Abstract
To perform a meta-analysis to help resolve the controversy of whether the Angiogenin (ANG) rs11701 polymorphism is associated with amyotrophic lateral sclerosis (ALS) risk. A literature search of PubMed, Embase, Web of Science, Chinese National Knowledge Infrastructure, Wanfang and SinoMed was conducted for eligible studies published up to Jun 5, 2015. The strength of the association between the polymorphism and ALS susceptibility was estimated by odds ratio (OR) and associated 95 % confidence interval (CI). The pooled ORs were assessed for the dominant model (TG + GG vs. TT), recessive model (GG vs. TG + TT), heterozygote model (TG vs. TT), homozygote model (GG vs. TT) and allele model (G vs. T). Ten eligible articles were identified, which reported 14 case–control studies and a total of 5807 cases and 3861 controls. Analysis of pooled ORs and 95 % CIs suggested lack of association between the ANG rs11701 polymorphism and risk for ALS, Familial ALS or Sporadic ALS (all p value for z test >0.05). A stratified analysis according to Caucasian or Han Chinese origin further showed that the rs11701 polymorphism was not associated with the disease risk in Caucasians or Han Chinese. There is no difference in the polymorphism frequencies between patients with FALS or SALS. The ANG rs11701 polymorphism was not associated with risk for ALS, FALS or SALS. There is no difference between the polymorphism frequencies in patients with FALS or SALS. Further well-designed studies with larger populations are required to validate these results.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amyotrophic lateral sclerosis (ALS) is an idiopathic, fatal neurodegenerative disorder clinically characterized by progressive upper and lower motor neuron degeneration. The etiology and pathogenesis underlying the disease remain unknown, although it appears to be a multifactorial disorder caused by genetic–environmental interactions [1]. The ANG gene, which encodes angiogenin (ANG) playing a significant role in the biological process of angiogenesis, is an interesting candidate gene for modifying ALS risk [2, 3].
Quite a few epidemiological studies have been performed to evaluate the association between the rs11701 polymorphism (T/G) and ALS risk, yet results of these researches failed to reach an agreement. Greenway et al. first reported that the individuals carrying the G allele of the rs11701 polymorphism had an increased risk for ALS and sporadic ALS (SALS) than those carrying the T allele in the Irish and Scottish populations [2, 4]. Later, it is observed that subjects carrying the G allele had a greater risk for ALS and Familial ALS (FALS) than those carrying the T allele in an Italian cohort [5]. However, other studies observed no association in the populations from the USA [4, 6], England and Sweden [4], Italy [7–9], French [10], Germany [11] or Han Chinese [12, 13].
In our attempt to help resolve these discrepancies, a meta-analysis of all available studies assessing the association between the ANG rs11701 polymorphism and risk for ALS, FALS or SALS was conducted. We also compared the polymorphism frequencies between patients with familial ALS (FALS) or sporadic ALS (SALS).
Methods
Literature search strategy
Eligible studies were identified by systematically searching PubMed, Embase, Web of Science, Chinese National Knowledge Infrastructure, Wanfang and SinoMed. We used the following search terms: amyotrophic lateral sclerosis or ALS and Angiogenin or ANG. No language restriction was imposed. Last literature search was conducted on June 5, 2015. To identify studies that may have been missed by the database search, reference lists of all articles that met the inclusion criteria and of relevant review articles were examined.
Selection criteria
To be included in the meta-analysis, studies had to: (1) evaluate the association between the ANG rs11701 polymorphism and ALS risk; (2) provide sufficient data for assessing an odds ratio (OR) with 95 % confidence interval (CI); and (3) apply a case–control or genome-wide association design. If multiple articles appeared to report on overlapping cohorts, only the study with the largest number of patients was included. Studies were excluded if they did not report original research or if they were published only as abstracts or letters to the editor. Studies were also excluded if the frequency of the rs11701 polymorphism was zero in both cases and controls, since such studies would automatically be excluded by Stata software during meta-analysis [14].
