Abstract
Rheumatoid arthritis (RA) is the most common autoimmune rheumatic disease, in which an epigenetic implication in the disease etiopathogenesis has been noted. Here in this meta-analysis, we attempted to investigate the pooled association of methylenetetrahydrofolate reductase (MTHFR) gene C677T and A1298C polymorphisms and susceptibility to RA risk. A systematic search was performed in the main databases, including MEDLINE and Scopus to search for studies assessing the association between MTHFR gene C677T and A1298C polymorphisms and the risk of RA prior to December 2019. In this meta-analysis, 15 studies with 2165 patients and 1751 healthy controls for C677T SNP and 14 studies containing 2021 patients and 1760 healthy controls for A1298C SNP were included. A significant positive association between C677T SNP and RA risk was recognized in the dominant, recessive, and allelic model, but not TT and CT genotypes. The results indicated that the risk of RA in African population was increased under all genotype models while these results were repeated in Asian population just for recessive model, allelic model, and TT genotype. Moreover, the analysis of A1298C SNP demonstrated a significant association in overall population according to only the recessive model and CC genotype. Subgroup analysis according to the genotyping method indicated that RFLP-PCR method could impress the results of association between MTHFR gene A1298C and C677T SNPs and RA risk. The outcome of this meta-analysis indicated that MTHFR gene C677T SNP was much possibly be associated with RA risk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is a most common systemic and autoimmune disorder characterized by synovial inflammation, erosive articular degradation, and joint destruction, leading joint deformity and movement limitation [1]. Although the main etiology of RA is yet to be elucidated, studies demonstrated that both environmental and genetic risk factors may play a major role in the onset and progression of the disease [2]. The prevalence of the disease is almost 0.5–1% among the world population and is more frequently occurred in women than in men (sex ratio 3:1) [3, 4]. According to the most recent studies, genetic factors may contribute to approximately 60% of the total risk in susceptibility to RA [5]. The class II Human leukocyte antigen (HLA) has been identified as the most powerful genetic factors in susceptibility to RA [6, 7]. On the other hand, a number of non-HLA genes, such as Cytotoxic T lymphocyte-associated protein 4 (CTLA4), TNF receptor associated factor 1 (TRAF1), Protein tyrosine phosphatase non-receptor type 22 (PTPN22), Peptidyl arginine deiminase 4 (PADI4), TNF alpha induced protein 3 (TNFAIP3), and Methylenetetrahydrofolate reductase (MTHFR) have been consistently associated with RA predisposition [8,9,10,11,12,13].
The folate biological function is to provide methyl groups required for metabolic processes, such as DNA methylation, synthesis, and repair. Therefore, folate deficiency can disrupt these processes [14]. MTHFR, an essential enzyme in folate homeostasis and metabolic pathway, catalyzes the irreversible reduction of 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate, which serves a methylation group toward the conversion of homocysteine to methionine with the precursor of S-adenosylmethionine (SAM) [15]. Reduced function of MTHFR results in hypomethylation of DNA and enhanced levels of homocysteine, which leads to increased secretion of proinflammatory cytokines that are implicated in the pathogenesis of RA [16]. Therefore, genetic variations of MTHFR could impress the susceptibility to RA. On the other hand, genetic variants of the MTHFR gene have been associated with hyperhomocysteinemia, which has been considered as risk factor for cardiovascular diseases in RA patients [17].
MTHFR gene is located on the chromosomal region 1p36.3, and several single nucleotide polymorphisms (SNPs) within this gene have been recognized in susceptibility to diverse autoimmune disorders [18]. Two common polymorphisms C677T (rs1801133) and A1298C (rs1801131) have been recognized. These polymorphisms are correlated with a reduction in MTHFR enzyme activity. The C677T mutation in the MTHFR gene causes alanine (A) to valine (V) amino acid substitution. This situation is an important cause of reduced enzyme activity and leads to a higher level of homocysteine levels [19]. Besides, the A to C change at position 1298 leads to glutamine to alanine substitution, resulting in decreased enzyme activity [20].
