Abstract
A total of eight strains of lactic acid bacteria were isolated from water kefir grains and assessed for their in vitro α-glucosidase inhibitory activity. Lactobacillus mali K8 demonstrated significantly higher inhibition as compared to the other strains, thus was selected for in vitro probiotic potential characterization, antibiotic resistance, hemolytic activity and adaptation to pumpkin fruit puree. L. mali K8 demonstrated tolerance to pH 2.5 and resisted the damaging effects of bile salts, pepsin and pancreatin, comparable to that of Lactobacillus rhamnosus GG ATCC 53103 (reference strain). Lack of hemolytic activity and susceptibility to the five standard antibiotics indicated the safety of the K8 strain. This strain showed singular properties to be used as starters in the pumpkin fruit puree fermentation. These preliminary in vitro tests indicated the safety and functionality of the K8 strain and its potential as a probiotic candidate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The definition for probiotics as “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” (FAO/WHO, 2002a). The most well-known probiotics are lactic acid bacteria (LAB) which are often found in fermented dairy, fruits, and vegetable-based food products (Corona et al., 2016; Koh et al., 2017; Plessas et al., 2017; Randazzo et al., 2016). Research has shown the correlation of numerous gastrointestinal health benefits with the probiotic intake which include antibacterial properties, alleviation of lactose intolerance, alleviation of irritable bowel syndrome and diarrhea (Park et al., 2016). Besides, probiotics consumption could improve postprandial hyperglycemia by a modest degree and help in controlling Type-2 diabetes mellitus (T2D) (Chen et al., 2014).
Emerging data demonstrated that postprandial hyperglycemia could predispose to the progression of T2D (Lee et al., 2012; van Dijk et al., 2011). Therefore, it is vital to keep the postprandial blood glucose level under control for the management of T2D. One method for improving postprandial hyperglycemia condition is through α-glucosidase enzyme inhibition which could retard the glucose absorption (Lee et al., 2012; Serra-Barcellona et al., 2017). The α-glucosidase enzyme is a hydrolase attached to the brush-border of intestinal cells. It hydrolyzes complex carbohydrates into glucose to be absorbed by the gut. Curbing the activity of intestinal α-glucosidase leads to a reduction in disaccharide hydrolysis which in turn can reduce glucose liberation and absorption, thereby decreasing blood glucose levels (Serra-Barcellona et al., 2017). Competitive inhibitors of intestinal α-glucosidase (e.g., miglitol, voglibose, and acarbose) play an essential role in managing T2D. However, these drugs commonly have various gastrointestinal side effects causing diarrhea and flatulence (Lee et al., 2012; Serra-Barcellona et al., 2017). In recent years, LAB strains have been reported to possess α-glucosidase inhibitory activity and might serve to alleviate the effects of T2D without adverse side effects (Chen et al., 2014; Muganga et al., 2015; Panwar et al., 2014; Zeng et al., 2016). However, to the best of authors’ knowledge, no study has dealt with the evaluation of α-glucosidase inhibitory activity of LAB isolated from water kefir grains for potential use as anti-hyperglycemic probiotics.
Water kefir is fermented by a consortium of yeasts, lactic and acetic acid bacteria embedded in exopolysaccharides matrix called water kefir grains (Koh et al., 2017). The microbial species from water kefir grains have high adaptability to different substrates thus could result in the development of novel probiotic products (Fiorda et al., 2017). Furthermore, water kefir has the potential to be used as anti-hyperglycemic and hypolipidemic supplements (Alsayadi et al., 2014; Koh et al., 2017). Therefore, in this study, several autochthonous LAB strains from water kefir grains were isolated, and their ability to inhibit α-glucosidase activity was evaluated, followed by the selection of the best strain based on the highest α-glucosidase inhibition. The best strain was then identified and examined for its safety as well as its in vitro probiotic characteristics. Its technological and microbiological properties were evaluated through the fermentation of pumpkin fruit puree.
