Abstract
Objective
To investigate the frequency of glutathione S-transferase (GST), catalase, and SOD2 genetic polymorphisms and their correlation with SLE.
Methods
A total of 290 females (patients = 151; controls= 139) were recruited. Multiplex PCR was performed for genotyping GSTM1 and GSTT1 genes, whereas real-time qPCR was used for determination of SNPs: CAT C262T, SOD2 C47T, GSTP1 A313G and GSTP1 IVS6 -C16T.
Results
Thiol levels are decreased in SLE patients (p<0.001), while MDA levels were significantly higher (p<0.001) and those carrying the polymorphisms had higher rates of oxidative stress. Patients with double null deletion GSTT1null/GSTM1null had a frequency almost five times higher than the controls (p<0.001, OR 4.81, CI 1.98–12.11). SLE patients had a lower wild-type frequency of SOD2CC allele compared to controls (12.4% vs 27.3%). Statistical significances were observed on the association between the GSTT1null and GSTM1null with SOD2mut (p<0.001, OR 0.15, CI 0.05–0.47), with GSTP1 A303G (p=0.012, OR 0.19, CI 0.05–0.69), and with GSTP1 IVS6 (p=0.008, OR 0.14, CI 0.03–0.63). The same was observed between SOD2 C47T with GSTP1 A303G (p=0.09, OR 0.27, CI 0.09–0.74) and GSTP1 IVS6 (p=0.036, OR 0.41, CI 0.18–0.92).
Conclusions
The deletion GSTT1null/GSTM1null may contribute to the increased of the oxidative stress in SLE patients. Isolated GSTP1 and CAT polymorphisms do not seem to influence the increased oxidative stress, neither SLE clinical manifestations. SOD2 47CT/TT allele may have greater oxidative stress due to structural change in the protein and decreased H2O2 production. The combination of polymorphic genes may be involved in the pathogenesis of the disease.
Key points • Major question of our paper: Many studies have shown that the antioxidant status levels are decreased in patients with SLE, especially in severe stages of disease. We believe that this paper will be of interest to the readership of your journal had the involvement of polymorphisms and mutations in several genes that contribute to the genetic etiology of SLE, suggesting that these may influence the mechanisms of disease. • Our results. Thiol level was significantly (p<.001) lower and MDA level significantly increased (p<.001) among SLE patients. Those carrying the polymorphisms had higher rates of oxidative stress. SLE Patients had a frequency almost five times higher of double null deletion GSTT1null/GSTM1null than the controls. SLE Patients had a lower wild type frequency of SOD2CC allele compared to controls (12.4% vs 27.3%). We believed the deletion GSTT1null/GSTM1null may contribute to the increased of the oxidative stress in SLE patients while carriers of the mutant SOD2 47CT/TT allele may have greater oxidative stress due to structural change in the protein and decreased H2O2 production. The combination of polymorphic genes may be involved in the pathogenesis of the disease. • Implications of our results: Evidence for the involvement of genetic factors in severe clinical to lupus is compelling. This manuscript shows genetic insights in pathogenic pathways that may lead to severe clinical implications to LES. Therefore, it is necessary to understand their impact on overall disease pathogenesis and prognosis in these patients. We understand from general consensus about environmental factors can modify disease, however, maybe just in individuals who have a permissive genetic background. Even that no single gene predisposes some individuals to LES, we believe the genetic factors described in this manuscript are important elements in susceptibility to severe clinical to LES |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic lupus erythematosus (SLE) is a chronic inflammatory autoimmune disease. The formation of immune complexes leads to multisystemic disorders, with specific signs and symptoms depending on the affected organ, characterized by periods of exacerbation and remission. The inflammatory nature of the disease implies the existence of increased oxidative stress in these patients and can contribute to immune dysfunction and damage of biological molecules such as DNA, RNA, proteins, and antioxidants. Although the etiology is not yet fully understood, studies show that combinations of polymorphic genes can predispose to the disease, allowing environmental factors to trigger immunopathologic responses in these patients [1].
Oxidative damage can be propagated through endogenous and exogenous pathways. Decreased levels of reduced glutathione, formation of superoxide anion (O2-) via mitochondrial dysfunction, and formation of hydroxyl radicals (OH-), for example, are endogenous causes of the oxidative stress. This state is also observed in the decrease of antioxidant defense, caused by the decrease or inactivity of the enzymes that participates in redox equilibrium [2, 3]. The products of these oxidative modification cascades, such as serum levels of malondialdehyde (MDA), resulting from lipid peroxidation, are associated with the activity, organ damage, and comorbidities in SLE. Inversely, decreased levels of serum thiol have been used as markers of oxidative stress [4]. Studies demonstrate that enzymes such as glutathione S-transferase (GST), catalase (CAT), and mitochondrial superoxide dismutase 2 (SOD2) are altered in SLE. These enzymes have polymorphic genes that alter the structure or activity, thus contributing to the increased oxidative stress [5, 6]. The mundial literature showed lupus nephropathy is a severe clinical of SLE. Recently, Bona and colleague (2020) showed imbalance in the redox status between lupus nephritis types, principally in active lupus nephritis patients, with potential lipid peroxidation and could also affect renal tubular function in these patients [7].
Glutathione S-transferase (GST) is a superfamily of phase II enzymes that act catalyzing the detoxification of hydrophobic and electrophilic compounds with reduced glutathione [8]. Products of GSTM1 and GSTT1 genes are involved in the detoxification of aromatic hydrocarbons, mutagens, and compounds such as MDA [8, 9]. Both genes have homozygous null variants (M1null and T1null) resulting in the absence of the enzymes and they are investigated in the susceptibility to diseases such as cancers, atherosclerosis, and hypertension [10,11,12]. Glutathione S-transferase P1 (GSTP1) is of importance for oxidative stress research because of its role mainly in detoxifying xenobiotics and metabolizing any drugs and because of its involvement in cell cycle and apoptosis regulation [13, 14]. Two common GSTP1 genetic polymorphisms are studied extensively. Many common GSTP1 genetic polymorphisms are studied extensively; however, functionally, the single nucleotide polymorphisms (SNP) rs1695 (A313G) and rs1871042 (IVS6 -C16T) express a protein with altered thermal instability and catalytic activity [15,16,17,18].