Several studies evaluated the associations between MTHFR polymorphisms (C677T and A1298C) and susceptibility to RA in different populations with variable frequency and conflicting results. This discrepancy might be owing to different sample sizes, ethnicity, clinical heterogeneity, and publication bias. To compensate these limitations, we conducted this most updated meta-analysis to investigate whether MTHFR gene polymorphisms play a role in RA proneness.
Methods
This meta-analysis follows the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement [21]. The current study is registered in the International Prospective Register of Systematic Reviews (PROSPERO).
Literature search
A comprehensive search was conducted through MEDLINE and Scopus databases to retrieve all potential publications considered the association between MTHFR (C677T and A1298C) gene polymorphism and susceptibility to RA. The following combinations of key words were searched: (“MTHFR” OR “methylenetetrahydrofolate reductase”) AND (“Rheumatoid Arthritis” OR “Arthritis” OR “RA”) AND (“single nucleotide polymorphism” OR “SNP” OR “polymorphisms” OR “mutation” OR “variation”). The reference list of all studies was cross-checked to find other potential studies which might miss during initial search.
Inclusion and exclusion criteria
We included studies in quantitative analysis if they met the following criteria: (i) all observational studies (cohort or case-control design) considered the association between MTHFR (C677T and A1298C) gene polymorphism and susceptibility to RA; (ii) studies reporting sufficient data to extract or calculate risk estimates with 95% CI; (iii) studies with sufficient information regarding numbers or genotype frequencies in cases and healthy controls. Duplicates, book chapters, letters to editor, animal study, case reports, review articles, and studies with repetitive subjects all were excluded. The application of these criteria recognized 16 and 14 eligible studies for C677T and A1298C SNPs, respectively.
Study selection criteria
The results of initial search were exported to Endnote software. Then two reviewers independently assessed titles and abstracts of all studies. Articles which not follow the eligibility criteria were excluded according to a hierarchical approach. The full-text examination was examined if we could not decide to include or exclude studies based on title and abstract. In particular conditions, if an author has published more than one study by the same case series, the most recently published study was included. Any disagreements were discussed and resolved by consensus.
Data extraction
The detailed data of all eligible studies were extracted according to a standardized extraction form including: the author’s name, journal and year of publication, country of origin, ethnicity, mean or range of age, genotyping method, allele frequency of cases and controls, total sample size of cases and controls, and the number of cases and controls for each genotype. Any discrepancy between two reviewers was solved by mutual discussion.
Quality assessment
Methodological quality of eligible studies was evaluated by Newcastle–Ottawa Scale (NOS), a validated scale for non-randomized studies in meta-analysis [22]. Accordingly, studies were categorized to high quality (7–9), intermediate quality (4–6), and low quality (1–3). Furthermore, chi-square tests were calculated to disclose potential deviation from Hardy–Weinberg equilibrium (HWE) for distribution of the allele frequencies in the control group.
Statistical analysis
The data analyses were carried out using STATA (version 14.0; Stata Corporation, College Station, TX) and SPSS (version 23.0; SPSS, Inc. Chicago, IL). In this study, crud OR and its 95% CI was calculated to estimate the association between MTHFR gene polymorphism and the risk of RA. Genotype models were defined as follows: MTHFR C677T, [dominant model (TT+CT vs. CC), recessive model (TT vs. CT+CC), allelic model (T vs. C), homozygote (TT vs. CC), heterozygote (CT vs. CC)] and MTHFR A1298C, [dominant model (CC+AC vs. AA), recessive model (CC vs. AC+AA), allelic model (C vs. A), homozygote (CC vs. AA), heterozygote (AC vs. AA)]. In consideration of the possible heterogeneity (between-study variability) across included studies, chi-square Q-test was used [23]. Additionally, to show heterogeneity quantitatively, the other index (I2) was calculated. There was significant heterogeneity if I2 values exceeded 50% or the Q statistic had a P value less than 0.1. In the presence of significant heterogeneity, the random-effects model (REM) (DerSimonian–Laird approach) was performed. Otherwise, the fixed-effects model (FEM) (Mantel–Haenszel approach) was performed for combination of data [24, 25]. We used sensitivity analysis to assess the stability of our results. Additionally, the Egger’s test and Begg’s test were applied for publication bias [26, 27].