Materials and methods
Isolation of LAB strain from water kefir grains
Commercial water kefir grains were obtained from Cultures for HealthSM (Morrisville, NC, USA) and were generated by inoculating them into brown sugar water (10% w/v) at 25 °C for 72 h. The grains were filtered from the fermented brown sugar water by using a plastic kitchen sieve and rinsed with distilled water. The same procedure was replicated for three subsequent days. Water kefir grains (16 g) were homogenized with 90 mL of Ringer’s solution (Merck, Darmstadt, Germany) in a sterile stomacher bag at high speed for 5 min. Subsequently, serial tenfold dilutions were made with phosphate buffer solution (PBS), and each dilution was streaked on De Man, Rogosa and Sharpe (MRS) agar supplemented with 150 μg/mL cycloheximide (Sigma-Aldrich, St. Louis, MO, USA). The plates were incubated at 37 °C for 72 h aerobically. After incubation, colonies from the plates with the dilutions (10−6 and 10−7) were isolated and purified. The comparison between colony morphology was conducted to quantify presumptive strain diversity, followed by light microscopy cell morphology observation. According to the size and shape of appeared colonies, eight colonies were isolated and successively streaked on MRS agar medium for three times, until a uniform colony was obtained. These eight isolates were Gram-positive and catalase positive; thus they were preliminarily identified as LAB (Park et al., 2016). The eight LAB isolates and the control Lactobacillus rhamnosus GG ATCC 53103 (LGG) strain were kept at − 80 °C in MRS broth (Merck) containing 20% (v/v) glycerol and sub-cultured every 6 months. Before every experiment, the isolates from frozen stocks were sub-cultured at least once in MRS broth at 37 °C for 24 h.
Evaluation of α-glucosidase inhibition
Stock cultures of 8 isolates and the control LGG strain were activated and incubated in MRS broth at 37 °C for 24 h. The cell concentrations obtained were between 1.2 × 109 and 2.7 × 109 CFU/mL. The cell-free supernatant (CFS) was prepared by centrifugation (12,000×g, 15 min, 4 °C), followed by filtration (0.22 µm membrane filter), and eventually, the pH of the solution was adjusted to 7.4 using sodium hydroxide (NaOH). For the preparation of cell-free extract (CFE), the intact cells were washed quickly with PBS (pH 7.4) and adjusted to 1.2 × 109 CFU/mL in PBS using McFarland standard. Consequently, the cells were disrupted by ultrasonication for 30 min in an ice bath (Elmasonic S 60 (H), Elma, Singen, Germany), followed by centrifugation (12,000×g, 20 min, 4 °C) and filter-sterilization (0.22 µm membrane filter). The final extract solution was kept at − 80 °C until use (Chen et al., 2014).
Rat intestinal acetone powder (I1630, Sigma-Aldrich) was dissolved in deionized water (10 mg/mL), and the solution was subjected to centrifugation (16,000×g, 30 min, 4 °C). The supernatant was kept at − 80 °C and used as rat intestinal α-glucosidase solution. Inhibition of α-glucosidase by the different LAB extracts (CFS and CFE) was assessed. The reaction mixture consisted of p-nitrophenyl-α-D-glucopyranoside (PNPG) (Sigma-Aldrich) (25 µL, 10 mM), PBS (pH 6.9, 25 µL, 0.1 M) and CFS/CFE samples (50 µL). Pre-incubation (37 °C, 10 min) of the reaction mixture was carried out in a 96-well plate. Subsequently, the reaction was started by adding rat intestinal α-glucosidase solution (50 µL) with ten times dilution in PBS (pH 6.9, 0.1 M) followed by incubation at 37 °C for 30 min. The reaction was stopped by adding sodium carbonate (100 µL, 0.1 M). The absorbance of p-nitrophenyl release was measured at 405 nm (Tecan microplate reader, Mannedorf, Switzerland). The absorbance values of the sample (enzyme reaction with sample) were rectified against sample blank (reaction with sample but without enzyme), positive control (reaction with enzyme without sample) and negative control (reaction without enzyme without sample). Percentage of α-glucosidase inhibition was calculated using the following formula (Zeng et al., 2016):
Identification of isolates
Genomic DNA from a single colony of the best isolate (with the highest α-glucosidase inhibition) was extracted using the NucleoSpin Tissue kit (Machery-Nagel, Duren, Germany) following the protocols specified by the manufacturer. The amount and purity of the extracted DNA were determined spectrophotometrically. The pure genomic DNA was sent to the Centre for Chemical Biology, Universiti Sains Malaysia, Penang, Malaysia to be identified by 16S rRNA sequencing and compared with the BLAST database (sequence-matching).
