Abstract
The aim of our study was to conduct a meta-analysis to assess whether combined evidence shows associations between C677T and A1298C polymorphisms of methylenetetrahydrofolate reductase (MTHFR) and genetic susceptibility to rheumatoid arthritis (RA). A total of 11 articles involving 20 comparisons were included, containing 12 comparisons for the MTHFR C677T polymorphism and 8 comparisons for the MTHFR A1298C polymorphism. Significant evidence was detected for the association of RA susceptibility with the MTHFR C677T polymorphism T allele under allelic contrast and dominant model in Asians (T versus C, OR = 1.300, 95 % CI = 1.104–1.531, p = 0.002; TT + CT versus CC, OR = 1.495, 95 % CI = 1.187–1.882, p = 0.001). Significant association between RA susceptibility and the MTHFR A1298C polymorphism A allele under recessive model was found in the overall meta-analysis (AA versus AC + CC, OR = 1.281, 95 % CI = 1.048–1.565, p = 0.016). Our meta-analysis results demonstrate that the MTHFR C677T polymorphism is involved in the genetic susceptibility of RA in Asians, and the MTHFR A1298C polymorphism is associated with genetic susceptibility to RA in the overall population. Given the paucity of studies, especially in non-Asian populations, further studies with larger sample sizes are required to elucidate the role of MTHFR polymorphisms in the genetic basis of RA in different ethnic populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is a common systemic autoimmune disease characterized by the proliferation of synovial cells, the infiltration of inflammatory cells and angiogenesis [1]. Although the etiology of RA has not been precisely demonstrated, it has been widely accepted that both environmental and genetic risk factors have been implicated in the onset and progression of this disorder. Available data shows that the prevalence of RA varies geographically, with much higher prevalence having been reported in North America and North Europe [2]. Overall, about three quarters of RA patients are female. The development of RA could occur in any age, but much more cases develop in their middle or old age [1]. Although great improvements have been achieved in the field of RA treatment, uncontrolled disease could result in disability, decreased life quality, and several severe comorbidities. During the past few decades, our understanding of the genetic basis of RA has been rapidly prompted through the application of numerous large-scale candidate gene studies and genome-wide association studies (GWASs), and multiple susceptible genes/loci associated with RA have been identified and confirmed [3].
Methylenetetrahydrofolate reductase (MTHFR) is a key enzyme involved in the folate metabolic pathway, catalyzing the irreversible conversion of 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate, which is a methylation group donor and has been implicated in the methylation of genomic DNA. Notably, 5-methyltetrahydrofolate acts as the primary methylation donor for the remethylation of homocysteine into methionine [4]. Thus, impaired MTHFR might result in hypomethylation of genomic DNA and hyperhomocysteinemia, and both of these events have been suggested to be involved in the pathogenesis of RA [5–7], making MTHFR as a candidate susceptible gene for RA. The gene encoding MTHFR has been mapped to the chromosomal region 1p36, and multiple single nucleotide polymorphisms (SNPs) within MTHFR have been identified [8]. Remarkably, the chromosomal region 1p36 has been previously been shown to be associated with genetic susceptibility to RA using genome-wide linkage studies [9–11]. Besides, a recent meta-analysis shows that SNPs within the PADI4 gene have also been revealed to be associated with RA genetic predisposition in Asians and Caucasians, and this gene also locates in the chromosomal region 1p36 [12]. Among the SNPs within the MTHFR gene, the two most commonly studied polymorphisms are C677T (rs1801133) and A1298C (rs1801131), which are both non-synonymous SNPs. The C to T change at nucleotide 677 leads to alanine to valine substitution at codon 222, rendering MTHFR more thermolabile and reducing its enzyme activity [13, 14]. Similar to the C677T polymorphism, the A to C change at nucleotide 1298 results in glutamine to alanine substitution, also leading to reduced enzyme activity [15–17]. The associations between C677T and A1298C polymorphisms within MTHFR and RA genetic susceptibility have been widely investigated in different populations with inconsistent results [18–35]. This discrepancy might be due to different sample sizes and genetic backgrounds, clinical heterogeneity, publication bias, etc. Meta-analysis is a useful tool to combine the results on the same topic to get a pooled estimation with increased statistical power [36]. Thus, the aim of our study was to conduct a meta-analysis to assess whether combined evidence shows associations between C677T and A1298C polymorphisms of MTHFR and genetic susceptibility to RA and to summarize the effect sizes of the polymorphisms associated with RA.
