Abstract
Catalytic cycle intermediates of a representative extradiol dioxygenase, homoprotocatechuate 2,3-dioxygenase (HPCD), have recently been characterized in crystallo by Kovaleva and Lipscomb. The structures of the identified species indicate that the process of inserting oxygen into the catechol ring occurs stepwise, and involves an Fe(II)-alkylperoxo intermediate and its O–O cleavage product: a gem diol species. In general, these findings corroborate the results of our previous computational studies; however, the fact that the gem diol species is stable enough to be observed in the crystal form seems to be at odds with the computational mechanistic data, which suggest that this intermediate should very readily and spontaneously convert to the epoxide species. The key question then becomes what is actually observed in the X-ray experiments. Here we report additional computational studies undertaken with the hope of clarifying this issue. The results obtained for active site models hosting both the native and the alternative (4-sulfonylcatechol) substrate indicate that the stability of the gem diol species is substantially increased if an electron and a proton are added. If this occurs somehow, the lifetime of the intermediate should be sufficient to observe it.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Extradiol dioxygenases are mononuclear metalloenzymes that host a single Fe(II) or Mn(II) ion at their active sites. The inorganic cofactor is coordinated by three protein ligands, forming the 2-His-1-carboxylate facial triad motif, whereas the three remaining coordination sites are used by the catecholic and O2 substrates. The catalytic reaction involves oxidative cleavage of the C–C bond adjacent to the diol group of the catecholic substrate, and leads to 2-hydroxymuconaldehyde acids.
Our previous computational studies of Fe- and Mn-dependent extradiol dioxygenases [1–3], and a related homogentisate dioxygenase [4], suggest that the catalytic reaction proceeds through the bidentate coordination of catecholic substrate to the metal ion and the subsequent trapping of dioxygen, which leads to a species with a peroxide bridge between the metal ion and the catecholic ring (species a in Fig. 1). In the subsequent step, the O–O bond is cleaved homolytically, yielding an alkoxyl radical species (b). This is predicted to be a short-lived intermediate that easily and spontaneously converts to the epoxide species (c). The exothermicity of this step (b → c) exceeds 10 kcal/mol. The final chemical steps of the catalytic reaction are the expansion of the oxirane ring via C–C bond cleavage (c → d) and then hydrolysis of the cyclic lactone.
Recently, Kovaleva and Lipscomb have characterized the crystal structures of several oxygenated intermediates of homoprotocatechuate 2,3-dioxygenase (HPCD) [5, 6]. In the first study, a slow alternative substrate (4-nitrocatechol) was used, and this allowed three intermediates to be trapped, including the alkylperoxo species with a peroxo bridge between the substrate molecule and iron (species a in Fig. 1) [5]. The next intermediate was trapped and characterized through the use of another alternative substrate, 4-sulfonylcatechol (4SC), and by introducing a single point mutation that alters the crystal packing (Glu323Leu) [6]. This species has a cleaved O–O bond and the structure of a gem diol (Fig. 2). Based on computational studies, it was previously proposed that such an intermediate participates in the catalytic cycle. An oxygen on the ring of this intermediate was found to have radical character; see species b in Fig. 1. However, the stability of this intermediate, which allows for successful X-ray structural determination, seems to be in conflict with the energetic data derived from the computations. More specifically, in the previous work devoted to the mechanism of extradiol dioxygenases, the calculated barrier to and exothermicity of the conversion of the gem diol intermediate into the epoxide species (b → c in Fig. 1) are around 0 and 13 kcal/mol, respectively [1, 3, 4]. Thus, this particular step of the catalytic cycle should proceed efficiently and practically irreversibly, which, in turn, should make it impossible to observe the gem diol intermediate.
Kovaleva and Lipscomb proposed three plausible reasons for this apparent discrepancy [6]: (1) at the stage of the gem diol intermediate, the active site provides a somewhat different environment than it does in the enzyme–substrate complex, whose structure was used as a starting point in the previous calculations; (2) 4SC has a different reactivity than homoprotocatechuate (HPCA), which was the substrate in the previous calculations; and (3) crystal packing forces, which were not accounted for in the computations, are important for stabilizing this gem diol intermediate. As well as these three suggestions, we can of course add that the method or model used in the previous calculations could have a very much larger error than usual. These possibilities, and others, will be discussed below.