Evaluation of hemolytic activity
Overnight-grown culture of the best isolate was diluted to a turbidity level identical to 0.5 McFarland Units and streaked on blood agar (BD Difco, Sparks, MD, USA) plates containing 5% defibrinated sheep blood (Thermo Fisher Scientific, Waltham, MA, USA), followed by incubation at 37 °C for 24–48 h. After incubation, the sheep blood agar plates were observed to evaluate the hemolytic reaction. Presence of a green zone suggested partial hydrolysis red blood cells (α-hemolysis), whereas a clear zone surrounding the bacterial colony indicated total hydrolysis of red blood cells (β-hemolysis). No changes in the agar surrounding and under the bacterial colony indicated the absence of a hemolytic reaction (γ-hemolysis) (Jovanović et al., 2015).
Evaluation of antibiotic susceptibility
Antibiotic susceptibility of the best isolate and LGG strain was performed using agar overlay disc diffusion method (da Ferrari, 2016). A total of six antimicrobial agents (Oxoid, Basingstoke, Hampshire, England): vancomycin (30 µg/disc), chloramphenicol (30 µg/disc), oxacillin (1 µg/disc), tetracyclin (30 µg/disc), penicillin G (10 units/disc) and ciprofloxacin (5 µg/disc) were used according to the Clinical and Laboratory Standards Institute (2012). The overnight-grown cultures of the best isolate and LGG strain were diluted to a turbidity level identical to 0.5 McFarland Units and spread onto MRS agar media. Subsequently, discs containing specific antibiotic concentration were laid on the agar, and the plates were incubated at 37 °C for 24 h. The zone inhibition diameters were interpreted as resistant, ≤ 15 mm; sensitive, ≥ 21 mm (CLSI, 2012).
Characterization of the probiotic potential in vitro
The probiotic properties (tolerance to low pH, resistant to bile salts, pepsin, and pancreatin) of the best isolate and LGG strain were assessed. The overnight-grown bacterial cell cultures were centrifuged (10,000×g, 5 min, 4 °C) and inoculated (109 CFU/mL) either into acidified MRS broth (pH 2.5) supplemented with hydrochloric acid (1 M) or into MRS broth supplemented with bile salts (Sigma-Aldrich) at 0.5% (w/v). Cell viability was determined at 0 and 3 h of incubation at 37 °C with hydrochloric acid (pH 2.5) and at 0 and 4 h of incubation at 37 °C with bile salts. The viable cell counts are presented as Log10CFU/mL. In a parallel experiment, the supernatant of overnight-grown cultures of the best isolate and LGG strain was also inoculated either in PBS solution (pH 2.0) consisting of 3 mg/mL pepsin from porcine gastric mucosa (≥ 2500 units/mg, Sigma-Aldrich) or in PBS solution (pH 8.0) consisting of 1 mg/mL pancreatin from porcine pancreas (Sigma-Aldrich). The viable cells on MRS agar plates at 0 and 3 h of incubation at 37 °C with pepsin and at 0 and 4 h of incubation at 37 °C with pancreatin were enumerated. The reason of applying the aforementioned different incubation times was to closely reflect the duration (time) spent by the food in the stomach (gastric digestion) and small intestine (pancreatic digestion), respectively (Plessas et al., 2017).
