Abstract
The revised 4th edition of the 2016 World Health Organization Classification of Tumors of the Central Nervous System (2016 CNS WHO) has introduced the integrated diagnostic classification that combines molecular and histological diagnoses for diffuse gliomas. In this study, we evaluated the molecular alterations for consecutive 300 diffuse glioma cases (grade 2, 56; grade 3, 62; grade 4, 182) based on this classification. Mutations in the isocitrate dehydrogenase (IDH) genes were common in lower grade glioma (LGG: grade2–3), and when combined with 1p/19q status, LGGs could be stratified into three groups except for four cases (Astrocytoma, IDH-mutant: 44; Oligodendroglioma, IDH-mutant and 1p/19q codeleted: 37; Astrocytoma, IDH-wildtype: 33). 1p/19q-codeleted oligodendrogliomas were clinically the most favorable subgroup even with upfront chemotherapy. In contrast, IDH-wildtype astrocytomas had a relatively worse prognosis; however, this subgroup was more heterogeneous. Of this subgroup, 11 cases had TERT promoter (pTERT) mutation with shorter overall survival than 12 pTERT-wildtype cases. Additionally, a longitudinal analysis indicated pTERT mutation as early molecular event for gliomagenesis. Therefore, pTERT mutation is critical for the diagnosis of molecular glioblastoma (WHO grade 4), regardless of histological findings, and future treatment strategy should be considered based on the precise molecular analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent revised WHO classification established integrated diagnosis for diffuse gliomas based on the combination of histological, molecular findings and clinical factors [1]. IDH1/2 mutation is considered one of the most crucial genetic alterations, which divide lower grade glioma (LGG) into two molecular trajectories during the early stage of gliomagenesis [2,3,4]. 1p/19q codeletion is another essential molecular alteration, which classified IDH-mutant LGGs into astrocytic and oligodendroglial tumors [5, 6]. IDH-wildtype LGG is considered to be a more aggressive genotype [2, 3]; however, it is a heterogeneous subgroup that should be further stratified [7]. Treatment strategy, including a surgical procedure, should be considered based on the integrated diagnosis [8,9,10,11] and optimal genetic analysis is recommended for the precise molecular classification. The Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy-Not Official WHO (cIMPACT-NOW) has provided novel information for clinical application of WHO classification via updates 1 through 7, published for the next WHO classification of CNS tumors [12,13,14,15,16,17,18]. The cIMPACT-NOW update 3 indicated that the tumor with molecular features of GBM, so-called “molecular GBM”, exists within the IDH-wildtype LGG [14]. These cases also showed a clinical course similar to that of IDH-wildtype GBMs. Furthermore, the cIMPACT-NOW update 6 proposed that one of the three genetic alterations (EGFR amplification, Combined whole chromosome 7 gain and whole chromosome 10 loss (+ 7/−10), TERT promoter (pTERT) mutation) is sufficient to define lower grade astrocytoma as IDH-wildtype GBM, grade 4 [17]. IDH-wildtype astrocytoma with pTERT mutation exhibited a worse prognosis similar to IDH-wildtype GBM, even when these tumors did not show the typical radiological findings of histologically defined GBM [19]. The cIMPACT-NOW update 4 indicated that IDH-wildtype diffuse gliomas are more heterogeneous and complex, especially in pediatric and young adults [15, 20]. The cIMPACT-NOW update 5 improved the grading of IDH-mutant astrocytomas based on CDKN2A/B homozygous deletion [16]. Based on these updates, the grade of diffuse glioma can be determined by specific molecular alterations regardless of histological findings in some situations. Here we evaluated molecular alterations in 300 diffuse glioma cases and summarized these molecular characteristics to determine the future direction of practical molecular testing algorism for the next WHO classification.
Materials and methods
Tumor samples
Tumor samples were obtained from consecutive 300 patients diagnosed with diffuse glioma, who were initially treated at Kyushu University Hospital between 2002 and 2019. Tumor tissues were saved for histopathological examination, and also snap-frozen in liquid nitrogen and stored at − 80 °C. Tumors were histologically diagnosed by two expert neuropathologists (SOS, TI).