The active site models used in this study are based on the crystal structure of the enzyme–gem diol intermediate complex (PDB code: 3ECK), and thus the first plausible reason given above can be eliminated. Moreover, the computations were performed for both native (HPCA) and alternative (4SC) substrates, and by comparing the results it is possible to estimate the extent to which 4SC differs in reactivity from HPCA (see point 2 above).
The first question addressed was: is it possible that another, similar, species is observed in the experiments, rather than the radical intermediate obtained in the previous calculations? Computations were therefore first performed for that radical and for the subsequent epoxide species, and the results were compared to the ones obtained by adding either one electron, or one electron and one proton, or two electrons and one proton to the radical. The radical gem diol and epoxide species are the ones encountered in the native catalytic cycle. In the radical +e− form, one additional electron has reduced the alkoxyl radical to an alkoxyl anion. In the radical +e− +H+ variant, the alkoxyl anion was protonated to form a charge-neutral hydroxyl group, whereas the radical +2e− +H+ species features an OH group and a high-spin Fe(II), as opposed to other forms where iron is present as the high-spin Fe(III) ion.
The results reported here were obtained with the hybrid B3LYP functional [7, 8] using the Jaguar [9] and Gaussian 03 [10] programs. The models and methods employed in this study are described in depth in “Computational details.”
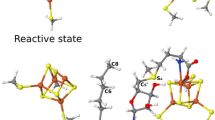
The calculated energy difference between the epoxide and the gem diol species and the distance between the two atoms derived from dioxygen are reported in Table 1 for both the native and the alternative (4-sulfonylcatechol) substrates. The optimized structures of the radical +e− +H+ forms of the gem diol and epoxide species for the 4-sulfonylcatechol substrate are presented in Fig. 3.
The results presented in Table 1 show that there are no big differences between active sites hosting 4SC or HPCA concerning the relative stabilities of the epoxide and gem diol species. For both substrates in the radical and radical +e− forms, it is the epoxide intermediate which is more stable; for the radical +e− +H+ and radical +2e− +H+ variants, the gem diol species has a lower energy than the epoxide. This means that one of the suggestions made in the experimental paper for why the gem diol species is observed for 4SC only is not supported by the calculations.
In the crystal structure of the gem diol, the distance between the two atoms derived from dioxygen is 2.68 Å, which may suggest that a hydrogen bond is present between these two oxygens. In the models for the gem diol in the radical +e− +H+ and radical +2e− +H+ forms, there is indeed such a hydrogen bond present, and the O–O distance ranges from 2.51 to 2.56 Å, which is somewhat shorter than the X-ray value; this is a typical discrepancy observed when using the present basis set.
Concerning the relative energies, the data gathered in Table 1 show that for the “native” and reduced forms obtained without protonating the alkoxyl oxygen (i.e., radical and radical +e−), the gem diol species is markedly less stable than the epoxide intermediate. The energy difference is larger than 10 kcal/mol, which is in line with the results obtained previously for the radical forms produced during the catalytic cycle [1, 3, 4]. Importantly, for the reduced forms that feature a neutral hydroxyl group (i.e., radical +e− +H+), the transformation of the gem diol into the epoxide species is an endothermic reaction. This is also true for the radical +2e− +H+ species for the natural substrate, but not for the 4-sulfonylcatechol substrate, which makes this double reduction less likely. As can be seen in Fig. 3, in this case the closure of the epoxide ring requires that a proton of the gem diolate hydroxide is shuttled to the hydroxide ligand bound to iron. For the radical +e− +H+ form, the gem diol species are more than 5 kcal/mol more stable than the epoxide. This energy difference is sufficient to make gem diol the prevalent species observed in the X-ray structure.
To check that the method employed is not biased towards the epoxide or the gem diol species, a benchmark computation was performed for a small monoprotonated model derived from catechol only. In the case of the standard DFT method, the computed energy difference is −0.5 kcal/mol, whereas the very accurate G2 scheme [11] yields +0.2 kcal/mol. This good numerical agreement shows that a big error in the DFT method is quite unlikely in the present case.