Adaptation to pumpkin fruit puree
The pumpkin fruit puree was prepared by blending steam-cooked pumpkin fruit with distilled water [2:3 (m/v)]. The prepared pumpkin fruit puree was employed directly as a substrate for the best isolate inoculation. For the inoculum preparation, the overnight-grown isolate cell cultures were centrifuged (12,000×g, 3 min) and the pellets were suspended in pumpkin fruit puree. A level of 108 viable cells was used for inoculation into the pumpkin fruit puree sample. The sample was incubated in the optimized fermentation condition (Koh et al., 2017), and the fermentation was stopped when the pH value reached 4.5. The fermented pumpkin fruit puree sample was stored at 4 °C until further analysis of their technological and microbiological properties.
For the evaluation of the technological properties, a high-performance liquid chromatography (HPLC) system (Waters, Milford, MA, USA) coupled with UV/visible and refractive index chromatograms were used to monitor the lactic acid and residual sugar concentrations, respectively (Park et al., 2017). Ethanol concentration was measured by headspace gas chromatography using a flame ionization detector (Shimadzu GC-17A, Kyoto, Japan) (Braga et al., 2013). The pH value was measured at 25 °C using a pH meter (Mettler-Toledo, Schwerzenbach, Switzerland). For the evaluation of the microbiological properties, the viability of LAB in the fermented pumpkin fruit puree sample was enumerated by using MRS agar plates. A serial decimal dilution was prepared using Ringer’s solution (Sigma-Aldrich) and spread on the MRS agar, followed by incubation at 37 °C for 72 h. The results are presented as Log10CFU/mL.
Statistical analysis
SPSS 21.0 software (IBM, Armonk, NY, USA) was used for statistical analyses. One-way analysis of variance and Tukey’s test (α = 0.05) were performed to identify significant differences among the samples. The paired sample t test was used to compare the samples before and after treatment in the in vitro probiotic potential characterization.
Results and discussion
Assessment of the α-glucosidase inhibitory activity of isolated strain
The microbial strain isolated from water kefir grains could be a potential probiotic strain (Fiorda et al., 2017). In this study, eight LAB strains were isolated from commercial water kefir grains; whereas the commercial LGG strain, which has the antidiabetic properties (Kim et al., 2013), was used as a reference probiotic strain in all in vitro characterization of new probiotic candidates. Firstly, these LAB strains were assessed for their inhibitory activity against α-glucosidase.
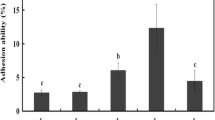
Previous studies have reported that α-glucosidase inhibition by LAB could be beneficial in the regulation of glycemia (Barreto et al., 2014; Koh et al., 2017; Muganga et al., 2015; Panwar et al., 2014; Zeng et al., 2016). Moreover, certain LAB strains were reported to be capable of generating bioactive components that could inhibit α-glucosidase activity in vitro (Muganga et al., 2015; Panwar et al., 2014; Zeng et al., 2016). As observed in Fig. 1, the α-glucosidase inhibition by CFS ranged from 5.2 to 39.4%, with the highest value for the K8 strain (39.4%) followed by LGG strain (37.9%). The α-glucosidase inhibition by CFE ranged from 2.3 to 15.5%, with the highest value for the K8 strain (15.5%) followed by LGG strain (13.7%). The K8 strain exhibited the best inhibitory activity among the eight strain tested, and the value was higher than that of the reference strain; thus the K8 strain was used for the subsequent analyses. The National Center for Biotechnology Information (NCBI) basic local alignment search tool was used to BLAST the sequence of the K8 strain. It was identified as Lactobacillus mali with GenBank accession number NR_112691.1 and 96% homology. The 16S rRNA sequencing is a suitable technique for identification of probiotic candidates (FAO/WHO, 2002a). The Lactobacillus mali species was proven to be present in water kefir grains (Hsieh et al., 2012), and a diet supplemented with Lactobacillus mali APS1 isolated from water kefir grains was efficacious in maintaining the blood glucose level in diet-induced obese mice (Lin et al., 2016). Furthermore, the CFS of L. mali K8 which demonstrated approximately 39% of α-glucosidase inhibition (Fig. 1) suggested its potential to alleviate postprandial hyperglycemia in T2D patients.