The tumor DNA and corresponding constitutional DNA from peripheral blood leukocytes were extracted using the QIAamp DNA Mini Kit and DNA Blood Kit (Qiagen Science, Germantown, MD, USA), respectively. This study was approved by the ethics committee of Kyushu University.
Evaluation of 1p/19q codeletion and chromosome10 loss
Loss of heterozygosity (LOH) on chromosomes 1p, 19q and 10 was detected by microsatellite analysis of blood and tumor DNA. We designed 20 microsatellite makers for covering the chromosome 1p, 10 and 19q13 regions as follow: D1S2667, D1S2647, D1S2734 (located on 1p36), D1S2797 (1p32), D1S2766, D1S435 (1p22); D10S537, D10S1649 (10p15), D10S213 (10p11), D10S196 (10q11), D10S1652 (10q21), D10S537 (10q22), D10S1765 (10q23, near PTEN region), D10S587, D10S216, D10S1655 (10q26); D19S420, D19S219, D19S921 (19q13.3), D19S418 (19q13.4). PCR and fluorescence labeling were performed according to previously described methods [21, 22]. Capillary electrophoresis was performed using 310 or 3730 Prism Genetic Analyzer (Applied Biosystems). Raw electrophoresis data were analyzed with GeneMapper analysis software (Applied Biosystems). Allelic status was assessed based on criteria established in a previous study [21].
Evaluation of IDH1/2, H3F3A, and pTERT mutation
The main driver genes (IDH1/2, H3F3A) were evaluated by high-resolution melting (HRM) analysis using DNA extracted from the frozen tissue as previously described [23]. TERT promoter mutations were retrospectively analyzed by direct sequencing, because it is difficult to detect these mutations due to the large cytosine-phosphate-guanine island promoter region [24].
Statistical analysis
Progression free survival (PFS) and overall survival (OS) were estimated by the Kaplan–Meier method. The log-rank test was used to compare the survival distribution of each molecular subgroup. The statistical analysis was performed using JMP 16.0 (SAS Institute, Cary, NC, USA).
Results
A total of 169 glioblastomas (GBM), 13 diffuse midline gliomas (DMG), H3 K27M-mutant and 118 LGGs (WHO grade 2–3) were diagnosed according to 2016 WHO CNS classification [1]. Amongst the 169 GBM, 153 IDH-wildtype GBM and 9 IDH-mutant GBM cases were identified, while 4 cases were not fully analyzed for the WHO CNS 2016 criteria and diagnosed as GBM-“not otherwise specified (NOS)”. Among the 153 IDH-wildtype GBM cases, 3 pediatric cases showed H3.3 G34R mutation. Of the 118 LGG, 33 cases showed IDH wildtype, including 13 diffuse astrocytomas and 20 anaplastic astrocytomas. Eighty-one cases showed IDH mutation including 22 diffuse astrocytoma, 22 anaplastic astrocytoma, 18 oligodendroglioma 1p19q codeleted, 19 anaplastic oligodendroglioma 1p19q codeleted, 2 oligodendroglioma NOS, 1 anaplastic oligodendroglioma NOS. Only one diffuse astrocytoma was diagnosed as “diffuse astrocytoma NOS” due to incomplete molecular testing. Among the 182 WHO grade 4 glioma patients, 14 were under 18 years old and 121 were over 55 years old. Among the 118 patients with LGG, 6 were under 18 years old and 27 were over 55 years old, while the remaining 85 cases ranged between 19 and 54 years old. The youngest patient with IDH mutation is 19 years old; thus all of the patients under the age of 18 were IDH-wildtype glioma. Among the 33 IDH-wildtype LGGs, 14 cases are over 55 years old. All IDH-mutant diffuse gliomas over 55 years old showed R132H IDH1 mutation, which is consistent with a previous report [25]. Among the IDH-mutant diffuse gliomas, the age distribution of 1p/19q-codeleted oligodendroglioma is higher than that of astrocytoma regardless WHO grading. On the other hand, IDH-wildtype anaplastic astrocytoma (grade 3) showed higher age distribution compared with IDH-wildtype diffuse astrocytoma (grade 2) (Table 1).
