis was identified by light microscopy of iodine stained smears of faecal concentrates obtained by a modified formol-ether-concentration technique (FECT), utilising ethyl-acetate instead of ether [33]. Seventy-two Blastocystis positive samples were collected randomly during 2008 and the first few months of 2009. As a precautionary measure prior to DNA extraction all faecal samples were divided into two parts, one part was frozen without additives, and one part with approximately 1 ml faeces fixed in 4 ml of 70% ethanol was stored at +4°C, the latter according to a protocol for PCR examination for Entamoeba in faeces by Lebbad and Svärd [34].
DNA extraction
For each sample, DNA was extracted from approximately 200 mg stool using the QiaAMP DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, and eluted in 200 μl buffer AE (Qiagen). Frozen samples were not thawed before extraction. Samples fixed in ethanol were washed three times in phosphate-buffered saline (PBS) before DNA extraction.
PCR
The method described by Scicluna et al. [26] was used for subtype analysis. The method includes a conventional PCR with the primers RD5 (ATCTGGTTGATCCTGCCAGT) and BhRDr (GAGCTTTTTAACTGCAACAACG) amplifying an approximately 600 base pair (bp) fragment of the 1,800 bp small subunit ribosomal RNA gene (SSU-rDNA). This region of the SSU-rDNA has been shown to provide sufficient information for differentiating subtypes of Blastocystis [26]. One μl of extracted DNA was added to an amplification mixture of primers and BioMix™ (Bioline, London, UK) containing Taq DNA polymerase. The amplification profile consisted of 30 cycles of denaturation, annealing and extension at 94°C, 59°C and 72°C (1 min for each step), with a final extension step of 2 min at 72°C. Primarily, DNA extracted from frozen stool samples was used for the PCR. Where no PCR product was obtained, DNA was extracted from the ethanol fixed samples. Positive PCR products were purified using GeneJET™ Genomic DNA Purification Kit (Fermentas, London, UK) according to the manufacturer’s recommendations.
Sequencing and sequence analysis
Purified PCR products were sequenced using the primer BhRDr, ABI BigDye sequencing kit version 3.1 (Applied Biosystems, Warrington, UK) and an ABI 3730 sequencer. The resulting chromatograms were analysed and edited in the computer software Chromas version 2.33 (Technelysium Pty. Ltd., Australia). The sequences obtained were compared to Blastocystis sequences in GenBank by BLAST searches at the National Center for Biotechnology Information (NCBI) [35]. Subtypes were identified by determining the exact match or closest similarity according to the classification by Stensvold et al. [22]. Sequences were aligned with SSU-rDNA sequences representing ST1–9 from GenBank using the software ClustalW [36] and MEGA version 4 [37] for comparative analysis of the sequence data.
Sequences obtained in the study are available as accession numbers JN003684-JN003686 and JN587541-JN587549 in the GenBank database.
Phylogenetic analysis of Blastocystis
To infer phylogenetic relationships among the sequences, a phylogenetic tree was constructed with the neighbor-joining (NJ) method [38] using the software MEGA version 4 [37]. The flagellate Proteromonas lacertae (accession number U37108) was used as the outgroup as in previous phylogenetic studies [39, 40]. Bootstrap confidence values for the branching reliability were calculated with 1,000 replicates.
Statistical analysis
For comparison of subtype prevalence of different groups the χ2 test (with collapsed categories when needed) was used. A P-value less than 0.05 was considered statistically significant. The software PASW version 18 (SPSS Inc, Chicago, IL, USA) was used for statistical analysis.
Results
Blastocystis was identified by light microscopy in 72 faecal samples from 68 patients (30 females and 38 males). The age of the patients ranged from 1 to 70 years; the median age was 30.5 years (interquartile range 16–46) in females and 38.5 years (interquartile range 26–49) in males. The age distribution is displayed in Fig. 1. In 63 of the 68 patients the expected 600 bp fragment of the SSU-rDNA was amplified and successfully sequenced. In four of the 68 patient samples no PCR product was produced despite repeated DNA extractions and numerous attempts towards optimising PCR conditions. In one sample a PCR product of the correct size was obtained but the resulting sequence chromatogram showed double peaks, which may indicate a mixed infection of two or more Blastocystis subtypes.