Results
Study characteristics
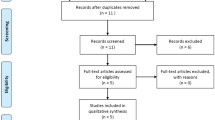
The four-phase search and screening process based on the PRISMA statement is presented in Fig. 1. After the removal of duplicates (47 studies), 290 studies were remained. Of these, 247 studies were excluded based on title and abstract and 43 studies excluded by full-text evaluation. Ultimately, 16 studies were qualified and included in quantitative analysis (Fig. 1) [28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43]. All included studies were conducted between 2004 and 2019 and had good methodological score. RFLP-PCR and Taq-man genotyping methods were used by most of included studies. Tables 1 and 2 summarized the characteristics and genotype frequency of the included studies.
Quantitative analysis
Meta-analysis of C677T and RA risk
Overall, 15 studies with 2165 patients and 1751 healthy controls were included in quantitative analysis of MTHFR gene C677T polymorphism and RA risk. The analysis of overall population revealed a significant positive association between C677T SNP and RA risk across dominant model (OR = 1.28, 95% CI, 1.01–1.65, P = 0.04), recessive model (OR = 1.47, 95% CI, 1.15–1.89, P < 0.001), allelic model (OR = 1.28, 95% CI, 1.06–1.56, P = 0.01), but not TT vs. CC model (OR = 1.31, 95% CI, 0.98–1.75, P = 0.07), and CT vs. CC model (OR = 1.12, 95% CI, 0.96–1.32, P = 0.15) (Fig. 2). Additionally, subgroup analysis was performed to evaluate ethnicity-specific effect on the association of C677T SNP and RA risk. Since there was only one study for Jewish, African-American and Mexican ethnicity these studies were excluded from subgroup analysis. The results indicated that the risk of RA in African population increases under all genotype models while these results were repeated in Asian population just for recessive model (OR = 2.18, 95% CI, 1.40–3.41, P < 0.001), allelic model (OR = 1.32, 95% CI, 1.06–1.64, P = 0.01), and TT vs. CC model (OR = 2.07, 95% CI, 1.16–3.69, P = 0.01). No significant association was detected in causations (Fig. 3). Furthermore, we stratified studies according to genotyping method and found that utilization of RFLP-PCR method can affect the results of association between C677T SNP and RA risk (Fig. 4).
Meta-analysis of A1298C and RA risk
In the quantitative analysis of MTHFR A1298C polymorphism and RA risk, 14 studies containing 2021 patients and 1760 healthy controls were included. The pooled results revealed a significant association in overall population across recessive model (OR = 1.58, 95% CI, 1.21–2.06, P < 0.001) and CC vs. AA model (OR = 1.34, 95% CI, 1–1.78, P = 0.04) (Fig. 2). The results of subgroup analysis by ethnicity rejected potential association between A1298C SNP and RA risk in all three populations (Africans, Asians, Causations) (Fig. 3). Moreover, subgroup analysis based on genotyping method highlighted the influence of RFLP-PCR on the association between A1298C SNP and RA risk (Fig. 4).
Evaluation of heterogeneity and publication bias
During the meta-analysis of MTHFR gene polymorphism, evidence of moderate heterogeneity was detected in some models. However, partial heterogeneity was resolved while the data were stratified by genotyping method and ethnicity. Publication bias was evaluated by funnel plot, Begg’s test, and Egger’s test. Subsequently, there was no obvious evidence of asymmetry according to the funnel plots (Fig. 5), and all P values of Begg’s test and Egger’s test were > 0.05, indicating no evidences of publication biases.
Sensitivity analysis
The leave-one-out method was used in the sensitivity analysis to explore the effect of individual data on the pooled ORs (Table 3). The significance of ORs was not altered through omitting any single study in the dominant model for C677T and A1298C SNPs, indicating that our results were statistically robust (Fig. 6).