pTERT mutations were evaluated retrospectively for 278 of the cases. pTERT mutation was common molecular alteration among IDH-wildtype GBM and 1p/19q-codeleted oligodendrogliomas, 85/145 (58.6%) and 35/36 (97.2%) respectively. Within IDH-wildtype astrocytoma, four out of 13 diffuse astrocytomas and seven out of 18 anaplastic astrocytomas showed this mutation. Among IDH-mutant astrocytic tumors, one IDH-mutant GBM and one IDH-mutant anaplastic astrocytoma showed pTERT mutation. On the other hand, all of the 3 cases with oligodendroglioma/anaplastic oligodendroglioma-NOS showed pTERT mutation; however, the “not elsewhere classified (NEC)” diagnoses would apply according to cIMPACT-NOW update1 at present [12].
Survival analysis was performed for 101 adult LGGs including 35 1p/19q-codeleted oligodendroglioma, 43 IDH-mutant astrocytoma, 11 IDH-wildtype/pTERT-mutant astrocytoma and 12 IDH-wildtype/pTERT-wildtype astrocytoma. The median PFS for 1p/19q-codeleted oligodendroglioma, IDH-mutant astrocytoma, IDH-wildtype/pTERT-mutant astrocytoma and IDH-wildtype/pTERT-wildtype astrocytoma are 112, 36.6, 11.8, and 77.4 months, respectively. The median OS for IDH-mutant astrocytoma and IDH-wildtype/pTERT-mutant astrocytoma are 82 and 36.6 months, respectively. The median OS was not reached for the other two subtypes.
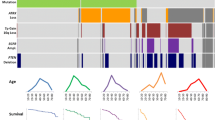
Survival analysis revealed that the most favorable outcome was with 1p/19q-codeleted oligodendrogliomas. Notably, both PFS and OS of IDH-wildtype LGGs were separated based on their pTERT mutation (Fig. 1).
Furthermore, three IDH-wildtype LGG cases underwent repeat surgery, and more than two samples of each case are analyzed longitudinally. Notably, the longitudinal analysis revealed that the IDH-wildtype LGG case with pTERT mutation gradually extended ch10 loss and finally showed total ch10 loss (Fig. 2). In contrast, the two cases without pTERT mutations never showed additional genetic alteration of the three parameters in repeat surgery.
A 44-year-old male diagnosed as Molecular GBM: The patient underwent four surgeries. MRI showed a non-enhanced mass in the right temporal lobe. First diagnosis is a diffuse astrocytoma, grade 2 without any LOH region (a). After repeat surgeries and adjuvant chemo-radiotherapy, MRI showed a heterogeneous enhanced mass. Final diagnosis is glioblastoma, grade 4 with ch10 loss (b). PFS and OS of this case are 4.0 and 26.6 months, respectively
Discussion
IDH1/2 mutation is now considered an early genetic event for gliomagenesis, and a critical genetic marker for diffuse glioma stratification [2, 3]. Combined with 1p/19q codeletion, diffuse glioma is classified into three major subgroups (IDH-mutant astrocytoma, IDH-mutant and 1p/19q-codeleted oligodendroglioma, and IDH-wildtype astrocytoma). IDH-wildtype GBM and 1p/19q-codeleted oligodendroglioma are common genotype well characterized by several previous clinical studies [8, 9, 26, 27].
Since 2002, we selected upfront chemotherapy and repeat surgeries for patients with 1p/19q-codeleted oligodendroglioma to prevent cognitive dysfunction [10, 11]. Precise detection of 1p/19q total loss caused by unbalanced translocation is crucial for selecting the less intensive treatments [28,29,30]. The cIMPACT-NOW update 2 proposed that histological astrocytic findings and alpha-thalassemia/mental retardation Syndrome X-linked helicase (ATRX)/p53 immunohistochemical results were sufficient for the diagnosis of “Astrocytoma, IDH-mutant” without 1p/19q molecular testing [13]. In our institution, however, the upfront chemotherapy has been selected only for the patients with 1p/19q codeletion confirmed by molecular analysis. Combined with the IDH1/2 mutation, we can detect this molecular subgroup more precisely, because some IDH-wildtype GBM showed apparent 1p/19q codeletion as the part of chromosomal alterations [31]. pTERT mutation is another important molecular marker for this subgroup. In this study, all 1p/19q-codeleted oligodendroglioma except one case showed pTERT mutation and favorable clinical course. However, the patient with 1p/19q-codeleted oligodendroglioma without pTERT mutation showed a relatively worse prognosis, while three patients with 1p/19q-intact oligodendroglioma with pTERT mutation showed better prognosis. A recent report also emphasized the implication of pTERT mutation regardless of 1p/19q status in IDH-mutant LGGs [32]. Further molecular estimation is needed for the case showing a discrepancy between 1p/19q codeletion and pTERT mutation.