Data extraction
Data were extracted by two of the authors (L-s. Pan and Z. Wang), and discrepancies were resolved by discussion with a third reviewer (D. Ding). The following data were extracted: first author’s name, year of publication, country or region, ethnicity of study population, gender distribution, family history, mean age at onset with standard deviation (SD), initial involvement (spinal or bulbar onset), SOD 1 mutation, sample size and genotype or allele distributions in cases and controls.
Statistical analysis
All data analyses were conducted using Stata 12.0 (http://www.stata.com). The strength of the association between the polymorphism and ALS susceptibility was estimated by OR and associated 95 % CI. The pooled ORs were assessed for the dominant model (TG + GG vs. TT), recessive model (GG vs. TG + TT), heterozygote model (TG vs. TT), homozygote model (GG vs. TT) and allele model (G vs. T). Subgroup analysis was conducted by stratification of population according to ethnic origin. A p value equal to or less than 0.05 was considered the threshold for statistical significance in all analyses.
Prior to meta-analysis, genotype distributions in each study were checked using the Hardy–Weinberg equilibrium test. Heterogeneity among studies was evaluated using the Q text and was quantified using I 2 [14]. An I 2 value below 25 % was considered to indicate homogeneity; values of 25 % to just under 50 %, to indicate low heterogeneity; values of 50 % to just under 75 %, moderate heterogeneity, and values of at least 75 %, substantial heterogeneity [15]. We planned to use a fixed-effect model to meta-analyze pooled data classified as homogeneous or of low heterogeneity, and a random-effect model to meta-analyze data classified as of moderate or substantial heterogeneity [14]. Publication bias was assessed using Egger’s and/or Begg’s tests [14, 16]. Sensitivity analysis was conducted by removing one single study each time [14].
Results
Literature search and included studies
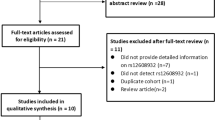
After screening titles or abstracts of 301 potentially eligible publications, 19 articles were retained for read in full. Through retrieving the full-text version of the above 19 articles, we excluded 9 articles because the authors did not examine the possible correlation between the ANG rs11701 polymorphism and ALS risk [17–20], because the study did not apply a case–control design [21, 22], because the frequency of the ANG rs11701 polymorphism was zero in both cases and controls [23, 24], or because the study [2] involved a population that overlapped with that in a larger study [4] that was included in the meta-analysis. Of the ten remaining publications, one [4] reported separate analyses for samples from five countries or regions were treated as five independent case–control studies in our meta-analysis. Therefore, the final meta-analysis included 14 case–control studies from ten publications [4–13] (Fig. 1).
Characteristics of included studies
Tables 1, 2, and 3 summarize key characteristics of the studies included in the meta-analysis. Of the 14 studies, 12 were from Europe or America (5564 cases and 3660 controls) and the other 2 were from mainland China (243 cases and 201 controls). Results of three studies (total 854 ALS patients) [4, 5] showed that individuals carrying the G allele of the rs11701 polymorphism had an increased ALS risk than those carrying the T allele, while no association was observed in the remaining 11 studies. ALS is more common in males than in females, which may reflect hormones and higher proportion of smoking and drinking histories in males than in females as possible risk factors for ALS risk [25]. Spinal onset ALS was more frequent than bulbar onset disease, which may reflect the greater proportion of the bulbar onset form in females than in males [25]. There was no indication of disequilibrium except for two studies of which data were not sufficient for calculation [6, 7].