Discussion
Dysregulated function of MTHFR may lead to hypomethylation of DNA and hyperhomocysteinemia, both have been implicated in RA etiopathogenesis [44], conferring MTHFR as a candidate RA predisposing gene. Several MTHFR gene SNPs have been identified [18], and genome-wide association studies (GWASs) have identified this gene to be associated with genetic susceptibility to RA [45]. In addition, numerous studies have reported that MTHFR gene C677T and A1298C polymorphisms might be contributing genetic factor to RA risk. Although several studies in different ethnic groups have tried to divulge the plausible association between MTHFR gene C677T and A1298C SNPs and RA risk, the observations are still controversial and an apprehensive meta-analysis seems to be indispensable to disclose the bona fide association of these variations with RA disease. Hence, this meta-analysis was performed to reveal the approximation of MTHFR gene C677T and A1298C SNPs and RA risk. According to the pooled analysis, both polymorphisms increased the risk of RA.
MTHFR plays a vital role in the folate metabolism and is involved in the conversion of 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate [46]. As a structural analog of folic acid, methotrexate (MTX) and sulphasalazine (SSZ) has been prescribed for the treatment of RA according to its well-known efficacy and less toxicity [47]. These drugs competitively inhibit dihydrofolate reductase (DHFR), an enzyme that participates in the tetrahydrofolate synthesis and leads to increased plasma homocysteine level. However, polymorphisms in the genes of the folate pathway are potentially responsible for interpatient variation in MTX and sulphasalazine efficacy. Studies demonstrated patients homozygous for the mutation in the MTHFR gene had significantly higher baseline homocysteine and heterozygous MTHFR genotype induced significantly higher plasma homocysteine compared with no mutation. Taken together, these findings illustrate the critical role of MTHFR polymorphisms in increasing homocysteine levels [48, 49].
There are two meta-analyses about these hypothesis; Fisher et al. [50] showed that the C677T polymorphism (not A1298C polymorphism) is associated with increased toxicity (OR 1.71, 95% CI 1.32–2.21, P < 0.001). In consistent with them, Song et al. revealed a significant association between the MTHFR C677T and A1298C polymorphisms and MTX toxicity. They suggested that modulation in the activity of MTHFR enzyme as the result of SNPs in the MTHFR gene might be expected reason [51,52,53].
Studies have disclosed the role of MTHFR gene C677T and A1298C polymorphisms in reduced activity of the enzyme [54], which may impress the RA pathogenesis through three major pathways. First, dysfunction of MTHFR enzyme may cause DNA hypomethylation, which has been implicated to be involved in the pathogenesis of RA [55]. Second, MTHFR plays a role in donation of methyl group for methylation of homocysteine to generate methionine; hence, dysregulation of MTHFR might lead to high homocysteine level [16], which has been observed in RA patients compared with healthy subjects [56]. In addition, homocysteine has been shown to trigger the proinflammatory transcription factor of nuclear factor (NF)-κB, which has been shown to be involved in inflammatory settings in RA [44]. Third, GWASs has reported that the chromosomal region harboring the MTHFR gene is associated with genetic susceptibility to RA risk [57, 58]. Hence, it is rational to study the RA pathogenesis with respect to genetic diversity and impaired function of MTHFR.
In this most recent systematic review and meta-analysis, 15 studies with 2165 patients and 1751 healthy controls were included in quantitative analysis of MTHFR gene C677T polymorphism and RA risk as well as 14 studies containing 2021 patients and 1760 healthy controls were included for evaluation of MTHFR gene A1298C polymorphism and RA risk. In addition, we performed a subgroup analysis with respect to the technique applied for genotyping (RFLP-PCR), which had not been performed in the previous meta-analyses. Our analysis indicated a significant positive association between C677T SNP and RA risk in the dominant model (OR = 1.28), recessive model (OR = 1.47), allelic model (OR = 1.28), but not TT and CT genotypes. The results indicated that the risk of RA in African population was increased under all genotype models while these results were repeated in Asian population just for recessive model (OR = 2.18), allelic model (OR = 1.32), and TT genotype (OR = 2.07). However, no significant association was observed in the causations. The analysis of another SNP, namely A1298C, revealed a significant association in overall population according to the recessive model (OR = 1.58) and CC genotype (OR = 1.34). However, the results of subgroup analysis by ethnicity did not show association between A1298C SNP and RA risk in all three populations, including Africans, Asians, and Causations. That notwithstanding, the previous meta-analysis, applying less studies and including less patients and controls, indicated that the recessive model of MTHFR gene C677T polymorphism was not associated with RA risk [59], unlike our meta-analysis that indicated a statistically significant association of this polymorphism with RA risk. Moreover, only the recessive model of MTHFR gene A1298C polymorphism was associated with RA risk in the late meta-analysis that was associated with RA risk in recessive model and CC genotype in the current meta-analysis. This difference originates from the number of studies and subjects included in each meta-analysis.