Mitochondrial superoxide dismutase 2 along with catalase comprises the primary enzyme system defense against reactive oxygen species (ROS) [19], whereas SOD2 acts in the conversion of O2 anion to the less toxic hydrogen peroxide (H2O2) and then catalase catalyzes the conversion of H2O2 into water and oxygen [20]. The SOD2 C47T SNP (rs4880) alters the structure of the enzyme, decreasing the efficiency of entry into the mitochondrial matrix and consequently decreases the production of H2O2 and dismutation. Catalase is expressed in all tissue types, predominantly in the liver, kidney, and erythrocytes [21]. The CAT C262T SNP (rs1001179) is found in the promoter region of the gene and is associated with low enzyme activity [22].
Studies have demonstrated the involvement of polymorphisms and mutations in several genes that contribute to the genetic etiology of SLE. This suggests that SNPs and deletions can influence the mechanisms of disease [23, 24]. Recent studies have shown that the antioxidant status levels are decreased in patients with SLE, especially in severe stages of disease [25], and an Egyptian study demonstrated a strong association of the SOD2 rs2758332 not GSTP1 rs1695 polymorphism with the risk of SLE disease [26]. The aim of our study was to investigate the polymorphisms of glutathione s-transferases (T1null, M1null, P1 A313G, and P1 IVS6 -C16T), superoxide dismutase (C47T), catalase (C-262T), and their correlation with SLE.
Materials and methods
Study population
This was a case-control study conducted in the period from 2009 to 2014. A total of 348,187 patients and 195 healthy volunteers (controls) attending Rheumatology Department of Federal University of Amazon (UFAM), Brazil, were evaluated. All the patients as well as controls were only females. The control group consisted of people without consanguinity with patients, non-smoking, without inflammatory symptoms, and diseases. Patients and controls were excluded if they exhibited diseases, condition, or behavior that influences the rise of ROS, such as infectious diseases, primary diabetes, HIV, smoking, and pregnancy. The protocol was approved by the Institutional Ethical Committee from UFAM (CAAE n° 0286.0.115.000-11) and confirms to the ethical guidelines of Declaration of Helsinki 1975, as revised in 2000.
Diagnosis of SLE was based on clinical and laboratory criteria of the American College of Rheumatology (ACR) and was evaluated according to disease activity index SLEDAI at the time of sample collection and divided into three groups: inactive lupus (IL) (n=71) consisted of individuals who did not show lupus activity (SLEDAI = 0) for at least 1 year and without laboratory results characteristic of kidney injury; active lupus (AL) (n = 47) composed of active lupus patients (SLEDAI ≥ 4) without renal manifestation; and lupus nephritis (LN) (n=69) of active lupus patients (SLEDAI ≥ 4) diagnosed with kidney injury (persistent proteinuria greater than 500mg/24h or ≥3 + in urinary sediment) [27]. It is important to emphasize that 85% of our LN patients were classified of group IV, patients with diffuse nephritis [28].
According to a 2013 genetic study, the ancestry of the inhabitants of Manaus is 45.9% European, 37.8% Native American, and 16.3% African [29]. The authors do not speculate about the race of the participants due to the strong regional ancestral mix found in the Amazonian caboclos, which originate with the arrival of Caucasians and blacks in indigenous lands. However, in the interview and signing of the informed consent form, we asked each patient/volunteer (control) to answer variables such as weight, disease time, and skin color (white, black, Brown, and indigenous). Brown color was the most frequent in both groups, with 82% in patients and 78% in controls, followed by 12 and 17% for white, and 6 and 5% black, respectively. No indigenous people participated in the study.
Sample collection and clinical and laboratory analysis
Peripheral blood samples (2–3 ml) were collected in tubes containing EDTA for hematological and molecular analysis and in tube without anticoagulant for serum analysis. All the hematological, molecular, and biochemical analysis were carried out at the Laboratory of Hematology and Molecular Biology (LAEBM), Faculty of Pharmaceutical Sciences (FCF-UFAM), Manaus, Amazonas, Brazil.
Serum biomarkers
Serum urea, creatinine, urea, uric acid, and total proteins were quantified using Cobas Mira Plus® (Roche Diagnostic Systems®, Inc., Branchburg, NJ). Quantitative measurement thiols and MDA were performed as described previously [4]. Total ROS stress index and total antioxidant capacity quantification were performed using established methods [30, 31].
Sample size and power analysis
To estimate sample size for our study, we chose the total serum thiols as the central study variable. From previous results of our group [4], it was demonstrated a detectable of 80±50μmol/L in serum thiol levels. It was then defined a significance of 0.05 and power at 90%, we calculated an effect size of 1.98 and estimated a sample size of 108 patients for each study group. Sample size calculation and power analysis were performed by G*Power: Statistical Power Analyses v3.1.9.2 [32].
DNA extraction and genotyping
The DNA was extracted from leukocytes using Brazol® technique according to the manufacturer’s instructions. Homozygous null deletion polymorphisms in M1 and T1 genes were determined by multiplex PCR using specific primers [33]. SNP genotyping was conducted using TaqMan® SNP genotyping assays (Applied Biosystems, Foster City, CA) on a StepOnePlus™ real-time PCR system (Applied Biosystems). The probes used were GSTP1 A303G (rs1695), GSTP1 IVS -16CT (rs1871042), CAT C267T (rs1001179), and SOD2 C47T (rs4880).
Statistical analysis
Analysis of the distribution of variables was performed using the Kolmogorov-Smirnov test together with ANOVA parametric tests or nonparametric Kruskal-Wallis tests. The ANOVA parametric test was used to analyze the distribution of the means of quantitative variables with normal distribution within categories. The nonparametric Kruskal-Wallis test was used for the off-normal distributions.