Heterogeneity test
The heterogeneity test revealed obvious heterogeneity among studies in the dominant model (TG + GG vs. TT), heterozygote model (TG vs. TT) and allele model (G vs. T), and the heterogeneity still existed in the subgroup analysis by stratification of population according to Caucasian or Han Chinese origin (Tables 4, 5, 6). The heterogeneity altered most after omission of the study of Irish conducted by Greenway et al. [4] (I 2: 54.6 vs. 75.4 % in the dominant model) followed by omission of another study from Scottish [4] (I 2: 66.8 vs. 75.4 %, dominant model). And it nearly disappeared (I 2 = 26.2 %) after omission of both of these two studies [4].
Meta-analysis results
Tables 4, 5, 6 and 7 summarize key results of the meta-analysis. Overall, the meta-analysis results suggested lack of association between ANG rs11701 polymorphism and ALS risk in all genetic models (TG + GG vs. TT: OR = 1.18, 95 % CI = 0.95–1.47, p = 0.136; GG vs. TG + TT: OR = 1.12, 95 % CI = 0.80–1.58, p = 0.515; GG vs. TT: OR = 1.17, 95 % CI = 0.83–1.65, p = 0.362; TG vs. TT: OR = 1.17, 95 % CI = 0.95–1.46, p = 0.143; G vs. T: OR = 1.13, 95 % CI = 0.94–1.35, p = 0.183; Table 4). Similar null results were observed upon the association between the polymorphism and risk for FALS or SALS (all p value for z test >0.05; Tables 5, 6). A stratified analysis according to Caucasian or Han Chinese origin further showed that the polymorphism was not associated with the disease.
We also performed a meta-analysis on the topic for the four studies from Italy (1372 cases and 1492 controls) [5, 7–9], and the results revealed null association between the rs11701 polymorphism and ALS risk (data not shown). In addition, after excluding the two aforementioned studies [4] as the potential heterogeneity source, the meta-analysis results still showed no association (data not shown). We further conducted a meta-analysis after excluding the two studies [6, 7] of which data were not sufficient for assessing Hardy–Weinberg equilibrium and which were included in the meta-analysis of allele model (Table 1), and the results were not significantly altered (G vs. T: OR = 1.15, 95 % CI = 0.95–1.40, p = 0.168, random-effect model).
There is no difference in the polymorphism frequencies between patients with FALS or SALS (TG + GG vs. TT: OR = 0.97, 95 % CI = 0.76–1.23, p = 0.799; GG vs. TG + TT: OR = 1.52, 95 % CI = 0.81–2.83, p = 0.191; GG vs. TT: OR = 1.48, 95 % CI = 0.79–2.77, p = 0.223; TG vs. TT: OR = 0.93, 95 % CI = 0.72–1.20, p = 0.574; G vs. T: OR = 0.95, 95 % CI = 0.58–1.55, p = 0.845).
Assessment of publication bias and sensitivity analysis
Neither Egger’s test nor Begg’s test showed significant risk of publication bias (Table 4). The results of sensitivity analysis indicated no significant differences after removing any single study (figures not shown).
Discussion
We performed the present meta-analysis to address the differences in the studies on whether the ANG rs11701 polymorphism is associated with ALS risk [4–13]. With a combined larger sample size, the findings should be particularly useful because of greater statistical power than separate studies they included [15] or than meta-analysis with smaller populations. In the previous meta-analysis (with 1839 ALS patients and 1494 controls) conducted by Del Bo et al., it has been suggested that carriers of the G allele may have an increased risk for SALS through the fixed-effect model (TG + GG vs. TT: I 2 = 71.6 %; OR = 1.37, 95 % CI = 1.16–1.61, p < 0.001) while no statistically significant association was observed by the random-effect model (OR = 1.31, 95 % CI = 0.96–1.78, p = 0.089) [8]. Based on the principles defined by Higgins et al. [15] (see “Methods”), it seems that the random-effect model should be applied in the meta-analysis by Del Bo et al. [8] due to the obvious heterogeneity (p value for the heterogeneity test = 0.004, I 2 = 71.6 %) and therefore it seems to be a null result. With a combined larger sample size, our meta-analysis results suggest no association between the rs11701 polymorphism and the risk for ALS (n = 5807), FALS (n = 430) or SALS (n = 3775) (3861 controls, Tables 4, 5, 6). In fact, the association between the polymorphism and ALS risk was observed in only 3 [4, 5] of 14 studies (854 of 5807 ALS patients) included in the meta-analysis (Table 3). However, we cannot exclude that the null association between the polymorphism and FALS risk could be due to small FALS population (n = 430) and the mismatch between the sample size of FALS and controls (n = 2111). In addition, there is no evidence of a difference in the frequency of the rs11701 polymorphism between patients with FALS (n = 399) or SALS (n = 2130) (all p > 0.05; Table 7), which may reflect the small FALS population and the mismatch between the sample size of FALS or SALS.