In this meta-analysis, the subgroup analysis was also conducted based on genotyping technique to further explore the influence of genotyping method on the associations. Subgroup analysis according to the genotyping method indicated that RFLP-PCR method could impress the results of association between MTHFR gene A1298C and C677T SNPs and RA risk. Therefore, the differences in the genotyping method may have significant impressions on the overall observations of the association between MTHFR gene A1298C and C677T SNPs and RA susceptibility.
Despite our attempt to carry out the best possible meta-analysis and conclusion of the available information, a number of limitations can be raised toward this meta-analysis. First, it was not possible to analyze the role of gender, age, lifestyle, drugs, and other genetic variations on the adjusted association of MTHFR gene polymorphisms and RA risk. Therefore, further research in respective of the gene–gene and gene–environment interactions is still required to come up with a more apprehensive estimation of MTHFR gene polymorphisms with RA risk. Second, we only searched the publications with English language. Third, the number of studies available for the subgroup analysis based on the ethnicity was fairly small. This issue did not allow to perform comprehensive subgroup analysis across all populations.
In conclusion, the results of pooled analysis supported significant association between MTHFR gene C677T SNP and RA risk in the dominant, recessive, and allelic model, but not TT and CT genotypes. Nonetheless, the analysis of A1298C SNP revealed a significant association in overall population according to only the recessive model and CC genotype. However, the results of subgroup analysis by ethnicity did not show association between A1298C SNP and RA risk in all three populations, including Africans, Asians, and Causations. The major drawback of the valid estimation of the association of MTHFR gene SNPs with RA risk in this meta-analysis stems from the insufficient amount of original data, which requires further investigations in the future. Furthermore, the role of life style, age, and gender should be taken into consideration in the stratification analyses.
References
Chua JR, Jamal S, Riad M, Castrejon I, Malfait AM, Block JA, Pincus T (2019) Disease burden in osteoarthritis (OA) is similar to rheumatoid arthritis (RA) at initial rheumatology visit and significantly greater 6-months later. Arthritis Rheumatol 71(8):1276–1284
Conigliaro P, Triggianese P, De Martino E, Chimenti MS, Sunzini F, Viola A, Canofari C, Perricone R (2019) Challenges in the treatment of rheumatoid arthritis. Autoimmun Rev 18(7):706–713
Tobón GJ, Youinou P, Saraux A (2010) The environment, geo-epidemiology, and autoimmune disease: rheumatoid arthritis. Autoimmun Rev 9(5):A288–A292
Oliver JE, Silman AJ (2009) Why are women predisposed to autoimmune rheumatic diseases? Arthritis Res Ther 11(5):252
Orozco G, Barton A (2010) Update on the genetic risk factors for rheumatoid arthritis. Expert Rev Clin Immunol 6(1):61–75
Holoshitz J (2010) The rheumatoid arthritis HLA-DRB1 shared epitope. Curr Opin Rheumatol 22(3):293–298
Razi B, Reykandeh SE, Alizadeh S, Amirzargar A, Saghazadeh A, Rezaei N (2019) TIM family gene polymorphism and susceptibility to rheumatoid arthritis: systematic review and meta-analysis. PLoS One 14(2):e0211146
Flores-Borja F, Jury EC, Mauri C, Ehrenstein MR (2008) Defects in CTLA-4 are associated with abnormal regulatory T cell function in rheumatoid arthritis. Proc Natl Acad Sci 105(49):19396–19401
Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR (2007) TRAF1–C5 as a risk locus for rheumatoid arthritis—a genomewide study. N Engl J Med 357(12):1199–1209
Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM (2004) A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 75(2):330–337
Takizawa Y, Sawada T, Suzuki A, Yamada R, Inoue T, Yamamoto K (2005) Peptidylarginine deiminase 4 (PADI4) identified as a conformation-dependent autoantigen in rheumatoid arthritis. Scand J Rheumatol 34(3):212–215
Matmati M, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, Geboes L, Louagie E, Mc Guire C, Vereecke L (2011) A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat Genet 43(9):908–912
Owen S, Lunt M, Bowes J, Hider S, Bruce I, Thomson W, Barton A (2013) MTHFR gene polymorphisms and outcome of methotrexate treatment in patients with rheumatoid arthritis: analysis of key polymorphisms and meta-analysis of C677T and A1298C polymorphisms. Pharmacogenomics J 13(2):137–147
Moores CJ, Fenech M, O’Callaghan NJ (2011) Telomere dynamics: the influence of folate and DNA methylation. Ann N Y Acad Sci 1229(1):76–88
Pietrzik K, Bailey L, Shane B (2010) Folic acid and L-5-methyltetrahydrofolate. Clin Pharmacokinet 49(8):535–548
Fujimaki C, Hayashi H, Tsuboi S, Matsuyama T, Kosuge K, Yamada H, Inoue K, Itoh K (2009) Plasma total homocysteine level and methylenetetrahydrofolate reductase 677C> T genetic polymorphism in Japanese patients with rheumatoid arthritis. Biomarkers 14(1):49–54
Palomino-Morales R, Gonzalez-Juanatey C, Vazquez-Rodriguez TR, Rodriguez L, Miranda-Filloy JA, Fernandez-Gutierrez B, Llorca J, Martin J, Gonzalez-Gay MA (2010) A1298C polymorphism in the MTHFR gene predisposes to cardiovascular risk in rheumatoid arthritis. Arthritis Res Ther 12(2):R71
Goyette P, Pai A, Milos R, Frosst P, Tran P, Chen Z, Chan M, Rozen R (1998) Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR). Mamm Genome 9(8):652–656
Kalemi TG, Lambropoulos AF, Gueorguiev M, Chrisafi S, Papazisis KT, Kotsis A (2005) The association of p53 mutations and p53 codon 72, Her 2 codon 655 and MTHFR C677T polymorphisms with breast cancer in Northern Greece. Cancer Lett 222(1):57–65
van der Put NM, Gabreëls F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel LP, Blom HJ (1998) A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet 62(5):1044–1051
Moher D, Liberati A, Tetzlaff J, Altman DG (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151(4):264–269
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25(9):603–605
Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, Botella J (2006) Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods 11(2):193–206
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22(4):719–748
Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics:1088–1101
Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. Bmj 315(7109):629–634
Berkun Y, Levartovsky D, Rubinow A, Orbach H, Aamar S, Grenader T, Atta IA, Mevorach D, Friedman G, Ben-Yehuda A (2004) Methotrexate related adverse effects in patients with rheumatoid arthritis are associated with the A1298C polymorphism of the MTHFR gene. Ann Rheum Dis 63(10):1227–1231
Hughes LB, Beasley TM, Patel H, Tiwari HK, Morgan SL, Baggott JE, Saag KG, McNicholl J, Moreland LW, Alarcón GS (2006) Racial or ethnic differences in allele frequencies of single-nucleotide polymorphisms in the methylenetetrahydrofolate reductase gene and their influence on response to methotrexate in rheumatoid arthritis. Ann Rheum Dis 65(9):1213–1218
Rubini M, Padovan M, Baricordi O, Fotinidi M, Govoni M, Trotta F (2008) The c. 1298A> C polymorphism in the methylenetetrahydrofolate reductase gene is associated with rheumatoid arthritis susceptibility in Italian patients. Clin Exp Rheumatol 26(1):163