Method
Identification of eligible studies and data extraction
The present meta-analysis is performed in accordance with the preferred reporting items for systematic reviews and meta-analyses (PRISMA) [37]. An exhaustive search on studies examining the association of MTHFR C677T and A1298C polymorphisms with RA susceptibility was performed. The literature search was made using PubMed, China National Knowledge Infrastructure (CNKI) database, and Wanfang database to identify relevant articles, applying the following medical subject heading (MeSH) terms and/or text words: ‘methylenetetrahydrofolate reductase,’ ‘MTHFR,’ ‘polymorphism,’ ‘polymorphisms,’ ‘rheumatoid arthritis,’ and ‘RA.’ No language restrictions were applied. The references in these studies were also reviewed to identify additional studies. The inclusion criteria were as follows: (a) being published before October 2015; (b) using case–control study design; (c) allele or genotype frequencies among RA patients and controls available, providing enough data to calculate odds ratio (OR); and (d) the genotype distribution among control group should conform to Hardy–Weinberg equilibrium (HWE), since deviation from HWE among controls could imply some potential bias in control selection or genotyping errors. When two or more studies published by the same authors contained overlapped data, we extracted data from the study with the largest sample size. When a study described the results in different subgroups, we treated them independently. Studies in which family members had been studied were excluded because its analysis was based on linkage consideration. The following information from each study was extracted: first author’s name, year of publication, country, ethnicity, the number of cases and controls, and allele/genotype distribution in cases and controls. Two authors independently extracted these data, and discrepancy was addressed by discussion.
Evaluation of the statistical association
Allele frequencies of the MTHFR C677T and A1298C polymorphisms from each study were determined by the allele counting method. Chi-square test was applied to assess whether the observed genotype frequencies in control group conformed to HWE. In this study, we performed meta-analysis on (a) allelic contrast, (b) recessive model, and (c) dominant model. The heterogeneity between studies was assessed by Cochran’s Q statistic, as well as I 2 statistic, which was used to quantify the effect of heterogeneity (I 2 = 100 % × (Q − df) / Q), measuring the proportion of total variation in study estimates due to heterogeneity [38]. Finally, the pooled estimate of risk was obtained by a random effects (DerSimonian–Laird) or a fixed effects model (Mantel–Haenszel) in the presence (P ≤ 0.1 or I 2 > 50 %) or absence (P > 0.1 and I 2 ≤ 50 %) of heterogeneity, respectively. Due to the genetic heterogeneity among different ethnicities, we performed subgroup meta-analysis stratified by ethnicity. Statistical analysis for this meta-analysis was performed by Stata version 10.0 (Stata Corporation, College Station, TX, USA).
Evaluation of publication bias
We assessed potential publication bias by funnel plot [39] and utilized the Egger’s linear regression test to evaluate the funnel plot asymmetry, which is an approach to measure funnel plot asymmetry on the natural logarithm scale of the OR [40]. The significance of the intercept was assessed by the t test suggested by Egger, and the p value less than 0.05 was considered significant publication bias.
Results
Studies included in the meta-analysis
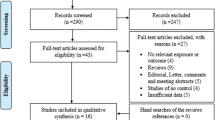
As shown in Fig. 1, a total of 18 relevant articles examining the association of MTHFR C677T polymorphism and/or A1298C polymorphism with genetic susceptibility to RA were identified through PubMed, CNKI database, and Wanfang database search and a review of the references [18–35], and seven articles were excluded [18, 20, 27, 29, 31–33]. Five articles were excluded since the detailed genotype, and allele information is unavailable [18, 20, 27, 29, 31]. Two articles were published by the same research group, so only the article with the largest sample size was included [26] and the other one was excluded [32]. One study in which the genotype distribution of MTHFR C677T polymorphism did not conform to HWE was excluded [33]. One article contained data on two different subgroups [34], and we treated them independently. Among the 11 eligible articles, there are 2 articles in which only MTHFR C677T polymorphism has been investigated [22, 23] and the other 9 articles in which both MTHFR C677T and A1298C polymorphisms have been studied [19, 21, 24–26, 28, 34, 35]. Among two eligible articles containing both MTHFR C677T and A1298C polymorphisms, the genotype distribution of MTHFR A1298C polymorphism deviated from HWE in the control group, so the data of MTHFR A1298C polymorphism in these two articles was excluded [19, 30]. Thus, we analyzed 20 separate comparisons in total, including 12 comparisons for the MTHFR C677T polymorphism and 8 comparisons for the MTHFR A1298C polymorphism. The patients with RA in all eligible studies were diagnosed according to the American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis [41]. The characteristics of the selected studies are summarized in Table 1 (detailed information about genotype and allele frequencies for each polymorphism in selected studies is available in Supplementary Tables 1 and 2).