These computational results suggest that the first two possibilities proposed in the experimental paper to rationalize the discrepancy between theory and experiment are both unlikely. A simple structural argument can be used to counter the third suggestion: the transformation of the gem diol into the epoxide intermediate requires only a rather small change in geometry of the substrate molecule and the active site residues hardly interfere with this process. Moreover, in the present calculations, the backbone atoms were frozen from the X-ray structure where the intermediate was observed, and any strain effect from crystal packing should therefore be included in the computed results. It can be added that a large strain effect on the relative energies of these quite similar species (the gem diol and the epoxide) of more than 10 kcal/mol is required to yield enough stability for the gem diol. Such a large effect has never been observed in model calculations for a case like this.
The above calculations have thus answered the main questions raised in the present study, and the most likely answer is that somehow a reduction and protonation has occurred. The question of how this has occurred is probably best answered by expert experimentalists. X-ray reduction is a common phenomenon in X-ray structural studies [14]. Another tentative suggestion is that the gem diol species is obtained in the radical +e− +H+ form by first forming a hydrogen peroxide that binds to iron. Such a reduced and protonated peroxo species is calculated to be markedly less stable than the gem diol for 4SC and HPCA; it lies 37.6 and 34.2 kcal/mol higher in energy, respectively. Thus, for the radical +e− +H+ redox form, the gem diol is more stable than both the preceding and subsequent intermediate, which could lead to its observation in the X-ray experiment.
Another possibility could be that the reduced species is formed by first oxidizing the diol substrate to an orthoquinone, and then hydrating one of the carbonyls. This is a process commonly observed for biomimetic species [15]. One problem with this explanation is that it is difficult to see why this only happens for the 4SC substrate, not for the native HPCD one.
In summary, based on the computed energy difference between the gem diol and epoxide species, which has been calculated for various redox forms, we propose that the experimentally observed gem diol intermediate is obtained by reducing and protonating some intermediate of the “native” catalytic cycle. The observed species should not therefore be an intermediate of that cycle itself, at least not according to any mechanism suggested so far.
Computational details
The models of the homoprotocatechuate 2,3-dioxygenase active site are based on the crystal structure solved for the (mutated) enzyme complexed with 4-sulfonylcatechol (PDB code: 3ECK, chain C). Two models were used: one with 4-sulfonylcatechol, and the other with the native substrate: homoprotocatechuate. Besides the substrate, the models included four histidine side chains (His155, His200, His214 and His248), modeled with imidazoles; Glu267 and Asn157, represented by acetate and acetamide, respectively; and Tyr257 and Trp192, truncated to water and indol.
Restrained optimizations were performed with several terminal atoms (marked with asterisks in the first coordinate list given in the “Electronic supplementary material”) constrained to their positions in the crystal structure. In this way, the rigidity of the protein backbone was taken into account in the model.
Calculations were performed with two programs, Gaussian 03 [10] and Jaguar [9], and they employed hybrid DFT with the B3LYP exchange-correlation functional [7, 8], whereas in the benchmark computations for a small model the G2 extrapolation scheme was also used [11]. Geometric optimizations were done with a valence double-zeta basis set coupled with an effective core potential describing the innermost electrons in iron. This particular basis set is labeled “lacvp” in Jaguar. For the optimized structures, the electronic energy was computed with a bigger basis set of triple-zeta quality with polarization functions on all atoms except iron (lacv3p + for iron and cc-pVTZ(-f) for other atoms). The solvent corrections were calculated with the self-consistent reaction field method implemented in Jaguar [12, 13]. Two dielectric constants of 4 and 15 together with a probe radius of 1.4 Å were used to model the protein surrounding the active site. The smaller value corresponds to buried and hydrophobic active site surroundings, whereas the larger value corresponds to a more solvent-exposed site. Visual inspection of the protein structure reveals that the active site is buried, with none of the amino acid side chains depicted in Fig. 2 being exposed to solvent. This observation is corroborated by the results of solvent-accessible surface area calculations performed with the program Surface Racer 5 [16]. For seven residues, the calculated solvent-accessible surface area is zero; only for one, namely His248, is it 0.5 Å2 (Richards van der Waals radii together with a probe radius of 1.4 Å were used in this calculation). Thus, the results obtained with a low dielectric constant should be reliable.