The analysis of qualitative or categorical variables of three or more groups was performed by nonparametric chi-square (χ2), corrected by the Mantel-Haenszel test and Yates. The analysis values of less than 4 were performed by Fisher’s exact test. Genotype frequencies of the SNPs observed were analyzed by χ2 test for association analysis of the variables (SLE × controls). The odds ratio (OR) was calculated to estimate the risk, and the adopted confidence interval was 95%. When values less than 4 were obtained, a Fisher’s exact test was used.
The data analysis was performed using IBM SPSS Statistics, version 22.0; GraphPad Prism version 5.0; and Epi Info™ 7.1.5.2. A p value of less than 0.05 was considered statistically significant. The genotypic frequencies for polymorphisms were analyzed in accordance with Hardy-Weinberg equilibrium.
Results
The genotypic frequencies of polymorphisms were 34.5% to GSTT1null and 15.8% to GSTM1null; 44.4% to GSTP1 313AG and 16.9% to GSTP1 313GG; 27.8% to GSTP1 IVS6CT and 6.7% to GSTP1 IVS6TT; 48.9% to SOD 47CT and 33.8% to SOD 47TT; and 14.8% to CAT 262CT and 0.9% to CAT 262TT.
All individuals in our study (patients and controls) were women. The average age was 32.7± 6.8 in the control group ranging from 18 to 54 years and 33.7 ± 9.8 in patients ranging from 15 to 64 years. Photosensitivity was prevalent in most patients (88.4%), followed by erythematous rash (72.5%), renal disease (46.7%), hypertension (high blood pressure) (41.7%), oral ulcers (34.8%), fibromyalgia (14.9%), and antiphospholipid syndrome (12.5%). Other clinics as diabetes, cardiac failure, and asthma had a frequency below 5%. Demographic data of patients and controls and the two most frequent medications used by patients are showed in Table 1. It is important to note that patients involved in this study were being treated with prednisone (20–60 mg/day) and chloroquine (150–400 mg/day). Those patients who were being treated with concentrations above of these were excluded from the study. Corticosteroid use (prednisone) was categorized as follows: low (≤7.5mg/day), medium (7.5–30 mg/day), and high dose (>30mg/d) [34].
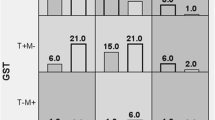
In Fig. 1, we can observe the involvement of the mutations on the oxidative stress index, while Table 2 demonstrates the hematological, biochemical, and oxidative stress parameters among SLE patients and controls. Hematocrit, hemoglobin, as well as platelet counts were found to be significantly lower (p<0.001) in patients as compared to controls.
Serum urea, creatinine, and uric acid levels were increased among patients, while the levels of total protein and albumin were decreased in the present study. We also evaluated serum oxidative markers such as thiol, ROS, and MDA between SLE patients and controls. Whiles thiol level was significantly (p<0.001) lower in SLE patients, MDA on the other hand was significantly increased (p<0.001) among SLE patients.
Frequencies of GSTM1 and GSTT1 genotypes between the study groups by index SLEDAI (IL, AL, and LN) were also evaluated (data not shown). We found that 20 individuals carrying the double null deletion (GSTT1null/GSTM1null) are in IL group. The comparison between the frequencies of GSTT1 and GSTM1 genotypes in patients and controls is shown in Table 3. No significant differences were observed in the frequencies of GSTT1null and GSTM1null genotypes between patients and controls. However, we observed significant differences among patients carrying GSTT1null/GSTM1null double null deletion and controls (OR 4.81, CI 1.98–12.11, p<0.001).
The SOD2 C47T genotypic frequency demonstrated that patients had a lower frequency of the wild allele than the controls (12.4% vs 27.3%, p=0.026). We found that frequency of the mutant allele was nearly two times higher in patients compared to wild-type allele.
Frequency analysis between two polymorphisms was performed in order to verify the joint involvement of the mutations with the disease (Table 4). Associations of protection of the combined GSTs and SOD2 genotypes were observed in controls when compared to SLE patients.
The possible influence of the polymorphisms, separate and in combination, with the levels of thiols was also evaluated (Fig. 2). It was shown that SLE patients with of mutation have a higher concentration of these antioxidants, suggesting a protective effect mainly changing by GSTT1null/GSTM1null patients. Figure 3 shows greater peripheral pancytopenia in patients with SNPs and SLE, mainly involving the GSTs genotypes Mu, Theta, and Pi.
Discussion
The majority frequency of women in our study is in agreement with the literature that demonstrates that SLE is predominant in women of fertile age in the proportion 1:9 in relation to men [35, 36]. The fact that SLE as well as other autoimmune diseases are predominant in women is not fully understood, but recent study has shown that the X chromosome through independent hormones mechanisms can be one of the genetic causes of SLE based on the comparison in subject people with X trisomy and carriers of the disease [37].
The had higher rates of oxidative stress were found in individuals carrying the polymorphisms, therefore we believe that the mutations mainly in combination contribute to increased oxidative stress. Although they did not correlate genetic markers, the same results were found in rheumatoid arthritis patients [38] and lupus nephritis patients [25], which demonstrated high levels of oxidative stress in these patients.
The GST polymorphisms are distributed heterogeneously in different populations around the world [39, 40]. GSTT1null frequency was similar to other studies in Brazilian populations ranging from 23 to 30% [41]. However, GSTM1null frequency was different from other regions of Brazil ranging from 31 to 40% [42] but similar to other studies, particularly among Brazilian Indians and South African populations. The distribution of A313G variants of GSTP1 shows data similar to a study conducted among indigenous populations of Amazonas; wild-type, heterozygous, and homozygous frequency were 35, 46, and 19%, respectively [43, 44]. Our study showed for the first time the frequency of GSTP1 IVS6 -C16T in the Brazilian population corroborating with databases like dbSNP (https://www.ncbi.nlm.nih.gov/snp/rs1871042).