Cai YMG (2009) Linkage study on methylenetetrahydrofolate reductase single nucleotide polymorphisms and methotrexate-related adverse effects in patients with rheumatoid arthritis. Chin J Prim Med Pharm 1155-1157:16
Taşbaş Ö, Borman P, Karabulut HG, Tükün A, Yorgancıoğlu R (2011) The frequency of A1298C and C677T polymorphisms of the methylentetrahydrofolate gene in Turkish patients with rheumatoid arthritis: relationship with methotrexate toxicity. Open Rheumatol J 5:30
Plaza-Plaza JC, Aguilera M, Canadas-Garre M, Chemello C, González-Utrilla A, Faus Dader MJ, Calleja MA (2012) Pharmacogenetic polymorphisms contributing to toxicity induced by methotrexate in the southern Spanish population with rheumatoid arthritis. Omics: a journal of integrative biology 16(11):589–595
Inanir A, Yigit S, Tekcan A, Tural S, Kismali G (2013) IL-4 and MTHFR gene polymorphism in rheumatoid arthritis and their effects. Immunol Lett 152(2):104–108
Boughrara W, Aberkane M, Fodil M, Benzaoui A, Dorgham S, Zemani F, Dahmani C, Petit Teixeira E, Boudjema A (2015) Impact of MTHFR rs1801133, MTHFR rs1801131 and ABCB1 rs1045642 polymorphisms with increased susceptibility of rheumatoid arthritis in the West Algerian population: a case-control study.Acta Reumatol Port 40(4):363–71
Saad MN, Mabrouk MS, Eldeib AM, Shaker OG (2015) Genetic case-control study for eight polymorphisms associated with rheumatoid arthritis. PLoS One 10(7):e0131960
Saleh MM, Irshaid YM, Mustafa KN (2015) Methylene tetrahydrofolate reductase genotypes frequencies: association with toxicity and response to methotrexate in rheumatoid arthritis patients. Int J Clin Pharmacol Ther 53(2):154–162
Hashiguchi M, Shimizu M, Hakamata J, Tsuru T, Tanaka T, Suzaki M, Miyawaki K, Chiyoda T, Takeuchi O, Hiratsuka J (2016) Genetic polymorphisms of enzyme proteins and transporters related to methotrexate response and pharmacokinetics in a Japanese population. J Pharm Health Care Sci 2(1):35
Shaker OG, El-Demellawy HH, Salem MN, Eesa NN (2016) Methylene tetrahydrofolate reductase (MTHFR) gene polymorphisms in rheumatoid arthritis patients: correlation with serum osteopontin levels and disease activity. The Egyptian Rheumatologist 38(4):283–288
González-Mercado MG, Rivas F, Gallegos-Arreola MP, Morán-Moguel MC, Salazar-Páramo M, González-López L, Gámez-Nava JI, Munoz-Valle JF, Medina-Coss y Leon R, González-Mercado A (2017) MTRR A66G, RFC1 G80A, and MTHFR C677T and A1298C polymorphisms and disease activity in mexicans with rheumatoid arthritis treated with methotrexate. Genet Test Mol Biomarkers 21(11):698–704
El-Aziz TAA, Mohamed RH (2017) Influence of MTHFR C677T gene polymorphism in the development of cardiovascular disease in Egyptian patients with rheumatoid arthritis. Gene 610:127–132
Premkumar B, Sivaraman J, Nandini R (2018) Methylenetetrahydrofolate reductase gene polymorphism MTHFR A1298C and rheumatoid arthritis in south indian population. Drug Invention Today 10(3)
Wang S, Zuo S, Liu Z, Ji X, Yao Z, Wang X (2019) Association of MTHFR and RFC1 gene polymorphisms with methotrexate efficacy and toxicity in Chinese Han patients with rheumatoid arthritis. J Int Med Res 1–11
Lazzerini P, Selvi E, Lorenzini S, Capecchi P, Ghittoni R, Bisogno S, Catenaccio M, Marcolongo R, Galeazzi M, Laghi-Pasini F (2006) Homocysteine enhances cytokine production in cultured synoviocytes from rheumatoid arthritis patients. Clin Exp Rheumatol 24(4):387–393
MacKay K, Eyre S, Myerscough A, Milicic A, Barton A, Laval S, Barrett J, Lee D, White S, John S (2002) Whole-genome linkage analysis of rheumatoid arthritis susceptibility loci in 252 affected sibling pairs in the United Kingdom. Arthritis Rheum 46(3):632–639