In view of the fact that the samples of the included studies were from nine different countries, we also performed a meta-analysis for the four studies from Italian (1372 cases and 1492 controls) [5, 7–9], and the results also showed a null association between the rs11701 polymorphism and ALS risk (data not shown). Therefore, the fact that populations were from different countries may not be the potential confounding factor. In addition, the null results were found in the meta-analysis after excluding the two aforementioned studies [4] as the potential heterogeneity source (see “Results”). In fact, our sensitivity analysis showed that the pooled ORs and 95 % CIs were not significantly impacted by the studies that contribute to heterogeneity. Furthermore, no association was observed after excluding the two studies [7, 11] of which data were not sufficient for assessing Hardy–Weinberg equilibrium in the meta-analysis (see “Results”). These results suggest a high stability of our results.
While our meta-analysis offers the comprehensive evaluation of the rs11701 polymorphism and ALS risk with the combined largest sample size, the results should be interpreted with caution in view of several limitations. First, obvious heterogeneity among studies in the dominant, heterozygote and allele comparison models (TG + GG vs. TT, TG vs. TT, and G vs. T, Tables 4, 5, 6, 7) may influence the validity of the conclusion, though we applied a random effect for the meta-analysis in these three genetic models and our sensitivity analysis showed that the pooled ORs and 95 % CIs were not significantly influenced by the studies that contribute to heterogeneity. Second, the publication bias risk always exists, though we searched a range of international and Chinese databases without language constraints and the Egger’s and Begg’s tests suggested no significant risk of such bias. Finally, because of insufficiency of original information for each included subjects, data were not adjusted by risk factors of gender and initial involvement (spinal or bulbar onset) which may modify the association between the rs11701 polymorphism and ALS risk.
Future studies should verify our findings in larger populations, particularly in Han Chinese subjects, with larger groups of patients with FALS. Studies should also assess the rs11701 polymorphism in other ethnicities.
Conclusion
The ANG rs11701 polymorphism was not associated with risk for ALS, FALS or SALS. There is no difference between the polymorphism frequencies in patients with FALS or SALS.