The genotypic frequencies of SOD2 C47T and CAT C262T are also in accordance with data found in different Brazilian populations [45]. Taufer et al. (2005) demonstrated an approximated frequency of 60.6 and 24.4%, respectively, for CT and TT genotypes of SOD2 and it was a mixed ethnicity population, like our study [46].
Lower hematocrit and hemoglobin concentrations are expected in SLE patients. Studies have shown a correlation between low levels of hematocrit with the inflammatory response and disease activity. Also, thrombocytopenia is reported to be present in approximately 33% of SLE patients; however, levels below 100.000/mm3 were demonstrated only in 5% of cases [47, 48].
Thiol level results corroborated with earlier studies that suggest decrease in thiol levels may be an important biomarker of disease activity [3, 49]. The MDA marker is in accordance with the study by Pérez et al. (2012), which demonstrated a significant increase of MDA in SLE patients (OR 12,5; IC 3,7–41,5; p<0.001), indicating increased oxidative stress by lipid peroxidation in these patients [4].
In our study 86.8% of patients used prednisone as immunosuppressive (p<.001) and the high frequence of the double null deletion (GSTT1null/GSTM1null) found in IL group patients suggests the possibility that this double deletion is involved in a better response to treatment. In a study of patients with acute lymphoblastic leukemia, a better prognostic factor was the initial response to treatment with prednisone and, although it was not statistically significant (p=.071) GSTT1null deletion gave a decrease of 6.7-fold the risk of poor response to prednisone compared to wildtype individuals, while, GSTM1 and GSTP1 genotypes did not show any association [50]. Recent study, Salime and col. (2015) in an Iranian population showed high frequency to GSTT1 null in SLE patients than controls. This same study, the combination of GSTT1 null/ GSTM1 null genotypes as a risk factor for SLE (OR 2.8; CI 1.2-6.4, p=.014) [51]. We know that glutathione (GSH) is an important antioxidant, peforming concomitantly with glutathione S-transferase, important role in the detoxification via catalysis. This way, we understand that antioxidant activity of these enzymes exercise significant reduction of the formation of conjugates with endogenous dangerous compounds, avoiding the increased oxidative stress and facilitating metabolic detoxification of this xenobiotics.
Deletions on the GST genes are widely studied for associations with diseases [52, 53]. Although some studies have shown the involvement of GSTT1null and GSTM1null deletions in SLE, the results are different in each population, while in our study, the influence of the polymorphisms was not observed separately; other studies have reported the risk of at least one of the genotypes in susceptibility to SLE. Rupasree et al. (2013) demonstrated that GSTT1null deletion may be associated with risk of SLE (OR 1.83, CI 1.11–3.01) as well as the association between polymorphisms of the enzymes GSTM1, GSTT1, and catecholamine methyltransferase (phase II enzyme) (OR 3.33, CI 2.30–4.82) [54]. Zhang et al. (2010) in a Chinese population showed significant differences between carriers of the GSTM1null, concluding the participation of the polymorphism on susceptibility to SLE [55]. Kang et al. (2005) did not observe any association between GST genes and susceptibility to SLE [56]. The authors suggest that the deletions may influence some clinical manifestations, the GSTM1null was associated with low frequency of hematological disorders (p=0.042), and GSTT1null was associated with low frequencies of discoid rash (p=0.018) and nephritis (p=0.033).
In present study, we also evaluated the influence of genotypes on clinical manifestations but no positive correlations were observed (data not shown). Fraser et al. (2003) evaluated the polymorphisms of GST in SLE patients with workers exposed to solar radiation. The authors in their study suggested GSTM1null can modify the effects of occupational exposure and risk to SLE in Caucasians. Similarly, our findings suggest that the double deletion can be a risk factor for SLE; however, our results show risk among African Americans (OR 3.9) and not in Caucasians (OR 0.8) [57].
Sobkowiak et al. (2008) observed association between SOD2TT and Raynaud’s phenomenon (OR 12.0, CI 2.31–62.19, p=0.001) and immunologic manifestations (OR 2.9, CI 1.20–7.24, p=0.022) in patients with SLE, which suggests the participation of mutant homozygotes in clinical manifestations of the disease [58]. Despite the significant results found in our study, there were no correlations between the clinical manifestations and SOD2 C47T polymorphism (data not shown). SOD2 in C allele carriers are able to cross the two mitochondrial membranes, whereas T variant is incorporated only in the inner membrane of the mitochondrial matrix; therefore, carriers of TT genotype become 30 to 40% less effective in production of H2O2, favoring oxidative stress in these patients.
There was no positive correlation between SNPs of GSTP1 (A313G and IVS6 -C16T) and the risk to SLE (data not shown). Unlike our results, Glesse et al. (2014) demonstrated the protective role of heterozygous variant GSTP1 313 AG in European descent patients [58, 59]. We had the purpose of verifying the participation of haplotypes of GSTP1 A313G in patients with SLE. Unfortunately, few studies so far found in the literature describing possible correlations between GSTP1 IVS6 -C16T polymorphism and diseases (cognitive ability, asthma, allergies, hypertension, and acute myocardial infarction) [60,61,62,63]; however, there were no positive correlations, and our results are the first to cite these correlations.
Also, in present study, no positive correlation was observed between the CAT C262T polymorphism and SLE patients (data not shown). These observations are supported by the study which suggested that the polymorphism is associated more with lower enzyme activity rather than the risk of the disease, hence confirming our results [64]. The same was observed by Eny et al. (2005) and D’Souza et al. (2008), suggesting that this polymorphism does not have a central role in the pathogenesis of SLE; however, the authors urge the need for studies with combinations of genotypes to determine whether combined mutations can influence the development of disease [65, 66].