Eight lactic acid bacteria strain from water kefir grains showing their inhibition percentage (%) of α-glucosidase activity. Error bars represent the standard deviation of all trials. Different superscripts have mean ranks that are significant different from each other. Mean values with different uppercase alphabets (A–G) indicate significant difference among cell-free extract (CFE) samples; mean values with different lowercase alphabets (a–g) indicate significant difference among cell-free supernatant (CFS) samples. The probiotic Lactobacillus rhamnosus GG ATCC 53103 (LGG) served as a reference strain
Evaluation of hemolytic activity
Some lactic acid bacteria cultures exhibited pervasive hemolytic activity that could attack the host and stimulate lysis of red blood cells as well as penetration of toxic pathogenic agents, resulting in cytotoxic or severe bloody diarrhea (Hwang and Park, 2015; Jovanović et al., 2015). Therefore, it is vital to determine whether the hemolysin was produced by the probiotic candidate in blood agar media. In the present study, no clear lysis zone was observed surrounding the colonies of the L. mali K8 strain (Fig. 2). Hence, L. mali K8 strain was considered as γ-hemolytic. A similar finding conducted by using Brazilian kefir grains showed that the isolated LAB was non-hemolytic (Leite et al., 2015).
Evaluation of antibiotic susceptibility
As observed in Table 1, L. mali K8 and LGG strain were susceptible to all antibiotics tested except vancomycin. Vancomycin resistance of Gram-positive Lactobacillus bacteria is considered to be a natural and intrinsic property which could be due to the antibiotic permeability, antibiotic enzymatic degradation, and efflux of antimicrobials from the cytosol (Jampaphaeng et al., 2017; Lim et al., 2011).
Characterization of the probiotic potential in vitro
Preliminary information regarding the probiotic properties of the isolated strain can be obtained through in vitro tests. Several tests recommended by FAO/WHO (2002b) include tolerance to gastrointestinal condition (acid, bile salts, pepsin and pancreatin resistance), where probiotics can survive, grow, and perform beneficial actions. As can be seen in Fig. 3, L. mali K8 strain displayed probiotic potential comparable to the LGG reference strain. More specifically, when the L. mali K8 strain was exposed to pH 2.5 and pepsin independently for 3 h, this strain presented satisfactory levels of viability within the range of 106–107 CFU/mL. Furthermore, the L. mali K8 strain demonstrated adequate viability when exposed to bile salts and pancreatin independently for 4 h within the range of 107–108 CFU/mL. The tolerance to low pH, resistance to bile salts, pepsin and pancreatin of microbial strain is strain-specific hence could not be generalized (Jovanović et al., 2015).
Assessment of viability of the best strain, Lactobacillus mali K8, and the reference strain, Lactobacillus rhamnosus GG ATCC 53103 (LGG) after exposure to low pH, pepsin, pancreatin and bile salts. Error bars represent the standard deviation of all trials. Different superscripts have mean ranks that are significantly different from each other. Mean values with different lowercase alphabets (a–b) indicate the significant difference before and after treatment; mean values with different uppercase alphabets (A–B) indicate the significant difference between K8 and LGG strain for each experiment
Adaptation to pumpkin fruit puree