References
Ludolph AC, Brettschneider J, Weishaupt JH (2012) Amyotrophic lateral sclerosis. Curr Opin Neurol 25(5):530–535
Greenway MJ, Alexander MD, Ennis S et al (2004) A novel candidate region for ALS on chromosome 14q11.2. Neurology 63(10):1936–1938
Aparicio-Erriu IM, Prehn JHM (2012) Molecular mechanisms in amyotrophic lateral sclerosis: the role of angiogenin, a secreted rnase. Front Neurosci 6:167
Greenway MJ, Andersen PM, Russ C et al (2006) ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet 38(4):411–413
Conforti FL, Sprovieri T, Mazzei R et al (2008) A novel Angiogenin gene mutation in a sporadic patient with amyotrophic lateral sclerosis from southern Italy. Neuromuscul Disord 18(1):68–70
Brown JA, Min J, Staropoli JF et al (2012) SOD1, ANG, TARDBP and FUS mutations in amyotrophic lateral sclerosis: a United States clinical testing lab experience. Amyotroph Lateral Scler 13(2):217–222
Corrado L, Battistini S, Penco S et al (2007) Variations in the coding and regulatory sequences of the angiogenin (ANG) gene are not associated to ALS (amyotrophic lateral sclerosis) in the Italian population. J Neurol Sci 258(1–2):123–127
Del Bo R, Scarlato M, Ghezzi S et al (2008) Absence of angiogenic genes modification in Italian ALS patients. Neurobiol Aging 29(2):314–316
Gellera C, Colombrita C, Ticozzi N et al (2008) Identification of new ANG gene mutations in a large cohort of Italian patients with amyotrophic lateral sclerosis. Neurogenetics 9(1):33–40
Paubel A, Violette J, Amy M et al (2008) Mutations of the ANG gene in French patients with sporadic amyotrophic lateral sclerosis. Arch Neurol 65(10):1333–1336
Fernandez-Santiago R, Hoenig S, Lichtner P et al (2009) Identification of novel Angiogenin (ANG) gene missense variants in German patients with amyotrophic lateral sclerosis. J Neurol 256(8):1337–1342
Zou ZY, Wang XN, Liu MS et al (2012) Identification of a novel missense mutation in angiogenin in a Chinese amyotrophic lateral sclerosis cohort. Amyotroph Lateral Scler 13(3):270–275
Zhang H, Zhang Y, Tang L et al (2015) The association between angiogenin gene variations and familial amyotrophic lateral sclerosis in Chinese Han patients. Chin J Intern Med 54(2):122–124
Zhang TS (2012) Zhong WZ Applied methodology for evidence-based medicine. Central South University Press, Changsha
Higgins JP, Thompson SG, Deeks JJ et al (2003) Measuring inconsistency in meta-analyses. BMJ 327(7414):557–560
Egger M, Smith GD, Phillips AN (1997) Meta-analysis: principles and procedures. BMJ 315(7121):1533–1537
Van Es MA, Schelhaas HJ, Van Vught PWJ et al (2011) Angiogenin variants in Parkinson disease and amyotrophic lateral sclerosis. Ann Neurol 70(6):964–973
van Es MA, Diekstra FP, Veldink JH et al (2009) A case of ALS-FTD in a large FALS pedigree with a K17I ANG mutation. Neurology 72(3):287–288
Millecamps S, Salachas F, Cazeneuve C et al (2010) SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: genotype-phenotype correlations. J Med Genet 47(8):554–560
Kirby J, Highley JR, Cox L et al (2013) Lack of unique neuropathology in amyotrophic lateral sclerosis associated with p. K54E angiogenin (ANG) mutation. Neuropathol Appl Neurobiol 39(5):562–571
Wu D, Yu W, Kishikawa H et al (2007) Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol 62(6):609–617
Tomiyama H, Kokubo Y, Sasaki R et al (2008) Mutation analyses in amyotrophic lateral sclerosis/parkinsonism-dementia complex of the Kii peninsula, Japan. Mov Disord 23(16):2344–2348
Chio A, Calvo A, Mazzini L et al (2012) Extensive genetics of ALS: a population-based study in Italy. Neurology 79(19):1983–1989
Kwon MJ, Baek W, Ki CS et al (2012) Screening of the SOD1, FUS, TARDBP, ANG, and OPTN mutations in Korean patients with familial and sporadic ALS. Neurobiol Aging 33(5):1017.e1017–1023
McCombe PA, Henderson RD (2010) Effects of gender in amyotrophic lateral sclerosis. Gend Med 7(6):557–570
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Rights and permissions
About this article
Cite this article
Pan, Ls., Deng, Xb., Wang, Z. et al. Lack of association between the Angiogenin (ANG) rs11701 polymorphism and amyotrophic lateral sclerosis risk: a meta-analysis. Neurol Sci 37, 655–662 (2016). https://doi.org/10.1007/s10072-015-2473-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-015-2473-x