Within LGGs, IDH-wildtype astrocytoma is a relatively small subgroup that is considered to be a more aggressive genotype compared with IDH-mutant LGGs. Recent reports have revealed that IDH-wildtype LGG is a more heterogeneous subgroup, and all of these patients do not show a dismal prognosis [7, 27, 33, 34]. Further stratification is required based on genetic alterations for IDH-wildtype LGG because pediatric-type diffuse gliomas demonstrate complex molecular alterations [15, 35,36,37]. In particular, for tiny biopsy specimens, appropriate genetic testing is mandatory for accurate diagnosis.
At our institution, we routinely evaluate the genetic alterations of IDH1/2, BRAF, H3F3A and pTERT, adding to LOH status of chromosomes 1p, 19q and 10 [22, 23, 31]. Using these molecular analyses, diffuse gliomas are diagnosed based on the 2016 CNS WHO classification. Combined with histological diagnosis, we identified 33 cases of IDH-wildtype LGG. According to the cIMPACT-NOW update 3, we detected 11 IDH-wildtype astrocytoma with molecular features of glioblastoma, WHO grade 4. Considering the molecular test algorithm for molecular GBM, the pivotal molecular parameter is pTERT mutation, which was the most sensitive for detecting this subtype [38]. More than 60% of molecularly diagnosed GBM cases can be identified from IDH-wildtype LGG by pTERT mutation analysis alone [19].
In this cohort, one case with WHO grade 2 diffuse astrocytoma suffered several recurrences, and finally became histologically diagnosed as GBM with whole ch10 loss (Fig. 2). Retrospective analysis revealed that pTERT mutation occurred in the initial tumor of this case. In contrast, two cases with IDH-wildtype LGG, which underwent repeat surgeries for recurrent lesion, did not show further genetic alterations of the three molecular markers (pTERT mutation, EGFR amplification, + 7/ − 10), when pTERT mutation was not identified in the initial operation. Furthermore, due to the high frequency of pTERT mutation in molecular GBM, pTERT mutation can be considered an earlier genetic event compared with the others (EGFRamplification, + 7/ − 10). Several reports also support that pTERT mutation precedes + 7/ − 10 in the molecular evolution of IDH-wildtype LGG [39,40,41]. In contrast, a recent report revealed that pTERT mutation was associated with the rapid tumor growth of IDH-wildtype GBM, while one or more chromosomal alterations (+ 7/ − 9p/ − 10) were required for the tumor initiation [42]. Nevertheless, pTERT mutation, similar to H3 K27M and G34R/V mutations, is considered to play an important role in the early stage of gliomagenesis [40, 43].
Diffuse glioma is well recognized as the tumor showing marked spatio-temporal heterogeneity. Recent longitudinal studies demonstrated the molecular evolution of diffuse glioma during disease progression [41, 42, 44]. Furthermore, the diversity of genetic/epigenetic states remains unclear due to the marked intratumoral heterogeneity [45, 46]. In particular, molecular heterogeneity is becoming more complicated for recurrent gliomas under therapy [47]. In the near future, heterogeneous molecular alterations will be accelerated under molecular target therapy such as tyrosine kinase inhibitors [41, 48]. More precise genetic/epigenetic characterization is required to overcome the marked spatio-temporal heterogeneity of diffuse glioma in the era of cancer genome medicine.