The majority of fermented probiotic products are dairy based thus are unsuitable for those with lactose intolerant and cholesterol issues (Fiorda et al., 2017). Researchers have been seeking dairy free fermented probiotic food products developments (Vijaya Kumar et al., 2015). Among the non-dairy foodstuff, pumpkin fruit puree was used as a substrate for the L. mali K8 fermentation in this study due to its potent anti-diabetic properties and α-glucosidase inhibitory activity (Baldi et al., 2009; Song et al., 2012). Figure 4 demonstrated the pH, LAB cells viability, residual sugar, lactic acid, and ethanol concentrations of the fermented pumpkin fruit puree during 4 weeks of refrigerated storage (4 °C). Increases in lactic acid concentration (0.2–0.9 g/L) and decreases in pH values (4.5–4.2) in fermented pumpkin fruit puree sample was observed, which could be attributed to post-acidification or the residual activity of the Lactobacillus strain that continue to produce acid during storage (Costa et al., 2013). Additionally, the residual sugar content decreased from 5.0 to 2.4 g/L during storage due to the activity of the Lactobacillus strain, which could utilize the residual sugar and produce lactic acid as a by-product (de Olmos et al., 2015; Koh et al., 2017). The formation of lactic acid could lower the pH and restrict the growth of the fermenting Lactobacilli during storage (Koh et al., 2017). As a result, the viability of the L. mali K8 strain in fermented pumpkin fruit puree maintained at 109 CFU/mL throughout the storage period, and it surpassed the minimum recommended probiotics level required for health benefits (106 CFU/mL) (Muganga et al., 2015). Furthermore, the ethanol content increased from 0.2 to 0.8% v/v during storage, which complied with the halal regulatory guideline (< 1% v/v ethanol in fermented food products) (Najiha et al., 2010). The increased ethanol content could be due to the heterofermentative metabolism of the Lactobacillus strain that fermented sugar present in the pumpkin fruit puree substrate thus produced ethanol and organic acids during storage (de Olmos et al., 2015; Koh et al., 2017). Therefore, the outcome of this investigation proved that L. mali K8 strain isolated from water kefir grains has the potential to serve as a probiotic in the development of functional fruit and vegetable-based food products.
pH value, lactic acid bacteria viability, residual sugar, lactic acid, and ethanol content, in the fermented pumpkin fruit puree inoculated with the best strain, Lactobacillus mali K8 during storage for 4 weeks at 4 °C. Error bars represent the standard deviation of all trials. Different superscripts have mean ranks that are significant different from storage time
References
Alsayadi M, Al Jawfi Y, Belarbi M, Soualem-Mami Z, Merzouk H, Sari, F, Sabri DC, Ghalim M. Evaluation of Anti-hyperglycemic and anti-hyperlipidemic activities of water kefir as probiotic on streptozotocin-induced diabetic Wistar rats. J. Diabetes Mellit. 4: 85–95 (2014)
Baldi A, Choudhary N, Maru J, Joshi R. Effect of pumpkin concentrate on alloxan induced diabetic rats. J. Glob. Pharma. Technol. 2: 24–27 (2009)
Barreto FM, Colado Simão AN, Morimoto HK, Batisti Lozovoy MA, Dichi I, Helena da Silva Miglioranza L. Beneficial effects of Lactobacillus plantarum on glycemia and homocysteine levels in postmenopausal women with metabolic syndrome. Nutrition. 30: 939–942 (2014)
Braga CM, Zielinski AAF, Da Silva KM, De Souza FKF, De Arruda Moura Pietrowski G, Couto M, Granato D, Wosiacki G, Nogueira A. Classification of juices and fermented beverages made from unripe, ripe and senescent apples based on the aromatic profile using chemometrics. Food Chem. 141: 967–974 (2013)
Chen P, Zhang Q, Dang H, Liu X, Tian F, Zhao J, Chen Y, Zhang H, Chen W. Screening for potential new probiotic based on probiotic properties and α-glucosidase inhibitory activity. Food Control. 35: 65–72 (2014)
CLSI. Performance standards for antimicrobial susceptibility testing: twenty two informational supplement. In: Clinical and Laboratory Standards Institute document M100-S22. Wayne, Michigan (2012)