Quantitative synthesis
Meta-analysis of the MTHFR C677T polymorphism with RA susceptibility
A total of 1744 cases and 1599 controls in 12 case–control studies were eligible for meta-analysis of association between the MTHFR C677T polymorphism and genetic susceptibility to RA (Table 2). Six studies were from Asian, two from European, one from African, one from African-American, one from Caucasian-American, and one from Jewish. Overall meta-analysis revealed that non-significant evidence for the association of the MTHFR C677T polymorphism with RA susceptibility was detected (T versus C, OR = 1.221, 95 % CI = 0.999–1.493, I 2 = 64.2 %, p = 0.051; TT + CT versus CC, OR = 1.271, 95 % CI = 0.975–1.658, I 2 = 61.7 %, p = 0.076; TT versus CT + CC, OR = 1.183, 95 % CI = 0.943–1.484, I 2 = 30.2 %, p = 0.146) (Figs. 2 and 3). When we combined results by ethnicity, significant evidence was detected for the association of RA susceptibility with the MTHFR C677T polymorphism T allele under allelic contrast and dominant model in Asians (T versus C, OR = 1.300, 95 % CI = 1.104–1.531, I 2 = 28.4 %, p = 0.002; TT + CT versus CC, OR = 1.495, 95 % CI = 1.187–1.882, I 2 = 0, p = 0.001) but not in Europeans (T versus C, OR = 1.150, 95 % CI = 0.587–2.255, I 2 = 83.4 %, p = 0.684; TT + CT versus CC, OR = 0.954, 95 % CI = 0.453–2.013, I 2 = 70.5 %, p = 0.902).
Meta-analysis of the MTHFR A1298C polymorphism with RA susceptibility
A total of 1094 cases and 1026 controls in 8 case–control studies were included for meta-analysis of association between the MTHFR A1298C polymorphism and RA susceptibility (Table 3). Four studies were from Asian, one from European, one from African-American, one from Caucasian-American, and one from Jewish. Overall meta-analysis showed that RA susceptibility was significantly associated with the MTHFR A1298C polymorphism A allele under recessive model (AA versus AC + CC, OR = 1.281, 95 % CI = 1.048–1.565, I 2 = 0, p = 0.016) (Fig. 4). However, non-significant evidence for the association of RA susceptibility with the MTHFR A1298C polymorphism A allele under allelic contrast or dominant model was found in the overall meta-analysis (A versus C, OR = 1.117, 95 % CI = 0.955–1.306, I 2 = 13.6 %, p = 0.167; AA + AC versus CC, OR = 0.973, 95 % CI = 0.572–1.657, I 2 = 49.4 %, p = 0.921) (Fig. 5). Only the number of study performed in Asian population was more than two, so we combined the results from Asian. However, the pooled results revealed non-significant association between RA susceptibility and the MTHFR A1298C polymorphism in Asians (A versus C, OR = 1.174, 95 % CI = 0.943–1.463, I 2 = 18.0 %, p = 0.152; AA + AC versus CC, OR = 0.997, 95 % CI = 0.589–1.685, I 2 = 41.4 %, p = 0.990; AA versus AC + CC, OR = 1.296, 95 % CI = 0.980–1.714, I 2 = 23.5 %, p = 0.069).
Evaluation of heterogeneity and publication bias
Heterogeneity was detected for the association of RA with the MTHFR C677T polymorphism T allele under allelic contrast and dominant model in the overall meta-analysis. When we pooled results according to the ethnicity of study populations, the heterogeneity disappeared in the subgroup analysis of Asians but remained significant in Europeans. In addition, heterogeneity was found for the association of RA with the MTHFR C677T polymorphism T allele under recessive model in the subgroup analysis of Europeans.