Recently the Japan Society of Brain Tumor Pathology proposed three levels of diagnoses for diffuse astrocytic and oligodendroglial tumors [49]. Especially for the subgroup of LGG, level 3A/B analysis (1p/19q codeletion and IDH1/2 mutation) is required for the precise diagnosis. In our institution, level 3A/B molecular analysis was applied as advanced medical care for diffuse glioma cases. After IDH1/2 wildtype is defined, the tumors with H3F3A or BRAF mutant should be excluded for the further analysis of molecularly GBM. The next step was to evaluate pTERT mutation, the most sensitive molecular marker for molecular GBM. For IDH-wildtype LGG without pTERT mutation, evaluation of EGFR amplification is required for a precise diagnosis. EGFR amplification is the most specific parameter within these three markers; however, its sensitivity is relatively low [38]. We planned to apply multiplex ligation-dependent probe amplification (MLPA) kit P105 (MRC-Holland, Amsterdam, The Netherlands) for detecting EGFR amplification, thus, we can detect the copy number of this region of ch7p and EGFR variant III simultaneously [50]. Evaluation of whole ch7 gain is needed for the rare cases showing pTERT wildtype, EGFR gain and whole ch10 loss. Furthermore, this MLPA kit can also detect CDKN2A homozygous deletion, which is a critical molecular marker for “Astrocytoma, IDH-mutant, grade 4” [16]. This step-by-step diagnostic procedure is recommended for daily routine diagnostics of diffuse gliomas, not only for molecular GBM. Based on our results, the test algorism following the level 3A/B analysis is proposed in Fig. 3. Future treatment strategies should be considered based on precise molecular analysis.
References
Louis DN, Ohgaki H, Wiestler OD et al (2016) WHO Classification of Tumours of the central nervous system. International Agency for Research on Cancer (IARC), Lyon
Yan H, Parsons DW, Jin G et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773. https://doi.org/10.1056/NEJMoa0808710
Cancer Genome Atlas Research N, Brat DJ, Verhaak RG et al (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498. https://doi.org/10.1056/NEJMoa1402121
Komori T (2020) Updating the grading criteria for adult diffuse gliomas: beyond the WHO2016CNS classification. Brain Tumor Pathol 37:1–4. https://doi.org/10.1007/s10014-020-00358-y
Cairncross JG, Ueki K, Zlatescu MC et al (1998) Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 90:1473–1479
Jiang H, Ren X, Cui X et al (2013) 1p/19q codeletion and IDH1/2 mutation identified a subtype of anaplastic oligoastrocytomas with prognosis as favorable as anaplastic oligodendrogliomas. Neuro Oncol 15:775–782. https://doi.org/10.1093/neuonc/not027
Aibaidula A, Chan AK-Y, Shi Z et al (2017) Adult IDH wild-type lower-grade gliomas should be further stratified. Neuro Oncol 19:1327–1337. https://doi.org/10.1093/neuonc/nox078
Cairncross G, Wang M, Shaw E et al (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 31:337–343. https://doi.org/10.1200/JCO.2012.43.2674
van den Bent MJ, Brandes AA, Taphoorn MJ et al (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 31:344–350. https://doi.org/10.1200/JCO.2012.43.2229
Hata N, Yoshimoto K, Hatae R et al (2016) Deferred radiotherapy and upfront procarbazine-ACNU-vincristine administration for 1p19q codeleted oligodendroglial tumors are associated with favorable outcome without compromising patient performance, regardless of WHO grade. Onco Targets Ther 9:7123–7131. https://doi.org/10.2147/OTT.S115911
Kuga D, Hata N, Akagi Y et al (2018) The effectiveness of salvage treatments for recurrent lesions of oligodendrogliomas previously treated with upfront chemotherapy. World Neurosurg 114:e735–e742. https://doi.org/10.1016/j.wneu.2018.03.069
Louis DN, Wesseling P, Paulus W et al (2018) cIMPACT-now update 1: not otherwise specified (NOS) and not elsewhere classified (NEC). Acta Neuropathol 135:481–484. https://doi.org/10.1007/s00401-018-1808-0