Our study suggests that polymorphisms concomitantly may influence susceptibility to SLE. Different mechanisms associated with each mutation may be able to promote an exacerbated immune response by increased oxidative stress or low antioxidant capacity in these patients. It was shown that SLE patients with of mutation have a higher concentration of these antioxidants, suggesting a protective effect mainly changing by GSTT1null/GSTM1null patients. Studies have shown that elevated levels of thiols may reduce cellular damage caused by oxidative stress [1, 2, 53].
We understand that these data are important in the follow-up and treatment of patients, since spinal cord hypoplasia is a frequent finding in these patients and that these polymorphisms may aggravate clinical and severity, especially in those who use corticosteroids. We believe that this finding is of fundamental importance, since they reiterate the relevance of the hematological alterations associated with systemic lupus erythematosus.
Conclusion
Our study showed that patients with SLE have increased oxidative stress evidenced by increased lipid peroxidation markers and decreased levels of thiols. The double null deletion GSTT1null/GSTM1null may contribute to the increased oxidative stress in SLE, but has no influence on disease activity or nephritis. The isolated polymorphisms of SOD2 C47T and CAT C262T do not seem to be important in the increased oxidative stress or clinical manifestations of the disease; however, further studies are needed to determine the actual participation of polymorphisms in these patients.
References
Zhang Q, Ye DQ, Chen GP, Zheng Y (2010 Apr) Oxidative protein damage and antioxidant status in systemic lupus erythematosus. Clin Exp Dermatol 35(3):287–294. https://doi.org/10.1111/j.1365-2230.2009.03437.x
Perl A (2013 Nov) Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat Rev Rheumatol 9(11):674–686. https://doi.org/10.1038/nrrheum.2013.147
Wang G, Pierangeli SS, Papalardo E, Ansari GA, Khan MF (2010 Jul) Markers of oxidative and nitrosative stress in systemic lupus erythematosus: correlation with disease activity. Arthritis Rheum 62(7):2064–2072. https://doi.org/10.1002/art.27442
Pérez YG, Pérez LCG, de Netto R CM, de Lima DSN, Lima ES (2012 Aug) Malondialdehyde and sulfhydryl groups as biomarkers of oxidative stress in patients with systemic lupus erythematosus. Rev Bras Reumatol 52(4):658–660
Sirivarasai J, Wananukul W, Kaojarern S, Chanprasertyothin S, Thongmung N, Ratanachaiwong W, Sura T, Sritara P (2013) Association between inflammatory marker, environmental lead exposure, and glutathione S-transferase gene. Biomed Res Int 2013:474963. https://doi.org/10.1155/2013/474963
Khodayari S, Salehi Z, Fakhrieh Asl S, et al (2013 May) Catalase gene C-262T polymorphism: importance in ulcerative colitis. J Gastroenterol Hepatol 28(5):819-822. https://doi.org/10.1111/jgh.12141
Bona N, Pezzarini E, Balbi B, Daniele SM, Rossi MF, Monje AL, Basiglio CL, Pelusa HF, Arriaga SMM (2020 Mar) Oxidative stress, inflammation and disease activity biomarkers in lupus nephropathy. Lupus 29(3):311–323. https://doi.org/10.1177/0961203320904784
Ye Z, Song H (2005 May) Glutathione s-transferase polymorphisms (GSTM1, GSTP1 and GSTT1) and the risk of acute leukaemia: a systematic review and metaanalysis. Eur J Cancer 41(7):980–989. https://doi.org/10.1016/j.ejca.2005.01.014
Thorn CF, Ji Y, Weinshilboum RM, Altman RB, Klein TE (2012 Aug) PharmGKB summary: very important pharmacogene information for GSTT1. Pharmacogenet Genomics 22(8):646–651. https://doi.org/10.1097/FPC.0b013e3283527c02
Mo Z, Gao Y, Cao Y, Gao F, Jian L (2009 May 1) An updating meta-analysis of the GSTM1, GSTT1, and GSTP1 polymorphisms and prostate cancer: a HuGE review. Prostate 69(6):662–688. https://doi.org/10.1002/pros.20907
Jia CY, Liu YJ, Cong XL, Ma YS, Sun R, Fu D, Lv ZW (2014 Apr) Association of glutathione S-transferase M1, T1, and P1 polymorphisms with renal cell carcinoma: evidence from 11 studies. Tumour Biol 35(4):3867–3873. https://doi.org/10.1007/s13277-013-1513-5
Pandey SN, Jain M, Nigam P, Choudhuri G, Mittal B (2006 May-Jun) Genetic polymorphisms in GSTM1, GSTT1, GSTP1, GSTM3 and the susceptibility to gallbladder cancer in North India. Biomarkers 11(3):250–261. https://doi.org/10.1080/13547500600648697
Cowell IG, Dixon KH, Pemble SE, Ketterer B, Taylor JB (1988) The structure of the human gltathione S-transferase pi gene. Biochem J 255(1):79–83. https://doi.org/10.1042/bj2550079