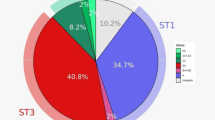
Five Blastocystis subtypes were identified in the study, ST1, ST2, ST3, ST4, and ST7 (Fig. 2). ST3 was the most prevalent subtype, found in 30/63 patients (47.6%) followed by ST4 in 13/63 patients (20.6%). One patient harboured Blastocystis ST7, a subtype that has been found mostly in birds [41]. For comparison, Blastocystis subtype distributions found in previous European studies are displayed in Table 1.
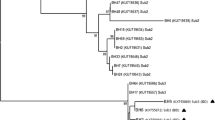
A phylogenetic tree comparing representative Blastocystis SSU-rDNA sequences generated in this study, designated ‘BL0--’, and reference sequences from GenBank, shown with accession numbers. The tree is inferred using the neighbor-joining method (with the maximum composite likelihood model) based on a hypervariable region at the 5′ end of the SSU-rRNA gene. Proteromonas lacertae (accession number U37108) is used as the outgroup. Bootstrap values (%) are indicated at the internal nodes (1,000 replicates). Bootstrap values of less than 50% are not shown. The subtype prevalence in the 63 samples is shown to the right as a percentage. Italicised subtypes were not found in the present study
There was a difference between men and women in the prevalence of ST3 and ST4 (Fig. 3). Although supported by a marginal significance, ST3 was more common in males compared to females (P = 0.049, collapsed categories). Among females ST4 was almost as common as ST3 but the difference in the relative frequency of ST4 among men and women was not statistically significant (P = 0.13) (Fig. 3).
As previously reported [26], the sequence variation in the 5′-end of the SSU-rRNA gene of Blastocystis is extensive. The isolate BL051 (accession number JN003686), assigned to ST7, differs from its closest match in GenBank (DQ232821) by seven nucleotides in the studied region of SSU-rDNA. However, this level of divergence falls within the range of variation identified among previously published ST7 sequences (Fig. 2). A similar level of diversity was seen among samples assigned to STs 1–3 identified in this study (ST1: up to 2 differences; ST2: up to 6 differences; ST3: up to 6 differences). In contrast, no variation was found among the 13 identified ST4 sequences. A BLAST search revealed that the ST4 sequences of this study were identical to most published 5´ fragments of ST4 SSU-rRNA genes, among them 17 from human isolates deposited by Scicluna et al. [26], three human and four rat ST4 sequences from France [21, 42] and three sequences from humans in Japan and Germany [43]. Single nucleotide polymorphisms (SNPs) differentiated these sequences from a group of five previously published ST4 sequences from rodents that all shared at least three of the SNPs [40, 44]. The sequence diversity leads to the generation of two clades of ST4 in the phylogenetic tree (Fig. 2).
Discussion
During the last few years several studies of the prevalence of Blastocystis subtypes have been published. This study is, to our knowledge, the first that describes the occurrence of different Blastocystis subtypes in Sweden. ST3 was the most common Blastocystis subtype found in the Stockholm area. The distribution of Blastocystis subtypes in human faecal samples varies from country to country. However, ST3 has dominated as the most prevalent subtype in most studies although the figures differ depending on populations studied [42].
ST4 was the second most prevalent subtype in our study, being found in 20.6% of the samples. This is in contrast to an average prevalence figure of 5.4% reported by Souppart et al. in 2009 in a summary of Blastocystis subtypes from different countries [42]. In most studies ST4 was the fourth most common subtype found in humans, after ST3, ST1 and ST2. However, when only studies from Europe are included in the comparison, and with additional data from the two last years included, the average prevalence for ST4 increases to 16.5% (Table 1). Using this figure, our findings are generally in concordance with the results from the rest of Europe. Since very few isolates of ST4 have been identified in studies from Asia, among them a large study of Blastocystis prevalence in China by Li et al. [8], the results may also indicate geographic differences in the prevalence of ST4. Interestingly, studies from Turkey, a country situated both in Europe and Asia, reports very low numbers of ST4 (1/337 samples) (Table 1). It should be kept in mind, however, that differences in study populations and methodology used may influence the results.
Blastocystis carriers in the present study were diagnosed primarily by light microscopy. Routine examination using FECT is considered to have a low sensitivity of approximately 30–50% for the detection of Blastocystis [4, 45]. The sensitivity has been shown to be lower for ST3 compared to the other subtypes [45]. Our selection method could therefore have underestimated the prevalence of ST3. The faecal samples in the present study were all from a diagnostic parasitology laboratory suggesting that the majority of the samples arise from patients with gastrointestinal symptoms. No PCR product could be amplified in the samples from four of the patients in this study. Possible reasons for this could be PCR inhibition, degradation of DNA due to long storage time or misidentification by light microscopy.