Louis DN, Giannini C, Capper D et al (2018) cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol 135:639–642. https://doi.org/10.1007/s00401-018-1826-y
Brat DJ, Aldape K, Colman H et al (2018) cIMPACT-NOW update 3: recommended diagnostic criteria for “diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV.” Acta Neuropathol 136:805–810. https://doi.org/10.1007/s00401-018-1913-0
Ellison DW, Hawkins C, Jones DTW et al (2019) cIMPACT-NOW update 4: diffuse gliomas characterized by MYB, MYBL1, or FGFR1 alterations or BRAF (V600E) mutation. Acta Neuropathol 137:683–687. https://doi.org/10.1007/s00401-019-01987-0
Brat DJ, Aldape K, Colman H et al (2020) cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol 139:603–608. https://doi.org/10.1007/s00401-020-02127-9
Louis DN, Wesseling P, Aldape K et al (2020) cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol 30:844–856. https://doi.org/10.1111/bpa.12832
Ellison DW, Aldape KD, Capper D et al (2020) cIMPACT-NOW update 7: advancing the molecular classification of ependymal tumors. Brain Pathol. https://doi.org/10.1111/bpa.12866
Tesileanu CMS, Dirven L, Wijnenga MMJ et al (2020) Survival of diffuse astrocytic glioma, IDH1/2 wildtype, with molecular features of glioblastoma, WHO grade IV: a confirmation of the cIMPACT-NOW criteria. Neuro Oncol 22:515–523. https://doi.org/10.1093/neuonc/noz200
Komori T (2021) The molecular framework of pediatric-type diffuse gliomas: shifting toward the revision of the WHO classification of tumors of the central nervous system. Brain Tumor Pathol 38:1–3. https://doi.org/10.1007/s10014-020-00392-w
Yoshimoto K, Iwaki T, Inamura T et al (2002) Multiplexed analysis of post-PCR fluorescence-labeled microsatellite alleles and statistical evaluation of their imbalance in brain tumors. Jpn J Cancer Res 93:284–290
Akagi Y, Yoshimoto K, Hata N et al (2018) Reclassification of 400 consecutive glioma cases based on the revised 2016 WHO classification. Brain Tumor Pathol 35:81–89. https://doi.org/10.1007/s10014-018-0313-4
Hatae R, Hata N, Yoshimoto K et al (2016) Precise detection of IDH1/2 and BRAF hotspot mutations in clinical glioma tissues by a differential calculus analysis of high-resolution melting data. PLoS ONE 11:e0160489. https://doi.org/10.1371/journal.pone.0160489
Hatae R, Hata N, Suzuki SO et al (2017) A comprehensive analysis identifies BRAF hotspot mutations associated with gliomas with peculiar epithelial morphology. Neuropathology 37:191–199. https://doi.org/10.1111/neup.12347
Chen L, Voronovich Z, Clark K et al (2014) Predicting the likelihood of an isocitrate dehydrogenase 1 or 2 mutation in diagnoses of infiltrative glioma. Neuro Oncol 16:1478–1483. https://doi.org/10.1093/neuonc/nou097
Ramirez C, Bowman C, Maurage CA et al (2010) Loss of 1p, 19q, and 10q heterozygosity prospectively predicts prognosis of oligodendroglial tumors–towards individualized tumor treatment? Neuro Oncol 12:490–499. https://doi.org/10.1093/neuonc/nop071
Eckel-Passow JE, Lachance DH, Molinaro AM et al (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508. https://doi.org/10.1056/NEJMoa1407279
Mizoguchi M, Kuga D, Guan Y et al (2011) Loss of heterozygosity analysis in malignant gliomas. Brain Tumor Pathol 28:191–196. https://doi.org/10.1007/s10014-011-0038-0
Jenkins RB, Blair H, Ballman KV et al (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Can Res 66:9852–9861. https://doi.org/10.1158/0008-5472.can-06-1796
Griffin CA, Burger P, Morsberger L et al (2006) Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol 65:988–994. https://doi.org/10.1097/01.jnen.0000235122.98052.8f
Mizoguchi M, Yoshimoto K, Ma X et al (2012) Molecular characteristics of glioblastoma with 1p/19q co-deletion. Brain Tumor Pathol 29:148–153. https://doi.org/10.1007/s10014-012-0107-z