The heterogeneity was significant in the overall meta-analysis for the association of RA susceptibility with the MTHFR A1298C polymorphism A allele under dominant model. When we performed subgroup meta-analysis according to the ethnicity, heterogeneity disappeared in the pooled analysis of Asians.
We performed publication bias analysis on the MTHFR C677T and A1298C polymorphisms. No obvious asymmetry was found according to the shapes of the funnel plots (data not shown). We applied Egger’s linear regression test to assess the funnel plot asymmetry, and the results indicated no publication bias (p > 0.05).
Discussion
MTHFR is a key enzyme involved in the metabolism of folate, which catalyzes the conversion of 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate, providing methylation group for its substrate [4]. Methotrexate (MTX), a structural analog of folic acid, has become the anchor drug for the treatment of RA based on its best-known efficacy–toxicity ratio and long-term affordability [42]. Although the actual mechanism of MTX used to treat RA has not been completely elucidated, available evidence suggests that MTX could act through inhibiting several enzymes implicated in the folate metabolism pathway. MTHFR is not inhibited by MTX directly but in view of the effect of MTHFR in folate pool, so the function of MTHFR might affect the outcomes (efficacy and toxicity) of patients with RA treated by MTX. Within the MTHFR gene, the two most commonly studied SNPs (C677T and A1298C) could cause the alteration of enzyme activity [13–17]. Up to now, multiple studies have been performed to test whether the MTHFR C677T and A1298C polymorphisms are associated with the outcomes of MTX among RA patients and related meta-analyses have been reported [43, 44].
Apart from the associations of the MTHFR C677T and A1298C polymorphisms with the outcomes of MTX in RA patients, associations between the C677T and A1298C polymorphisms of MTHFR and genetic susceptibility to RA have also been extensively investigated [18–35]. The MTHFR C677T polymorphism T allele and A1298C polymorphism C allele have both been reported to be associated with reduced enzyme activity [13–17]. The rationale of investigating the associations of the MTHFR C677T and A1298C polymorphisms with genetic susceptibility to RA is as follows: First, MTHFR could catalyze the conversion of 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate, which is indispensable for nucleic acid methylation, thus impaired MTHFR activity might lead to hypomethylation of genomic DNA, and altered methylation patterns have been detected in several cell types in RA [5]. Second, MTHFR could act as the primary methylation donor for remethylation of homocysteine into methionine, so impaired MTHFR function might result in been increased level of homocysteine, and this possibility has been documented by multiple studies [7, 13–15, 17]. Several studies have shown that the level of homocysteine in RA patients is elevated compared with health controls [6, 7, 45–47], and homocysteine could cause the elevation of inflammation in RA through activating the proinflammatory transcription factor NF-κB [6]. Third, the chromosomal region containing MTHFR has been shown to be associated with genetic susceptibility to RA by genome-wide linkage studies [9–11].
Although the associations between the C677T and A1298C polymorphisms of MTHFR and RA susceptibility have been examined in different populations, the results are controversial. To our knowledge, so far, no meta-analysis has been reported regarding the associations of RA susceptibility with the MTHFR C677T and A1298C polymorphisms. Thus, we performed this meta-analysis to assess whether the C677T and A1298C polymorphisms of MTHFR are associated with genetic predisposition of RA and to summarize the effect sizes of these polymorphisms.