Mannervik B, Danielson UH (1988) Glutathione transferases--structure and catalytic activity. CRC Crit Rev Biochem 23(3):283–337. https://doi.org/10.3109/10409238809088226
Spurdle AB, Webb PM, Purdie DM, Chen X, Green A, Chenevix-Trench G (2001 Jan) Polymorphisms at the glutathione S-transferase GSTM1, GSTT1 and GSTP1 loci: risk of ovarian cancer by histological subtype. Carcinogenesis 22(1):67–72. https://doi.org/10.1093/carcin/22.1.67
Gulubova M, Vlaykova T (2010 Dec) Expression of the xenobiotic- and reactive oxygen species-detoxifying enzymes, GST-pi, Cu/Zn-SOD, and Mn-SOD in the endocrine cells of colorectal cancer. Int J Colorectal Dis 25(12):1397–1405. https://doi.org/10.1007/s00384-010-1041-3
Sánchez-Gómez FJ, Díez-Dacal B, García-Martín E, Agúndez JA, Pajares MA, Pérez-Sala D (2016) Detoxifying enzymes at the cross-roads of inflammation, oxidative stress, and drug hypersensitivity: role of glutathione transferase P1-1 and aldose reductase. Front Pharmacol 7:237. Published 2016 Aug 4 https://doi.org/10.3389/fphar.2016.00237
Moyer AM, Salavaggione OE, Wu TY, Moon I, Eckloff BW, Hildebrandt MA, Schaid DJ, Wieben ED, Weinshilboum RM (2008 Jun 15) Glutathione s-transferase p1: gene sequence variation and functional genomic studies. Cancer Res 68(12):4791–4801. https://doi.org/10.1158/0008-5472.CAN-07-6724
Crawford A, Fassett RG, Coombes JS, Kunde DA, Ahuja KD, Robertson IK, Ball MJ, Geraghty DP (2011 Sep) Glutathione peroxidase, superoxide dismutase and catalase genotypes and activities and the progression of chronic kidney disease. Nephrol Dial Transplant 26(9):2806–2813. https://doi.org/10.1093/ndt/gfq828
Xu Z, Zhu H, Luk JM, Wu D, Gu D, Gong W, Tan Y, Zhou J, Tang J, Zhang Z, Wang M, Chen J (2012 Nov 15) Clinical significance of SOD2 and GSTP1 gene polymorphisms in Chinese patients with gastric cancer. Cancer 118(22):5489–5496. https://doi.org/10.1002/cncr.27599
Crawford A, Fassett RG, Geraghty DP, Kunde DA, Ball MJ, Robertson IK, Coombes JS (2012 Jun 15) Relationships between single nucleotide polymorphisms of antioxidant enzymes and disease. Gene 501(2):89–103. https://doi.org/10.1016/j.gene.2012.04.011
dos Santos KG, Canani LH, Gross JL, Tschiedel B, Souto KE, Roisenberg I (2006) The catalase -262C/T promoter polymorphism and diabetic complications in Caucasians with type 2 diabetes. Dis Markers 22(5–6):355–359. https://doi.org/10.1155/2006/983408
Crispín JC, Hedrich CM, Tsokos GC (2013 Aug) Gene-function studies in systemic lupus erythematosus. Nat Rev Rheumatol 9(8):476–484. https://doi.org/10.1038/nrrheum.2013.78
Rhodes B, Vyse TJ (2008 Nov) The genetics of SLE: an update in the light of genome-wide association studies. Rheumatology (Oxford) 47(11):1603–1611. https://doi.org/10.1093/rheumatology/ken247
Lalwani P, de Souza GK, de Lima DS, Passos LF, Boechat AL, Lima ES. Serum thiols as a biomarker of disease activity in lupus nephritis. PLoS One. 2015 Mar 23;10(3):e0119947. https://doi.org/10.1371/journal.pone.0119947
Abd El Azeem RA, Zedan MM, Saad EA, Mutawi TM, Attia ZR (2021 Feb) Single-nucleotide polymorphisms (SNPs) of antioxidant enzymes SOD2 and GSTP1 genes and SLE risk and severity in an Egyptian pediatric population. Clin Biochem 88:37–42. https://doi.org/10.1016/j.clinbiochem.2020.11.010
Uribe AG, Vilá LM, McGwin G Jr, Sanchez ML, Reveille JD, Alarcón GS (2004 Oct) The systemic lupus activity measure-revised, the Mexican Systemic Lupus Erythematosus Disease Activity Index (SLEDAI), and a modified SLEDAI-2K are adequate instruments to measure disease activity in systemic lupus erythematosus. J Rheumatol 31(10):1934–1940
Weening JJ, D'Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, Balow JE, Bruijn JA, Cook T, Ferrario F, Fogo AB, Ginzler EM, Hebert L, Hill G, Hill P, Jennette JC, Kong NC, Lesavre P, Lockshin M, Looi LM, Makino H, Moura LA, Nagata M, International Society of Nephrology Working Group on the Classification of Lupus Nephritis, Renal Pathology Society Working Group on the Classification of Lupus Nephritis (2004 Feb) The classification of glomerulonephritis in systemic lupus erythematosus revisited. Kidney Int 65(2):521–530. https://doi.org/10.1111/j.1523-1755.2004.00443.x Erratum in: Kidney Int. 2004 Mar;65(3):1132
Saloum de Neves Manta F, Pereira R, Vianna R, Rodolfo Beuttenmüller de Araújo A, Leite Góes Gitaí D, Aparecida da Silva D, de Vargas Wolfgramm E, da Mota Pontes I, Ivan Aguiar J, Ozório Moraes M, Fagundes de Carvalho E, Gusmão L (2013 Sep 20) Revisiting the genetic ancestry of Brazilians using autosomal AIM-Indels. PLoS One 8(9):e75145. https://doi.org/10.1371/journal.pone.0075145