A difference in the occurrence of ST3 and ST4 between men and women was found. The higher relative frequency of ST3 in men was statistically significant, although the sample size in each subtype group was small. Differences in the relative proportions of subtypes between the sexes could indicate distinct pathways of transmission or exposure to the organism. Further studies are needed to confirm the gender difference in the distribution of subtypes of Blastocystis.
Overall the amplified fragment of Blastocystis SSU-rDNA exhibited extensive sequence variability. Although a limited number of Blastocystis were examined, variability was found within subtypes. This further supports the suggestion that the diversity in Blastocystis STs may qualify them to be classified as separate species, since the ribosomal RNA genes in most organisms are generally considered as too conserved to discriminate between subspecies variants. The recent genome sequence of a Blastocystis ST7 strain [46] showed that some sequence variation does exist among individual copies of the SSU-rRNA gene in the genome. However, intra-strain variation does not interfere with assignment of Blastocystis samples to subtypes as the sequence obtained from the PCR reaction represents only the dominant sequence in the sample. The assignment of Blastocystis isolates to subtypes generated by the SSU-rRNA gene may be insufficient to clarify questions about pathogenic potential and transmission patterns. A further subdivision of the subtypes with molecular techniques that have a higher discriminatory capacity, for example, multilocus sequence typing (MLST), will possibly aid in investigations of the epidemiology of Blastocystis and possible associations between subtypes and pathogenicity. Aspects of zoonotic potential and transmission patterns may also be revealed by better discrimination among the subtypes. MLST has been used in many bacterial species but has recently also been used for subdivision of assemblages in the intestinal parasitic protozoan Giardia intestinalis [47, 48].
An interesting observation was the low genetic variability found in the ST4 sequences compared to what was seen among the other common subtypes identified in the study. In a BLAST search these Blastocystis sequences were identical to human ST4 sequences from different parts of the world in addition to four Blastocystis sequences from rodent faeces with origins in France [21]. Another group of rodent isolates with at least three SNPs in common constituted another lineage among the ST4 sequences that was revealed in the phylogenetic tree (Fig. 2). A similar branching among ST4 sequences has been shown by Noël et al. when examining mostly rodent isolates [21]. If the data indicating a low variability among human ST4 sequences can be confirmed by examination of other genes this may indicate that the ST4 lineage has expanded more recently on an evolutionary scale. In epidemiological studies a low grade of variability within a subgroup of isolates can also be explained by a selection of a specific lineage caused by advantageous properties for colonization or infection [49, 50]. Since the epidemiological context of the sequences compared in this study is unknown no such conclusions can be drawn. However, in relation to this hypothesis it is interesting that 76% of Blastocystis positive patients with acute diarrhoea harboured ST4 isolates in a recently published Danish study [31], indicating that ST4 may have a pathogenic potential.
In summary, Blastocystis of ST1, ST2, ST3, ST4, and ST7 were identified in 63 Swedish patient faecal samples with a dominance of ST3 (47.6%), followed by ST4 (20.6%). Intra-subtype sequence variations were identified in all the subtypes with the exception of ST4, which showed low variability in the 5′-end of SSU-rRNA. A comparison with recent studies in Europe indicates that the relative prevalence of ST4 in Europe may be higher than in other geographic regions.