Our meta-analysis results revealed that non-significant association of the MTHFR C677T polymorphism T allele with RA susceptibility was detected. Since heterogeneity was found in the overall meta-analysis of the MTHFR C677T polymorphism T allele with RA susceptibility, we performed stratified meta-analysis by ethnicity, and the heterogeneity disappeared in the subgroup analysis of Asians but remained significant in Europeans, and significant association between RA susceptibility and the MTHFR C677T polymorphism T allele was observed in Asians but not in Europeans. Overall meta-analysis revealed non-significant association between RA susceptibility and the MTHFR C677T polymorphism T allele under dominant model and recessive model. Heterogeneity was also detected for the overall meta-analysis of the association between RA susceptibility and the MTHFR C677T polymorphism T allele under dominant model, so subgroup meta-analysis by ethnicity was conducted, and the results showed that significant association was found in Asians but not in Europeans. Although no heterogeneity was detected for the overall meta-analysis of the association between RA susceptibility and the MTHFR C677T polymorphism T allele under recessive model, we also performed stratified meta-analysis according to the ethnicity. However, non-significant association was found in Asians or Europeans. Since the number of studies in non-Asians is small, further studies with larger sample sizes are required to examine the association between MTHFR C677T polymorphism and RA susceptibility in non-Asian populations. Our meta-analysis indicated that the MTHFR C677T polymorphism is involved in the genetic background of RA in Asians. The overall meta-analysis revealed that significant association between RA susceptibility and the MTHFR A1298C polymorphism A allele under recessive model was found, but non-significant association was detected for the association between RA susceptibility and the MTHFR A1298C polymorphism A allele under allelic contrast and dominant model. Only studies performed in Asians were eligible for subgroup meta-analysis, but non-significant association was found between RA susceptibility and the MTHFR A1298C polymorphism in Asians, and this might be due to the small number of eligible study and the relatively low power of each study caused by small sample size.
Besides the relationships between MTHFR polymorphisms and RA genetic susceptibility, a few studies have been performed to assess the associations of MTHFR C677T and A1298C polymorphisms with cardiovascular events (CVs) and osteoporosis among RA patients [27, 48–50]. Although the results are controversial, these two polymorphisms within MTHFR might be associated with CVs and osteoporosis, so comorbidities might contribute to the heterogeneity among eligible studies. However, information about comorbidity among eligible studies is unavailable; we could not determine whether comorbidity is an influencing factor of heterogeneity. In addition, it is also valuable to examine the relationships between MTHFR polymorphisms and clinical features of RA patients. However, among all the eligible studies, only one study has analyzed the association of MTHFR C677T polymorphism with ocular involvement and rheumatoid factor and the results are non-significant [22]. Studies are needed to clarify the relationships between MTHFR polymorphisms and clinical features of RA in the future.
Several limitations of the present meta-analysis should be mentioned. First, significant heterogeneity was detected among several comparisons, which might distort the meta-analysis. Second, the number of studies among subgroup analysis by ethnicity was small. Only two studies from European were available for the meta-analysis of the MTHFR C667T polymorphism with RA. In the subgroup meta-analysis for the MTHFR A1298C polymorphism, only four studies from Asian population were available and the number of study from non-Asian population was only one. Third, publication bias might exist, although the Egger’s test provided non-significant result. Fourth, this meta-analysis was based on uncorrected estimate, so a more precise analysis could be conducted if the potential confounding factors, such as age, sex, clinical characteristic, and environmental factors were available. Finally, due to the lack of related data, the associations of MTHFR polymorphisms with clinical features of RA could not be evaluated by meta-analysis.
In conclusion, our meta-analysis results demonstrate that the MTHFR C677T polymorphism is involved in the genetic susceptibility of RA in Asians, and the MTHFR A1298C polymorphism is associated with genetic susceptibility to RA in the overall population. Given the paucity of studies, especially in non-Asian populations, further well-designed studies with larger sample sizes are required to elucidate the role of MTHFR polymorphisms in the genetic basis of RA in different ethnic populations.