Aycicek A, Erel O, Kocyigit A (2005 Dec) Decreased total antioxidant capacity and increased oxidative stress in passive smoker infants and their mothers. Pediatr Int 47(6):635–639. https://doi.org/10.1111/j.1442-200x.2005.02137.x
Harma M, Harma M, Erel O (2005 Feb) Oxidative stress in women with preeclampsia. Am J Obstet Gynecol 192(2):656–657. https://doi.org/10.1016/j.ajog.2004.07.094
Faul F, Erdfelder E, Buchner A, Lang AG (2009 Nov) Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods 41(4):1149–1160. https://doi.org/10.3758/BRM.41.4.1149
Goncalves M, Neto J, Souza C, Melo P, Reis M (2009) Evaluating glutathione S-Transferase (GST) null genotypes (GSTT1 and GSTM1) as a potential biomarker of predisposition for developing leukopenia. Int J Lab Hematol 32:e49–e56. https://doi.org/10.1111/j.1751-553X.2009.01169.x
Buttgereit F, da Silva JA, Boers M, Burmester GR, Cutolo M, Jacobs J et al (2002) Standardised nomenclature for glucocorticoid dosages and glucocorticoid treatment regimens: current questions and tentative answers in rheumatology. Ann Rheum Dis 61(8):718–722
dos Santos KSC, de Almeida MF, Maciel TKP, de Lima DSN (jan./dez. - 2010) Perfil Clínico e Laboratorial de Pacientes com Lúpus Eritematoso Sistêmico (LES). Revista HUGV - Revista do Hospital Universitário Getúlio Vargas 9:1–2
Systemic Lupus Erythematosus (SLE) (2005) Clinical and laboratory profile of patients followed at the Onofre Lopes University Hospital (UFRN - Natal/Brazil) and early organ damage in patients with recently diagnosed disease. Rev Bras Reumatol [online] 45(6):339–342. ISSN 1809-4570. https://doi.org/10.1590/S0482-50042005000600002
Liu K, Kurien BT, Zimmerman SL et al (2016) X chromosome dose and sex bias in autoimmune diseases: increased prevalence of 47, XXX in systemic lupus erythematosus and sjögren's syndrome. Arthritis Rheumatol 68(5):1290–1300. https://doi.org/10.1002/art.39560
Altindag O, Karakoc M, Kocyigit A, Celik H, Soran N (2007 Feb) Increased DNA damage and oxidative stress in patients with rheumatoid arthritis. Clin Biochem 40(3-4):167–171. https://doi.org/10.1016/j.clinbiochem.2006.10.006
Zhong S, Huang M, Yang X, Liang L, Wang Y, Romkes M, Duan W, Chan E, Zhou SF (2006 Oct) Relationship of glutathione S-transferase genotypes with side effects of pulsed cyclophosphamide therapy in patients with systemic lupus erythematosus. Br J Clin Pharmacol 62(4):457–472. https://doi.org/10.1111/j.1365-2125.2006.02690.x
Karaca S, Karaca M, Cesuroglu T, Erge S, Polimanti R (2015 May-Jun) GSTM1, GSTP1, and GSTT1 genetic variability in Turkish and worldwide populations. Am J Hum Biol 27(3):310–316. https://doi.org/10.1002/ajhb.22671
Gattás GJF, Kato M, Soares-Vieira JA, Siraque MS, Kohler P, Gomes L et al (2004) Ethnicity and glutathione S-transferase (GSTM1/GSTT1) polymorphisms in a Brazilian population. Braz J Med Biol Res [online] 37(4):451–458. ISSN 1414-431X. https://doi.org/10.1590/S0100-879X2004000400002
Hatagima A, Klautau-Guimarães MN, Da Silva FP, Cabello PH (Dec. 2000) Glutathione S-transferase M1 (GSTM1) polymorphism in two Brazilian populations. Genet. Mol. Biol (São Paulo) 23(4). https://doi.org/10.1590/S1415-47572000000400003
De Oliveira C, De Oliveira SF, Hatagima A, Ferreira LB, Grisolia CK, Klautau- MDN (2007) Glutathione S-Transferase M1 and T1 polymorphisms in Brazilian African descendants. Hum Biol 79(1):131–140. Project MUSE, https://doi.org/10.1353/hub.2007.0025
Klautau-guimarães MDN, Hiragi CDO, Ascenção RFD, Oliveira SF, Grisolia CK, Hatagima A, et al (Jan./Mar. 2005) Distribution of glutathione S-transferase GSTM1 and GSTT1 null phenotypes in Brazilian Amerindians. Genet Mol Biol (São Paulo) 28(1). https://doi.org/10.1590/S1415-47572005000100005
de Hiragi C O, Miranda-Vilela AL, Rocha DMS, de Oliveira SF, Hatagima A, de Klautau-Guimarães MN (2011) Superoxide dismutase, catalase, glutathione peroxidase and gluthatione s-transferases M1 and T1 gene polymorphisms in three brazilian population groups. Genet Mol Biol [online]. 34(1):11-18. ISSN 1415-4757. https://doi.org/10.1590/S1415-47572010005000102
Taufer M, Peres A, de Andrade VM, de Oliveira G, Sá G, do Canto ME, dos Santos AR, Bauer ME, da Cruz IB (2005 Apr) Is the Val16Ala manganese superoxide dismutase polymorphism associated with the aging process? J Gerontol A Biol Sci Med Sci 60(4):432–438. https://doi.org/10.1093/gerona/60.4.432.