References
Stenzel DJ, Boreham PF (1996) Blastocystis hominis revisited. Clin Microbiol Rev 9:563–584
Stensvold CR, Nielsen HV, Mølbak K, Smith HV (2009) Pursuing the clinical significance of Blastocystis – diagnostic limitations. Trends Parasitol 25:23–29
Tan KS (2008) New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin Microbiol Rev 21:639–665
Rene BA, Stensvold CR, Badsberg JH, Nielsen HV (2009) Subtype analysis of Blastocystis isolates from Blastocystis cyst excreting patients. Am J Trop Med Hyg 80:588–592
Li LH, Zhang XP, Lv S, Zhang L, Yoshikawa H, Wu Z, Steinmann P, Utzinger J, Tong XM, Chen SH, Zhou XN (2007) Cross-sectional surveys and subtype classification of human Blastocystis isolates from four epidemiological settings in China. Parasitol Res 102:83–90
González-Moreno O, Domingo L, Teixidor J, Gracenea M (2011) Prevalence and associated factors of intestinal parasitisation: a cross-sectional study among outpatients with gastrointestinal symptoms in Catalonia, Spain. Parasitol Research 108:87–93
Roberts T, Barratt J, Harkness J, Ellis J, Stark D (2011) Comparison of microscopy, culture, and conventional polymerase chain reaction for detection of Blastocystis sp in clinical stool samples. Am J Trop Med Hyg 84:308–312
Amin OM (2002) Seasonal prevalence of intestinal parasites in the United States during 2000. Am J Trop Med Hyg 66:799–803
Stark D, van Hal S, Marriott D, Ellis J, Harkness J (2007) Irritable bowel syndrome: a review on the role of intestinal protozoa and the importance of their detection and diagnosis. Int J Parasitol 37:11–20
Boorom KF, Smith H, Nimri L, Viscogliosi E, Spanakos G, Parkar U, Li LH, Zhou XN, Ok UZ, Leelayoova S, Jones MS (2008) Oh my aching gut: irritable bowel syndrome, Blastocystis, and asymptomatic infection. Parasit Vectors 1:40
Idris NS, Dwipoerwantoro PG, Kurniawan A, Said M (2010) Intestinal parasitic infection of immunocompromised children with diarrhoea: clinical profile and therapeutic response. J Infect Dev Ctries 4:309–317
Dinleyici EC, Eren M, Dogan N, Reyhanioglu S, Yargic ZA, Vandenplas Y (2010) Clinical efficacy of Saccharomyces boulardii or metronidazole in symptomatic children with Blastocystis hominis infection. Parasitol Res 108:541–545
Vogelberg C, Stensvold CR, Monecke S, Ditzen A, Stopsack K, Heinrich-Gräfe U, Pöhlmann C (2010) Blastocystis sp. subtype 2 detection during recurrence of gastrointestinal and urticarial symptoms. Parasitol Int 59:469–471
Giacometti A, Cirioni O, Fiorentini A, Fortuna M, Scalise G (1999) Irritable bowel syndrome in patients with Blastocystis hominis infection. Eur J Clin Microbiol Infect Dis 18:436–439
Yakoob J, Jafri W, Jafri N, Khan R, Islam M, Beg MA, Zaman V (2004) Irritable bowel syndrome: in search of an etiology: role of Blastocystis hominis. Am J Trop Med Hyg 70:383–385
Yakoob J, Jafri W, Beg MA, Abbas Z, Naz S, Islam M, Khan R (2010) Blastocystis hominis and Dientamoeba fragilis in patients fulfilling irritable bowel syndrome criteria. Parasitol Res 107:679–684
Stensvold CR, Lewis HC, Hammerum AM, Porsbo LJ, Nielsen SS, Olsen KE, Arendrup MC, Nielsen HV, Mølbak K (2009) Blastocystis: unravelling potential risk factors and clinical significance of a common but neglected parasite. Epidemiol Infect 137:1655–1663
Taşova Y, Sahin B, Koltaş S, Paydaş S (2000) Clinical significance and frequency of Blastocystis hominis in Turkish patients with hematological malignancy. Acta Med Okayama 54:133–136
Tan TC, Ong SC, Suresh KG (2009) Genetic variability of Blastocystis sp. isolates obtained from cancer and HIV/AIDS patients. Parasitol Res 105:1283–1286
Clark CG (1997) Extensive genetic diversity in Blastocystis hominis. Mol Biochem Parasitol 87:79–83
Noël C, Dufernez F, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Ho LC, Singh M, Wintjens R, Sogin ML, Capron M, Pierce R, Zenner L, Viscogliosi E (2005) Molecular phylogenies of Blastocystis isolates from different hosts: implications for genetic diversity, identification of species, and zoonosis. J Clin Microbiol 43:348–355