References
Scott DL, Wolfe F, Huizinga TW (2010) Rheumatoid arthritis. Lancet 376:1094–1108
Li R, Sun J, Ren LM, Wang HY, Liu WH, Zhang XW, Chen S, Mu R, He J, Zhao Y, Long L, Liu YY, Liu X, Lu XL, Li YH, Wang SY, Pan SS, Li C, Wang HY, Li ZG (2012) Epidemiology of eight common rheumatic diseases in China: a large-scale cross-sectional survey in Beijing. Rheumatology (Oxford) 51:721–729
Viatte S, Plant D, Raychaudhuri S (2013) Genetics and epigenetics of rheumatoid arthritis. Nat Rev Rheumatol 9:141–153
Rosenblatt DS (2001) Methylenetetrahydrofolate reductase. Clin Invest Med 24:56–59
Klein K, Gay S (2015) Epigenetics in rheumatoid arthritis. Curr Opin Rheumatol 27:76–82
Lazzerini PE, Selvi E, Lorenzini S, Capecchi PL, Ghittoni R, Bisogno S, Catenaccio M, Marcolongo R, Galeazzi M, Laghi-Pasini F (2006) Homocysteine enhances cytokine production in cultured synoviocytes from rheumatoid arthritis patients. Clin Exp Rheumatol 24:387–393
Fujimaki C, Hayashi H, Tsuboi S, Matsuyama T, Kosuge K, Yamada H, Inoue K, Itoh K (2009) Plasma total homocysteine level and methylenetetrahydrofolate reductase 677C>T genetic polymorphism in Japanese patients with rheumatoid arthritis. Biomarkers 14:49–54
Goyette P, Pai A, Milos R, Frosst P, Tran P, Chen Z, Chan M, Rozen R (1998) Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR). Mamm Genome 9:652–656
Cornélis F, Fauré S, Martinez M, Prud’homme JF, Fritz P, Dib C, Alves H, Barrera P, de Vries N, Balsa A, Pascual-Salcedo D, Maenaut K, Westhovens R, Migliorini P, Tran TH, Delaye A, Prince N, Lefevre C, Thomas G, Poirier M, Soubigou S, Alibert O, Lasbleiz S, Fouix S, Bouchier C, Lioté F, Loste MN, Lepage V, Charron D, Gyapay G, Lopes-Vaz A, Kuntz D, Bardin T, Weissenbach J, ECRAF (1998) New susceptibility locus for rheumatoid arthritis suggested by a genome-wide linkage study. Proc Natl Acad Sci U S A 95:10746–10750
Shiozawa S, Hayashi S, Tsukamoto Y, Goko H, Kawasaki H, Wada T, Shimizu K, Yasuda N, Kamatani N, Takasugi K, Tanaka Y, Shiozawa K, Imura S (1998) Identification of the gene loci that predispose to rheumatoid arthritis. Int Immunol 10:1891–1895
MacKay K, Eyre S, Myerscough A, Milicic A, Barton A, Laval S, Barrett J, Lee D, White S, John S, Brown MA, Bell J, Silman A, Ollier W, Wordsworth P, Worthington J (2002) Whole-genome linkage analysis of rheumatoid arthritis susceptibility loci in 252 affected sibling pairs in the United Kingdom. Arthritis Rheum 46:632–639
Lee, YH, Bae, SC (2015) Association between susceptibility to rheumatoid arthritis and PADI4 polymorphisms: a meta-analysis. Clin Rheumatol [Epub ahead of print]
Kang SS, Zhou J, Wong PW, Kowalisyn J, Strokosch G (1988) Intermediate homocysteinemia: a thermolabile variant of methylenetetrahydrofolate reductase. Am J Hum Genet 43:414–421
Rozen R (1997) Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR). Thromb Haemost 78:523–526
van der Put NM, Gabreëls F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel LP, Blom HJ (1998) A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet 62:1044–1051
Weisberg I, Tran P, Christensen B, Sibani S, Rozen R (1998) A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab 64:169–172
Friedman G, Goldschmidt N, Friedlander Y, Ben-Yehuda A, Selhub J, Babaey S, Mendel M, Kidron M, Bar-On H (1999) A common mutation A1298C in human methylenetetrahydrofolate reductase gene: association with plasma total homocysteine and folate concentrations. J Nutr 129:1656–1661
Boughrara W, Aberkane M, Fodil M, Benzaoui A, Dorgham S, Zemani F, Dahmani C, Petit-Teixeira E, Boudjema A (2015) Impact of MTHFR rs1801133, MTHFR rs1801131 and ABCB1 rs1045642 polymorphisms with increased susceptibility of rheumatoid arthritis in the West Algerian population: a case-control study. Acta Reumatol Port 40:363–371