Corona O, Randazzo W, Miceli A, Guarcello R, Francesca N, Erten H, Moschetti G, Settanni L. Characterization of kefir-like beverages produced from vegetable juices. LWT Food Sci. Technol. 66: 572–581 (2016)
Costa MGM, Fonteles TV, De Jesus ALT, Rodrigues S. Sonicated pineapple juice as substrate for L. casei cultivation for probiotic beverage development: process optimisation and product stability. Food Chem. 139: 261–266 (2013)
da Ferrari IS, de Souza JV, Ramos CL, Costa MM da, Schwan RF, Dias FS. Selection of autochthonous lactic acid bacteria from goat dairies and their addition to evaluate the inhibition of Salmonella typhi in artisanal cheese. Food Microbiol. 60: 29–38 (2016)
de Olmos A, Bru E, Garro MS. Optimization of fermentation parameters to study the behavior of selected lactic cultures on soy solid state fermentation. Int. J. Food Microbiol. 196: 16–23 (2015)
FAO/WHO. Guidelines for the evaluation of probiotics in food. In: Food and Agriculture Organization of the United Nations and World Health Organization Working Group Report. Ontario, Canada (2002)
FAO/WHO. Health and nutritional properties and guidelines for evaluation. In: Food and Agriculture Organization of the United Nations and World Health Organization Expert Consultation on Probiotics in Food. Ontario, Canada (2002)
Fiorda FA, Vinicius de Melo Pereira G, Thomaz-Soccol V, Rakshit SK, Binder Pagnoncelli MG, Porto de Souza Vandenberghe L, Soccol CR, Vinicius G, Pereira DM, Assumpç F, Thomaz-Soccol V, Kumar S, Giovana M, Pagnoncelli B, Porto L, Vandenberghe DS, Ricardo C. Microbiological, biochemical, and functional aspects of sugary kefir fermentation—a review. Food Microbiol. 66: 86–95 (2017)
Hsieh HH, Wang SY, Chen TL, Huang YL, Chen MJ. Effects of cow’s and goat’s milk as fermentation media on the microbial ecology of sugary kefir grains. Int. J. Food Microbiol. 157: 73–81 (2012)
Hwang JY, Park JH. Distribution of six exotoxin genes and production of L2-HBL and nheA proteins in six Bacillus cereus isolates from infant formula and produce. Food Sci. Biotechnol. 24: 379–382 (2015)
Jampaphaeng K, Cocolin L, Maneerat S. Selection and evaluation of functional characteristics of autochthonous lactic acid bacteria isolated from traditional fermented stinky bean (Sataw-Dong). Ann. Microbiol. 67: 25–36 (2017)
Jovanović JN, Nikolić B, Šeatović S, Zavišić G, Mitić-Ćulafić D, Vuković-Gačić B, Knežević-Vukčević J. Characterization of some potentially probiotic Lactobacillus strains of human origin. Food Sci. Biotechnol. 24: 1781–1788 (2015)
Kim SW, Park KY, Kim B, Kim E, Hyun CK. Lactobacillus rhamnosus GG improves insulin sensitivity and reduces adiposity in high-fat diet-fed mice through enhancement of adiponectin production. Biochem. Biophys. Res. Commun. 431: 258–263 (2013)
Koh WY, Utra U, Rosma A, Effarizah ME, Rosli WIW, Park Y-H. Development of a novel fermented pumpkin-based beverage inoculated with water kefir grains: a response surface methodology approach. Food Sci. Biotechnol. (2017)
Koh WY, Uthumporn U, Rosma A, Irfan R, Park YH. Optimization of a fermented pumpkin-based beverage to improve Lactobacillus mali survival and α-glucosidase inhibitory activity: a response surface methodology approach. Food Sci. Hum. Wellness. (2017)
Lee SH, Park MH, Han JS, Jeong Y, Kim M, Jeon YJ. Bioactive compounds extracted from Gamtae (Ecklonia cava) by using enzymatic hydrolysis, a potent α-glucosidase and α-amylase inhibitor, alleviates postprandial hyperglycemia in diabetic mice. Food Sci. Biotechnol. 21: 1149–1155 (2012)
Leite AMO, Miguel MAL, Peixoto RS, Ruas-Madiedo P, Paschoalin VMF, Mayo B, Delgado S. Probiotic potential of selected lactic acid bacteria strains isolated from Brazilian kefir grains. J. Dairy Sci. 98: 3622–3632 (2015)