Stensvold CR, Suresh GK, Tan KS, Thompson RC, Traub RJ, Viscogliosi E, Yoshikawa H, Clark CG (2007) Terminology for Blastocystis subtypes – a consensus. Trends Parasitol 23:93–96
Stensvold CR, Alfellani MA, Nørskov-Lauritsen S, Prip K, Victory EL, Maddox C, Nielsen HV, Clark CG (2009) Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol 39:473–479
Parkar U, Traub RJ, Vitali S, Elliot A, Levecke B, Robertson I, Geurden T, Steele J, Drake B, Thompson RC (2010) Molecular characterization of Blastocystis isolates from zoo animals and their animal-keepers. Vet Parasitol 169:8–17
Kaneda Y, Horiki N, Cheng XJ, Fujita Y, Maruyama M, Tachibana H (2001) Ribodemes of Blastocystis hominis isolated in Japan. Am J Trop Med Hyg 65:393–396
Scicluna SM, Tawari B, Clark CG (2006) DNA barcoding of Blastocystis. Protist 157:77–85
Souppart L, Moussa H, Cian A, Sanciu G, Poirier P, El Alaoui H, Delbac F, Boorom K, Delhaes L, Dei-Cas E, Viscogliosi E (2010) Subtype analysis of Blastocystis isolates from symptomatic patients in Egypt. Parasitol Res 106:505–511
Dogruman-Al F, Dagci H, Yoshikawa H, Kurt Ö, Demirel M (2008) A possible link between subtype 2 and asymptomatic infections of Blastocystis hominis. Parasitol Res 103:685–689
Dogruman-Al F, Kustimur S, Yoshikawa H, Tuncer C, Simsek Z, Tanyuksel M, Araz E, Boorom K (2009) Blastocystis subtypes in irritable bowel syndrome and inflammatory bowel disease in Ankara, Turkey. Mem Inst Oswaldo Cruz 104:724–727
Domínguez-Márquez MV, Guna R, Muñoz C, Gómez-Muñoz MT, Borrás R (2009) High prevalence of subtype 4 among isolates of Blastocystis hominis from symptomatic patients of a health district of Valencia (Spain). Parasitol Res 105:949–955
Stensvold CR, Christiansen DB, Olsen KEP, Nielsen HV (2011) Blastocystis sp. subtype 4 is common in Danish Blastocystis-positive patients presenting with acute diarrhea. Am J Trop Med Hyg 84:883–885
Svenungsson B, Lagergren A, Ekwall E, Evengård B, Hedlund KO, Kärnell A, Löfdahl S, Svensson L, Weintraub A (2000) Enteropathogens in adult patients with diarrhea and healthy control subjects: a 1-year prospective study in a Swedish clinic for infectious diseases. Clin Infect Dis 30:770–778
Young KH, Bullock SL, Melvin DM, Spruill CL (1979) Ethyl acetate as a substitute for diethyl ether in the formalin-ether sedimentation technique. J Clin Microbiol 10:852–853
Lebbad M, Svärd SG (2005) PCR differentiation of Entamoeba histolytica and Entamoeba dispar from patients with amoeba infection initially diagnosed by microscopy. Scand J Infect Dis 37:680–685
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Arisue N, Hashimoto T, Yoshikawa H, Nakamura Y, Nakamura G, Nakamura F, Yano TA, Hasegawa M (2002) Phylogenetic position of Blastocystis hominis and of stramenopiles inferred from multiple molecular sequence data. J Eukaryot Microbiol 49:42–53
Noël C, Peyronnet C, Gerbod D, Edgcomb VP, Delgado-Viscogliosi P, Sogin ML, Capron M, Viscogliosi E, Zenner L (2003) Phylogenetic analysis of Blastocystis isolates from different hosts based on the comparison of small-subunit rRNA gene sequences. Mol Biochem Parasitol 126:119–123
Stensvold CR, Alfellani MA, Nørskov-Lauritsen S, Prip K, Victory EL, Maddox C, Nielsen HV, Clark CG (2009) Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int J Parasitol 39:473–479
Souppart L, Sanciu G, Cian A, Wawrzyniak I, Delbac F, Capron M, Dei-Cas E, Boorom K, Delhaes L, Viscogliosi E (2009) Molecular epidemiology of human Blastocystis isolates in France. Parasitol Res 105:413–421
Yoshikawa H, Wu Z, Kimata I, Iseki M, Ali IK, Hossain MB, Zaman V, Haque R, Takahashi Y (2004) Polymerase chain reaction-based genotype classification among human Blastocystis hominis populations isolated from different countries. Parasitol Res 92:22–29
Arisue N, Hashimoto T, Yoshikawa H (2003) Sequence heterogeneity of the small subunit ribosomal RNA genes among Blastocystis isolates. Parasitology 126:1–9