Saad MN, Mabrouk MS, Eldeib AM, Shaker OG (2015) Genetic case-control study for eight polymorphisms associated with rheumatoid arthritis. PLoS One 10(7):e0131960
Soukup T, Dosedel M, Pavek P, Nekvindova J, Barvik I, Bubancova I, Bradna P, Kubena AA, Carazo AF, Veleta T, Vlcek J (2015) The impact of C677T and A1298C MTHFR polymorphisms on methotrexate therapeutic response in East Bohemian region rheumatoid arthritis patients. Rheumatol Int 35:1149–1161
Saleh MM, Irshaid YM, Mustafa KN (2015) Methylene tetrahydrofolate reductase genotypes frequencies: association with toxicity and response to methotrexate in rheumatoid arthritis patients. Int J Clin Pharmacol Ther 53:154–162
Inanir A, Yigit S, Tekcan A, Tural S, Kismali G (2013) IL-4 and MTHFR gene polymorphism in rheumatoid arthritis and their effects. Immunol Lett 152:104–108
Shi YH, Zhou RH, Xu J, Zhu FX, Li BZ, Li LM, Mo HY (2013) Relationship between single nucleotide polymorphism of methylenetetrahydrofolate reductase gene and the side effect of low-dose methotrexate in the treatment of rheumatoid arthritis. Chin J New Clin Med 6:211–214
Plaza-Plaza JC, Aguilera M, Cañadas-Garre M, Chemello C, González-Utrilla A, Faus Dader MJ, Calleja MA (2012) Pharmacogenetic polymorphisms contributing to toxicity induced by methotrexate in the southern Spanish population with rheumatoid arthritis. OMICS 16:589–595
Taşbaş O, Borman P, Gürhan Karabulut H, Tükün A, Yorgancıoğlu R (2011) The frequency of A1298C and C677T polymorphisms of the methylenetetrahydrofolate gene in Turkish patients with rheumatoid arthritis: relationship with methotrexate toxicity. Open Rheumatol J 5:30–35
Xiao H, Xu JH, Zhou XM, Zhang ZH, Shen SH, Li YW (2011) Correlation of methylene tetrahydrofolate reductase gene single nucleotide polymorphisms and therapeutic effect of methotrexate in rheumatoid arthritis patients. Acta Univ Med Anhui 46:1157–1161
Palomino-Morales R, Gonzalez-Juanatey C, Vazquez-Rodriguez TR, Rodriguez L, Miranda-Filloy JA, Fernandez-Gutierrez B, Llorca J, Martin J, Gonzalez-Gay MA (2010) A1298C polymorphism in the MTHFR gene predisposes to cardiovascular risk in rheumatoid arthritis. Arthritis Res Ther 12:R71
Cai YM, Gong WX (2009) Linkage study on methylenetetrahydrofolate reductase single nucleotide polymorphisms and methotrexate-related adverse effects in patients with rheumatoid arthritis. Chin J Prim Med Pharm 16:1155–1157
Taraborelli M, Andreoli L, Archetti S, Ferrari M, Cattaneo R, Tincani A (2009) Methylenetetrahydrofolate reductase polymorphisms and methotrexate: no association with response to therapy nor with drug-related adverse events in an Italian population of rheumatic patients. Clin Exp Rheumatol 27:499–502
Rubini M, Padovan M, Baricordi O, Fotinidi M, Govoni M, Trotta F (2008) The c.1298A>C polymorphism in the methylenetetrahydrofolate reductase gene is associated with rheumatoid arthritis susceptibility in Italian patients. Clin Exp Rheumatol 26:163
Ghodke Y, Chopra A, Joshi K, Patwardhan B (2008) Are thymidylate synthase and methylene tetrahydrofolate reductase genes linked with methotrexate response (efficacy, toxicity) in Indian (Asian) rheumatoid arthritis patients? Clin Rheumatol 27:787–789
Zhou XM, Xu JH, Xu SQ (2008) Relationship between the single nucleotide polymorphism of 5,10-methylenetetrahydrofolate reductase gene and the treatment of methotrexate in rheumatoid arthritis. Chin J Rheumatol 12:598–602
Xu XY, Wang MM, Xiao CS (2007) The study of the relationship of MTHFR polymorphisms with rheumatoid diseases. J Southeast Univ (Med Sci Edi) 26:38–41
Hughes LB, Beasley TM, Patel H, Tiwari HK, Morgan SL, Baggott JE, Saag KG, McNicholl J, Moreland LW, Alarcón GS, Bridges SL Jr (2006) Racial or ethnic differences in allele frequencies of single-nucleotide polymorphisms in the methylenetetrahydrofolate reductase gene and their influence on response to methotrexate in rheumatoid arthritis. Ann Rheum Dis 65:1213–1218