Freedman JE (2008) Oxidative stress and platelets. Arterioscler Thromb Vasc Biol 28:s11–s16. https://doi.org/10.1161/ATVBAHA.107.159178
Yang M et al (2015) Hematocrit level could reflect inflammatory response and disease activity in patients with systemic lupus erythematosus. Clin Lab 61(7):801–807
Morgan PE, Sturgess AD, Davies MJ (2005 Jul) Increased levels of serum protein oxidation and correlation with disease activity in systemic lupus erythematosus. Arthritis Rheum 52(7):2069–2079. https://doi.org/10.1002/art.21130
Anderer G, Schrappe M, Brechlin AM, Lauten M, Muti P, Welte K, Stanulla M (2000 Nov) Polymorphisms within glutathione S-transferase genes and initial response to glucocorticoids in childhood acute lymphoblastic leukaemia. Pharmacogenetics 10(8):715–726. https://doi.org/10.1097/00008571-200011000-00006
Salimi S, Nakhaee A, Jafari M, Jahantigh D, Sandooghi M, Zakeri Z, Shahrakipour M, Naghavi A, Farajian-Mashhadi F (2015 Jun) Combination effect of GSTM1, GSTT1 and GSTP1 polymorphisms and risk of systemic lupus erythematosus. Iran J Public Health 44(6):814–821
Economopoulos KP, Sergentanis TN (2010 Jun) GSTM1, GSTT1, GSTP1, GSTA1 and colorectal cancer risk: a comprehensive meta-analysis. Eur J Cancer 46(9):1617–1631. https://doi.org/10.1016/j.ejca.2010.02.009
Tew MB, Reveille JD, Arnett FC, Friedman AW, McNearney T, Fischbach M, Ahn C, Tan FK (2001 Jun) Glutathione S-transferase genotypes in systemic sclerosis and their association with clinical manifestations in early disease. Genes Immun 2(4):236–238. https://doi.org/10.1038/sj.gene.6363756
Rupasree Y, Naushad SM, Rajasekhar L, Kutala VK (2013) Association of genetic variants of xenobiotic metabolic pathway with systemic lupus erythematosus. Indian J Biochem Biophys 50:447–452
Jufeng Z, Jingui D, David Z, Yanxin L, Li L, Qi W, Yong S, Jie Z, Hong Y, Bo Y, Jun W (2010) Association of GSTT1, GSTM1 and CYP1A1 polymorphisms with susceptibility to systemic lupus erythematosus in the Chinese population. Clin Chim Acta; Int J Clin Chem 411:878–881. https://doi.org/10.1016/j.cca.2010.03.007
Kang TY, El-Sohemy A, Comelis MC, Eny KM, Bae SC (2005) Glutathione S-transferase genotype and risk of systemic lupus erythematosus in Koreans. Lupus 14(5):381–384. https://doi.org/10.1191/0961203305lu2100oa
Fraser PA, Ding WZ, Mohseni M, Treadwell EL, Dooley MA, St Clair EW, Gilkeson GS, Cooper GS (2003) Glutathione S-transferase M null homozygosity and risk of systemic lupus erythematosus associated with sun exposure: a possible gene-environment interaction for autoimmunity. J Rheumatol
Sobkowiak A, Lianeri M, Wudarski M, Lacki JK, Jagodziński PP (2008 Jul) Manganese superoxide dismutase Ala-9Val mitochondrial targeting sequence polymorphism in systemic lupus erythematosus in Poland. Clin Rheumatol 27(7):827–831. https://doi.org/10.1007/s10067-007-0796-6
Glesse N, Rohr P, Monticielo OA, Rech TF, Brenol JC, Xavier RM, Kvitko K, Chies JA (2014 Sep) Genetic polymorphisms of glutathione S-transferases and cytochrome P450 enzymes as susceptibility factors to systemic lupus erythematosus in southern Brazilian patients. Mol Biol Rep 41(9):6167–6179. https://doi.org/10.1007/s11033-014-3496-8
Harris SE, Fox H, Wright AF, Hayward C, Starr JM, Whalley LJ, Deary IJ (2007 Jul 2) A genetic association analysis of cognitive ability and cognitive ageing using 325 markers for 109 genes associated with oxidative stress or cognition. BMC Genet 8:43. https://doi.org/10.1186/1471-2156-8-43
Melén E, Nyberg F, Lindgren CM, Berglind N, Zucchelli M, Nordling E, Hallberg J, Svartengren M, Morgenstern R, Kere J, Bellander T, Wickman M, Pershagen G (2008 Aug) Interactions between glutathione S-transferase P1, tumor necrosis factor, GSTP1and traffic-related air pollution for development of childhood allergic disease. Environ Health Perspect 116(8):1077–1084. https://doi.org/10.1289/ehp.11117
Joubert BR, Reif DM, Edwards SW, Leiner KA, Hudgens EE, Egeghy P, Gallagher JE, Hubal EC (2011 Feb 14) Evaluation of genetic susceptibility to childhood allergy and asthma in an African American urban population. BMC Med Genet 12:25. https://doi.org/10.1186/1471-2350-12-25
Levinsson A, Olin AC, Modig L, Dahgam S, Björck L, Rosengren A, Nyberg F (2014 Jun 10) Interaction effects of long-term air pollution exposure and variants in the GSTP1, GSTT1 and GSTCD genes on risk of acute myocardial infarction and hypertension: a case-control study. PLoS One 9(6):e99043. https://doi.org/10.1371/journal.pone.0099043
Ghaly MS, Ghattas MH, Labib SM (2012 Oct) Association of catalase gene polymorphisms with catalase activity and susceptibility to systemic lupus erythematosus in the Suez Canal area. Egypt Lupus 21(11):1244–1249. https://doi.org/10.1177/0961203312451505
Eny KM, El-Sohemy A, Cornelis MC, Sung YK, Bae SC (2005) Catalase and PPARgamma2 genotype and risk of systemic lupus erythematosus in Koreans. Lupus 14(5):351–355. https://doi.org/10.1191/0961203305lu2091oa
D'souza A, Kurien BT, Rodgers R, Shenoi J, Kurono S, Matsumoto H, Hensley K, Nath SK, Scofield RH (2008 Jul 7) Detection of catalase as a major protein target of the lipid peroxidation product 4-HNE and the lack of its genetic association as a risk factor in SLE. BMC Med Genet 9:62. https://doi.org/10.1186/1471-2350-9-62
Acknowledgments
The authors wish to thank all blood donors attending the FHEMOAM for contributing and participating in this study. Also, the authors thank the staff of the Molecular Biology Laboratory of the Federal University of Amazonas (UFAM) and Gonçalo Moniz Institute (IGM) (FIOCRUZ/BAHIA) for the technical support. Finally, the authors thank the Foundation for Research Support of the State of Amazonas (FAPEAM) for financial support. The sponsors of this study are public or nonprofit organizations that support science in general. They had no role in gathering, analyzing, or interpreting the data.
Funding
Financial support was provided by grants from the Fundação de Amparo à Pesquisa do Estado do Amazonas (FAPEAM)–Processo: 1094/2013-FAPEAM and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors alone are responsible for the content and writing of this article and report no conflicts of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
de Oliveira, M.A.A., Mallmann, N.H., de Souza, G.K.B.B. et al. Glutathione S-transferase, catalase, and mitochondrial superoxide dismutase gene polymorphisms modulate redox potential in systemic lupus erythematosus patients from Manaus, Amazonas, Brazil. Clin Rheumatol 40, 3639–3649 (2021). https://doi.org/10.1007/s10067-021-05680-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-021-05680-0