Stensvold CR, Arendrup MC, Jespersgaard C, Mølbak K, Nielsen HV (2007) Detecting Blastocystis using parasitologic and DNA-based methods: a comparative study. Diagn Microbiol Infect Dis 59:303–307
Denoeud F, Roussel M, Noel B, Wawrzyniak I, Da Silva C, Diogon M, Viscogliosi E, Brochier-Armanet C, Couloux A, Poulain J, Segurens B, Anthouard V, Texier C, Blot N, Poirier P, Ng GC, Tan KS, Artiguenave F, Jaillon O, Aury JM, Delbac F, Wincker P, Vivarès CP, El Alaoui H (2011) Genome sequence of the stramenopile Blastocystis, a human anaerobic parasite. Genome Biol 12:R29
Cacciò SM, Beck R, Lalle M, Marinculic A, Pozio E (2008) Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol 38:1523–1531
Lebbad M, Ankarklev J, Tellez A, Leiva B, Andersson JO, Svärd S (2008) Dominance of Giardia assemblage B in Leon, Nicaragua. Acta Trop 106:44–53
Enright MC, Spratt BG (1998) A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060
Sørensen UB, Poulsen K, Ghezzo C, Margarit I, Kilian M (2010) Emergence and global dissemination of host-specific Streptococcus agalactiae clones. mBio 1:e00178–10
Stensvold R, Brillowska-Dabrowska A, Nielsen HV, Arendrup MC (2006) Detection of Blastocystis hominis in unpreserved stool specimens by using polymerase chain reaction. J Parasitol 92:1081–1087
Stensvold CR, Nielsen SD, Badsberg JH, Engberg J, Friis-Møller N, Nielsen SS, Nielsen HV, Friis-Møller A (2011) The prevalence and clinical significance of intestinal parasites in HIV-infected patients in Denmark. Scand J Infect Dis 43:129–135
Poirier P, Wawrzyniak I, Albert A, El Alaoui H, Delbac F, Livrelli V (2011) Development and evaluation of a real-time PCR assay for detection and quantification of Blastocystis parasites in human stool samples: prospective study of patients with hematological malignancies. J Clin Microbiol 49:975–983
Böhm-Gloning B, Knobloch J, Walderich B (1997) Five subgroups of Blastocystis hominis from symptomatic and asymptomatic patients revealed by restriction site analysis of PCR-amplified 16S-like rDNA. Trop Med Int Health 2:771–778
Menounos PG, Spanakos G, Tegos N, Vassalos CM, Papadopoulou C, Vakalis NC (2008) Direct detection of Blastocystis sp. in human faecal samples and subtype assignment using single strand conformational polymorphism and sequencing. Mol Cell Probes 22:24–29
Scanlan PD, Marchesi JR (2008) Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J 2:1183–1193
Meloni D, Sanciu G, Poirier P, El Alaoui H, Chabé M, Delhaes L, Dei-Cas E, Delbac F, Luigi Fiori P, Di Cave D, Viscogliosi E (2011) Molecular subtyping of Blastocystis sp. isolates from symptomatic patients in Italy. Parasitol Res 109:613–619
Özyurt M, Kurt Ö, Mølbak K, Nielsen HV, Haznedaroglu T, Stensvold CR (2008) Molecular epidemiology of Blastocystis infections in Turkey. Parasitol Int 57:300–306
Eroglu F, Genc A, Elgun G, Koltas IS (2009) Identification of Blastocystis hominis isolates from asymptomatic and symptomatic patients by PCR. Parasitol Res 105:1589–1592
Dogruman-Al F, Yoshikawa H, Kustimur S, Balaban N (2009) PCR-based subtyping of Blastocystis isolates from symptomatic and asymptomatic individuals in a major hospital in Ankara, Turkey. Parasitol Res 106:263–268
Eroglu F, Koltas IS (2010) Evaluation of the transmission mode of B. hominis by using PCR method. Parasitol Res 107:841–845
Acknowledgements
The authors acknowledge the work involved in collecting the clinical samples and are thankful to Lillemor Karlsson, Christina Seeth Grönfors, Christine Stenström and Anna Dyrsmeds, all at Laboratory of Parasitology, Department of Clinical Microbiology, Karolinska University Hospital Solna, Stockholm, Sweden.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Forsell, J., Granlund, M., Stensvold, C.R. et al. Subtype analysis of Blastocystis isolates in Swedish patients. Eur J Clin Microbiol Infect Dis 31, 1689–1696 (2012). https://doi.org/10.1007/s10096-011-1416-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-011-1416-6