Berkun Y, Levartovsky D, Rubinow A, Orbach H, Aamar S, Grenader T, Abou Atta I, Mevorach D, Friedman G, Ben-Yehuda A (2004) Methotrexate related adverse effects in patients with rheumatoid arthritis are associated with the A1298C polymorphism of the MTHFR gene. Ann Rheum Dis 63:1227–1231
Egger M, Smith GD, Phillips AN (1997) Meta-analysis: principles and procedures. BMJ 315:1533–1537
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 6:e1000097
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21:1539–1558
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. (1988) The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 31:315–324
Favalli EG, Biggioggero M, Meroni PL (2014) Methotrexate for the treatment of rheumatoid arthritis in the biologic era: still an “anchor” drug? Autoimmun Rev 13:1102–1108
Song GG, Bae SC, Lee YH (2014) Association of the MTHFR C677T and A1298C polymorphisms with methotrexate toxicity in rheumatoid arthritis: a meta-analysis. Clin Rheumatol 33:1715–1724
Morgan MD, Al-Shaarawy N, Martin S, Robinson JI, Twigg S, YEAR Consortium, Magdy AA, Omar AS, Ghattas MH, Emery P, Barrett JH, Morgan AW (2014) MTHFR functional genetic variation and methotrexate treatment response in rheumatoid arthritis: a meta-analysis. Pharmacogenomics 15:467–475
Lopez-Olivo MA, Gonzalez-Lopez L, Garcia-Gonzalez A, Villa-Manzano AI, Cota-Sanchez AR, Salazar-Paramo M, Varon-Villalpando E, Cardona-Muñoz EG, Gamez-Nava JI (2006) Factors associated with hyperhomocysteinaemia in Mexican patients with rheumatoid arthritis. Scand J Rheumatol 35:112–116
Armstrong DJ, Quinn AD, McCausland EM, Finch MB, Wright GD (2007) Relationship between homocysteine and body mass index in rheumatoid arthritis. Scand J Rheumatol 36:243
Woolf K, Manore MM (2008) Elevated plasma homocysteine and low vitamin B-6 status in nonsupplementing older women with rheumatoid arthritis. J Am Diet Assoc 108:443–453
Davis LA, Cannon GW, Pointer LF, Haverhals LM, Wolff RK, Mikuls TR, Reimold AM, Kerr GS, Richards JS, Johnson DS, Valuck R, Prochazka A, Caplan L (2013) Cardiovascular events are not associated with MTHFR polymorphisms, but are associated with methotrexate use and traditional risk factors in US veterans with rheumatoid arthritis. J Rheumatol 40:809–817
Brambila-Tapia AJ, Durán-González J, Sandoval-Ramírez L, Mena JP, Salazar-Páramo M, Gámez-Nava JI, González-López L, Lazalde-Medina BB, Dávalos NO, Peralta-Leal V, Vázquez del Mercado M, Beltrán-Miranda CP, Dávalos IP (2012) MTHFR C677T, MTHFR A1298C, and OPG A163G polymorphisms in Mexican patients with rheumatoid arthritis and osteoporosis. Dis Markers 32:109–114
Saad MN, Mabrouk MS, Eldeib AM, Shaker OG (2015) Effect of MTHFR, TGFβ1, and TNFB polymorphisms on osteoporosis in rheumatoid arthritis patients. Gene 568:124–128
Acknowledgments
This work was supported by Zhejiang Provincial Key Laboratory of Pathophysiology, Nature Science Foundation of Ningbo city (Grant No. 2015A610205), Ningbo University Talent Project (F01256144702), School Research Foundation of Ningbo University (XKL14D2094, XKL14D2095), and K.C. Wong Magna Fund from Ningbo University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosures
None.
Additional information
Han Cen and Hua Huang contributed equally to this work and should be considered as co-first author.
Rights and permissions
About this article
Cite this article
Cen, H., Huang, H., Zhang, LN. et al. Associations of methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms with genetic susceptibility to rheumatoid arthritis: a meta-analysis. Clin Rheumatol 36, 287–297 (2017). https://doi.org/10.1007/s10067-016-3348-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-016-3348-0