Arita H, Matsushita Y, Machida R et al (2020) TERT promoter mutation confers favorable prognosis regardless of 1p/19q status in adult diffuse gliomas with IDH1/2 mutations. Acta Neuropathol Commun 8:201. https://doi.org/10.1186/s40478-020-01078-2
Aoki K, Nakamura H, Suzuki H et al (2018) Prognostic relevance of genetic alterations in diffuse lower-grade gliomas. Neuro Oncol 20:66–77. https://doi.org/10.1093/neuonc/nox132
Metellus P, Coulibaly B, Colin C et al (2010) Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol 120:719–729. https://doi.org/10.1007/s00401-010-0777-8
Qaddoumi I, Orisme W, Wen J et al (2016) Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol 131:833–845. https://doi.org/10.1007/s00401-016-1539-z
Ryall S, Zapotocky M, Fukuoka K et al (2020) Integrated molecular and clinical analysis of 1,000 pediatric low-grade gliomas. Cancer Cell 37:569-583.e565. https://doi.org/10.1016/j.ccell.2020.03.011
Zhang J, Wu G, Miller CP et al (2013) Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45:602–612. https://doi.org/10.1038/ng.2611
Stichel D, Ebrahimi A, Reuss D et al (2018) Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol 136:793–803. https://doi.org/10.1007/s00401-018-1905-0
Killela PJ, Reitman ZJ, Jiao Y et al (2013) TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci USA 110:6021–6026. https://doi.org/10.1073/pnas.1303607110
Barthel FP, Wesseling P, Verhaak RGW (2018) Reconstructing the molecular life history of gliomas. Acta Neuropathol 135:649–670. https://doi.org/10.1007/s00401-018-1842-y
Jonsson P, Lin AL, Young RJ et al (2019) Genomic correlates of disease progression and treatment response in prospectively characterized gliomas. Clin Cancer Res 25:5537–5547. https://doi.org/10.1158/1078-0432.CCR-19-0032
Korber V, Yang J, Barah P et al (2019) Evolutionary trajectories of IDH (WT) glioblastomas reveal a common path of early tumorigenesis instigated years ahead of initial diagnosis. Cancer Cell 35(692–704):e612. https://doi.org/10.1016/j.ccell.2019.02.007
Castel D, Philippe C, Calmon R et al (2015) Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol 130:815–827. https://doi.org/10.1007/s00401-015-1478-0
Barthel FP, Johnson KC, Varn FS et al (2019) Longitudinal molecular trajectories of diffuse glioma in adults. Nature 576:112–120. https://doi.org/10.1038/s41586-019-1775-1
Patel AP, Tirosh I, Trombetta JJ et al (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401. https://doi.org/10.1126/science.1254257
Suzuki H, Aoki K, Chiba K et al (2015) Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47:458–468. https://doi.org/10.1038/ng.3273
Wang J, Cazzato E, Ladewig E et al (2016) Clonal evolution of glioblastoma under therapy. Nat Genet 48:768–776. https://doi.org/10.1038/ng.3590
Furnari FB, Cloughesy TF, Cavenee WK et al (2015) Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer 15:302–310. https://doi.org/10.1038/nrc3918
Sonoda Y, Yokoo H, Tanaka S et al (2019) Practical procedures for the integrated diagnosis of astrocytic and oligodendroglial tumors. Brain Tumor Pathol 36:56–62. https://doi.org/10.1007/s10014-019-00337-y
Jeuken J, Sijben A, Alenda C et al (2009) Robust detection of EGFR copy number changes and EGFR variant III: technical aspects and relevance for glioma diagnostics. Brain Pathol 19:661–671. https://doi.org/10.1111/j.1750-3639.2009.00320.x
Acknowledgements
This study was supported by the Japanese Society for the Promotion of Science Grants-in-Aid for Scientific Research (JSPS KAKENHI) Grant No. JP21H03044, JP21K09128, JP20K09392, JP20K17972, 19K17673, and Fujita Memorial Fund for Medical Research (GAKF800362) and Ichiro Kanehara Foundation for the Promotion of Medical Sciences and Medical Care(GAKF800381)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mizoguchi, M., Hata, N., Kuga, D. et al. Clinical implications of molecular analysis in diffuse glioma stratification. Brain Tumor Pathol 38, 210–217 (2021). https://doi.org/10.1007/s10014-021-00409-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10014-021-00409-y