Lim SM, Jeong KS, Lee NG, Park SM, Ahn DH. Synergy effects by combination with lactic acid bacteria and frutooligosaccharides on the cell growth and antimicrobial activity. Food Sci. Biotechnol. 20: 1389–1397 (2011)
Lin YC, Chen YT, Hsieh HH, Chen MJ. Effect of Lactobacillus mali APS1 and L. kefiranofaciens M1 on obesity and glucose homeostasis in diet-induced obese mice. J. Funct. Foods. 23: 580–589 (2016)
Muganga L, Liu X, Tian F, Zhao J, Zhang H, Chen W. Screening for lactic acid bacteria based on antihyperglycaemic and probiotic potential and application in synbiotic set yoghurt. J. Funct. Foods. 16: 125–136 (2015)
Najiha AA, Tajul AY, Norziah MH, Wan Nadiah WA. A preliminary study on halal limits for ethanol content in food products. Middle-East J. Sci. Res. 6: 45–50 (2010)
Panwar H, Calderwood D, Grant IR, Grover S, Green BD. Lactobacillus strains isolated from infant faeces possess potent inhibitory activity against intestinal alpha- and beta-glucosidases suggesting anti-diabetic potential. Eur. J. Nutr. 53: 1465–1474 (2014)
Park S, Ji Y, Park HH, Lee K, Park HH, Beck BR, Shin H, Holzapfel WH. Evaluation of functional properties of lactobacilli isolated from Korean white kimchi. Food Control. 69: 5–12 (2016)
Park JB, Lim SH, Sim HS, Park JH, Kwon HJ, Nam HS, Kim MD, Baek HH, Ha SJ. Changes in antioxidant activities and volatile compounds of mixed berry juice through fermentation by lactic acid bacteria. Food Sci. Biotechnol. 26: 441–446 (2017)
Plessas S, Nouska C, Karapetsas A, Kazakos S, Alexopoulos A, Mantzourani I, Chondrou P, Fournomiti M, Galanis A, Bezirtzoglou E. Isolation, characterization and evaluation of the probiotic potential of a novel Lactobacillus strain isolated from Feta-type cheese. Food Chem. 226: 102–108 (2017)
Randazzo W, Corona O, Guarcello R, Francesca N, Germanà MA, Erten H, Moschetti G, Settanni L. Development of new non-dairy beverages from Mediterranean fruit juices fermented with water kefir microorganisms. Food Microbiol. 54: 40–51 (2016)
Serra-Barcellona C, Habib NC, Honoré SM, Sánchez SS, Genta SB. Enhydrin regulates postprandial hyperglycemia in diabetic rats by inhibition of α-glucosidase activity. Plant Foods Hum. Nutr. 72: 156–160 (2017)
Song Y, Zhang Y, Zhou T, Zhang H, Hu X, Li Q. A preliminary study of monosaccharide composition and α-glucosidase inhibitory effect of polysaccharides from pumpkin (Cucurbita moschata) fruit. Int. J. Food Sci. Technol. 47: 357–361 (2012)
van Dijk JW, Manders RJF, Hartgens F, Stehouwer CD, Praet SFE, van Loon LJC. Postprandial hyperglycemia is highly prevalent throughout the day in type 2 diabetes patients. Diabetes Res. Clin. Pract. 93: 31–37 (2011)
Vijaya Kumar B, Vijayendra SVN, Reddy OVS. Trends in dairy and non-dairy probiotic products—a review. J. Food Sci. Technol. 52: 6112–6124 (2015)
Zeng Z, Luo J, Zuo F, Zhang Y, Ma H. Screening for potential novel probiotic Lactobacillus strains based on high dipeptidyl peptidase IV and α-glucosidase inhibitory. J. Funct. Foods. 20: 486–495 (2016)
Acknowledgements
The authors are grateful to the ASEAN University Network (AUN) and Korea Association of the Southeast Asian Studies for providing financial support under ASEAN-ROK Academic Exchange Programme 2016/2017; Research Universiti Grant (1001/PTEKIND/811339) and Universiti Sains Malaysia Vice-Chancellor Award Scholarship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Koh, W.Y., Utra, U., Ahmad, R. et al. Evaluation of probiotic potential and anti-hyperglycemic properties of a novel Lactobacillus strain isolated from water kefir grains. Food Sci Biotechnol 27, 1369–1376 (2018). https://doi.org/10.1007/s10068-018-0360-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10068-018-0360-y