Rosenblatt DS (2001) Methylenetetrahydrofolate reductase. Clin Invest Med 24(1):56–59
Favalli EG, Biggioggero M, Meroni PL (2014) Methotrexate for the treatment of rheumatoid arthritis in the biologic era: still an “anchor” drug? Autoimmun Rev 13(11):1102–1108
Haagsma CJ, Blom HJ, van Riel PL, van’t Hof MA, Giesendorf BA, van Oppenraaij-Emmerzaal D, van de Putte LB (1999) Influence of sulphasalazine, methotrexate, and the combination of both on plasma homocysteine concentrations in patients with rheumatoid arthritis. Ann Rheum Dis 58(2):79–84
James HM, Gillis D, Hissaria P, Lester S, Somogyi AA, Cleland LG, Proudman SM (2008) Common polymorphisms in the folate pathway predict efficacy of combination regimens containing methotrexate and sulfasalazine in early rheumatoid arthritis. J Rheumatol 35(4):562–571
Fisher MC, Cronstein BN (2009) Metaanalysis of methylenetetrahydrofolate reductase (MTHFR) polymorphisms affecting methotrexate toxicity. J Rheumatol 36(3):539–545
Friedman G, Goldschmidt N, Friedlander Y, Ben-Yehuda A, Selhub J, Babaey S, Mendel M, Kidron M, Bar-On H (1999) A common mutation A1298C in human methylenetetrahydrofolate reductase gene: association with plasma total homocysteine and folate concentrations. J Nutr 129(9):1656–1661
Razi B, Imani D, Makoui MH, Rezaei R, Aslani S (2020) Association between MTHFR gene polymorphism and susceptibility to autism spectrum disorders: systematic review and meta-analysis. Res Autism Spectr Disord 70:101473
Song GG, Bae S-C, Lee YH (2014) Association of the MTHFR C677T and A1298C polymorphisms with methotrexate toxicity in rheumatoid arthritis: a meta-analysis. Clin Rheumatol 33(12):1715–1724
Weisberg I, Tran P, Christensen B, Sibani S, Rozen R (1998) A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab 64(3):169–172
Viatte S, Plant D, Raychaudhuri S (2013) Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol 9(3):141–153
Lopez-Olivo M, Gonzalez-Lopez L, Garcia-Gonzalez A, Villa-Manzano A, Cota-Sanchez A, Salazar-Paramo M, Varon-Villalpando E, Cardona-Muñoz E, Gamez-Nava J (2006) Factors associated with hyperhomocysteinaemia in Mexican patients with rheumatoid arthritis. Scand J Rheumatol 35(2):112–116
Cornélis F, Fauré S, Martinez M, Prud’homme J-F, Fritz P, Dib C, Alves H, Barrera P, De Vries N, Balsa A (1998) New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proc Natl Acad Sci 95(18):10746–10750
Shiozawa S, Hayashi S, Tsukamoto Y, Goko H, Kawasaki H, Wada T, Shimizu K, Yasuda N, Kamatani N, Takasugi K (1998) Identification of the gene loci that predispose to rheumatoid arthritis. Int Immunol 10(12):1891–1895
Cen H, Huang H, Zhang L-N, Liu L-Y, Zhou L, Xin X-F, Zhuo R-J (2017) Associations of methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms with genetic susceptibility to rheumatoid arthritis: a meta-analysis. Clin Rheumatol 36(2):287–297
Acknowledgments
We thank Dr. Shahab Alizadeh for comments that greatly improved the manuscript.
Author information
Authors and Affiliations
Contributions
ZB generated the idea. DI analyzed and interpreted the data. ZB and HY prepared the original draft. ZB, HY, and MA critically revised the paper. MA supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bagheri-Hosseinabadi, Z., Imani, D., Yousefi, H. et al. MTHFR gene polymorphisms and susceptibility to rheumatoid arthritis: a meta-analysis based on 16 studies. Clin Rheumatol 39, 2267–2279 (2020). https://doi.org/10.1007/s10067-020-05031-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-020-05031-5