The electronic state of the radical form is a spin quintet with the high-spin Fe(III) antiferromagnetically coupled with the organic radical. In the radical +e− form, the high-spin configuration on Fe(III) results in a sextet ground state; the spin quartet is around 15 kcal/mol higher in energy. Similarly, for the radical +e− +H+ form, the spin sextet is the ground state, with the quartet being at least 7 kcal/mol less stable. Finally, for the radical +2e− +H+ form, the ground state is a spin quintet with high-spin Fe(II).
References
Siegbahn PEM, Haeffner F (2004) Mechanism for catechol ring-cleavage by non-heme iron extradiol dioxygenases. J Am Chem Soc 126:8919–8932
Georgiev V, Borowski T, Siegbahn PEM (2006) Theoretical study of the catalytic reaction mechanism of MndD. J Biol Inorg Chem 11:571–585
Georgiev V, Borowski T, Blomberg MR, Siegbahn PEM (2008) A comparison of the reaction mechanisms of iron- and manganese-containing 2,3-HPCD: an important spin transition for manganese. J Biol Inorg Chem 13:929–940
Borowski T, Georgiev V, Siegbahn PEM (2005) Catalytic reaction mechanism of homogentisate dioxygenase: a hybrid DFT study. J Am Chem Soc 127:17303–17314
Kovaleva EG, Lipscomb JD (2007) Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates. Science 316:453–457
Kovaleva EG, Lipscomb JD (2008) Intermediate in the O–O bond cleavage reaction of an extradiol dioxygenase. Biochemistry 47:11168–11170
Becke ADJ (1993) Density-functional thermochemistry. III. The role of exact exchange. Chem Phys 98:5648–5652
Lee C, Yang W, Parr RG (1988) Development of the Colle–Salvetti correlation energy formula into a functional of the electron density. Phys Rev B37:785–789
Schrödinger, Inc. (2007) Jaguar 7. Schrödinger, Inc., Portland
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 03, Revision E.01. Gaussian Inc., Pittsburgh
Curtiss L, Raghavachari K, Redfern P, Pople J (1997) Assessment of Gaussian-2 and density functional theories for computation of enthalpies of formation. J Chem Phys 106:1063–1079
Tannor DJ, Marten B, Murphy R, Friesner RA, Sitkoff D, Nicholls A, Ringnalda M, Goddard WA III, Honig B (1994) Accurate first principles calculation of molecular charge distributions and solvation energies from ab initio quantum mechanics and continuum dielectric theory. J Am Chem Soc 116:11875–11882
Marten B, Kim K, Cortis C, Friesner RA, Murphy R, Ringnalda M, Sitkoff D, Honig B (1996) New model for calculation of solvation free energies: correction of self-consistent reaction field continuum dielelectric theory for short-range hydrogen-bonding effects. J Phys Chem 100:11775–11788
Yano J, Kern J, Irrgang K-D, Latimer MJ, Bergmann U, Glatzel P, Pushkar Y, Biesiadka J, Loll B, Sauer K, Messinger J, Zouni A, Yachandra VK (2005) X-ray damage to the Mn4 Ca complex in single crystals of photosystem II: a case study for metalloprotein crystallography. Proc Natl Acad Sci USA 102:12047–12052
Yamahara R, Ogo S, Masuda H, Watanabe Y (2002) (Catecholato)iron(III) complexes: structural and functional models for the catechol-bound iron(III) form of catechol dioxygenases. J Inorg Biochem 88:284–294
Tsodikov OV, Record MT Jr, Sergeev YV (2002) A novel computer program for fast exact calculation of accessible and molecular surface areas and average surface curvature. J Comput Chem 23:600–609
Acknowledgments
We are grateful to Sven de Marothy for providing us with his XYZ-Viewer program, which was used to produce Fig. 3. T.B. acknowledges support from the Polish State Committee for Scientific Research (Grant N301 093036).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Cartesian coordinates and calculated energies for all structures. This material is available free of charge via the Internet at http://xxx.
Rights and permissions
About this article
Cite this article
Borowski, T., Georgiev, V. & Siegbahn, P.E.M. On the observation of a gem diol intermediate after O–O bond cleavage by extradiol dioxygenases. A hybrid DFT study. J Mol Model 16, 1673–1677 (2010). https://doi.org/10.1007/s00894-010-0652-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0652-5