Abstract
Bioremediation in hypersaline environments is particularly challenging since the microbes that tolerate such harsh environments and degrade pollutants are quite scarce. Haloarchaea, however, due to their inherent ability to grow at high salt concentrations, hold great promise for remediating the contaminated hypersaline sites. This study aimed to isolate and characterize novel haloarchaeal strains with potentials in hydrocarbon degradation. A haloarchaeal strain IM1011 was isolated from Changlu Tanggu saltern near Da Gang Oilfield in Tianjin (China) by enrichment culture in hypersaline medium containing hexadecane. It could degrade 57 ± 5.2% hexadecane (5 g/L) in the presence of 3.6 M NaCl at 37 °C within 24 days. To get further insights into the mechanisms of petroleum hydrocarbon degradation in haloarchaea, complete genome (3,778,989 bp) of IM1011 was sequenced. Phylogenetic analysis of 16S rRNA gene, RNA polymerase beta-subunit (rpoB’) gene and of the complete genome suggested IM1011 to be a new species in Halorientalis genus, and the name Halorientalis hydrocarbonoclasticus sp. nov., is proposed. Notably, with insights from the IM1011 genome sequence, the involvement of diverse alkane hydroxylase enzymes and an intact β-oxidation pathway in hexadecane biodegradation was predicted. This is the first hexadecane-degrading strain from Halorientalis genus, of which the genome sequence information would be helpful for further dissecting the hydrocarbon degradation by haloarchaea and for their application in bioremediation of oil-polluted hypersaline environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Petroleum hydrocarbon pollution in saline/hypersaline ecosystem is prevalent due to industrial activities and accidental spill out. Crude oil production and refining industries are major contributor to such pollution (Lefebvre and Moletta 2006; Castillo-Carvajal et al. 2014). Approximately ten barrels of saline water (containing salt in the range of 1–250 g/L) are generated for every barrel of oil produced (Cuadros-Orellana et al. 2006). The salinity of waste water produced by oil industry vary to a great extent from fresh water up to three times the salinity of seawater (Díaz et al. 2002). Salinity imposes a barrier in bioremediation of pollutants as non-halophilic microorganisms cannot effectively carry out normal metabolic activities under such harsh conditions. In addition, physicochemical techniques to remove salt are costly and cumbersome. Therefore, an effective alternative means is to use halophiles that are attuned to grow under high salt conditions (Margesin and Schinner 2001; Le Borgne et al. 2008; Fathepure 2014).
Among halophiles, eubacteria once were considered better suited for pollution mitigation for their versatile metabolic capability and tolerance to fluctuating salt concentrations as compared to archaea (Oren et al. 1992). However, halophilic archaea on account of their inherent requirement of high salt concentrations for growth are better equipped for the treatment of effluents of hyper-salinity (≥ 20%). Notably, hydrocarbon degradation at higher salinities is much difficult to achieve, as high salt concentration inhibits solubilization of hydrocarbons as well as oxygen availability. Ward and Brock (1978) have found that at high salinities, hexadecane degradation is impaired possibly due to decreased metabolic rate of microorganisms. Recent literature has found that the optimum NaCl concentration of 2 M for hydrocarbon biodegradation was lower than that of 3.5–4.5 M for growth of two Haloferax strains on n-octadecane as substrate, and a probable explanation for this may be due to lower oxygen solubility at high salt concentration (Al-Mailem et al. 2010).
Alkanes are main component of petroleum hydrocarbons (Wang and Shao 2013). Among initial studies, a halophilic hydrocarbon-degrading archaeon EH4 was found to degrade tetradecane, hexadecane, eicosane, and pristane at relatively high rates in the presence of 3.5 M NaCl (Bertrand et al. 1990). Al-Mailem et al. (2010) reported Haloferax spp., Halobacterium sp. and Halococcus sp. competent of crude oil and octadecane degradation under 3 M NaCl. Heptadecane degradation has been demonstrated by Haloarcula sp. and Haloferax spp. in the presence of 22.5% NaCl (Tapilatu et al. 2010). In a study on sabkhas in Kuwait, the haloarchaeal species isolated by the culture-dependent method belonged to the genera Haloferax and Halobacterium, while the denaturing gradient gel electrophoresis (DGGE) fingerprinting of the 16S rDNA amplicons from the same samples showed that the haloarchaeal phylotypes were affiliated with the genera Halorussus, Halomicrobium, and Halorientalis (Al-Mailem et al. 2014). Amongst few description of archaea able to degrade alkane, information on enzymatic machinery and pathways involved is not available.
In the present study, a halophilic archaeon Halorientalis hydrocarbonoclasticus was isolated from the mix of salt crystal and sediment of saltern in Tianjin, China. The strain was able to grow and utilize hexadecane as carbon source. This is the first report on member of Halorientalis genus capable of alkane degradation. In addition, considering the potential of this strain, we sequenced the whole genome and this is the first complete genome of hexadecane-degrading haloarchaea. Analysis of genome revealed genes of alkane hydroxylase enzymes possibly involved in alkane degradation. Availability of first genome of hexadecane-degrading haloarchaeon is important since it will provide an opportunity to better understand the alkane degradation mechanism under hypersaline conditions.
Materials and methods
Microorganism and culture conditions
For isolation of halophilic archaea capable of degrading aliphatic hydrocarbons, samples including saline water, salt crystals, sediments, and the mix of salt and sediment were collected from Changlu Tanggu saltern near Da Gang Oilfield in Tianjin (China). The basic medium used for isolation was mineral salt basal medium (MSM) (per liter, 160 or 210 g NaCl, 20 g MgCl2·6H2O, 24 g MgSO4·7H2O, 0.5 g CaCl2, 5 g KCl, 0.2 g NaHCO3, 0.6 g NaBr, 2 g NH4Cl, 0.2 g KH2PO4, 0.008 g NH4 +-Fe(III) citrate, 1 mL trace element solution SL-6, 3 mL vitamin 10 stock solution, 8 g piperazine-N,N′-bis[2-ethanesulfonic acid] (PIPES), pH 7.0. The compositions of trace element solution SL-6 and vitamin 10 stock solutions are described in halohandbook (http://www.haloarchaea.com/resources/halohandbook). When preparing the solid MSM, 1.5% purified agar (BBI Life Sciences Corporation, Shanghai, China) was added.
To isolate and culture hydrocarbon-degrading haloarchaea, 5 g/L hexadecane (GC purity 99.0%, Haltermann, Germany) was added into liquid MSM or spread using a spreader onto solid MSM plate. The samples were inoculated into liquid MSM with hexadecane as almost sole carbon source (additionally only 0.008 g/L citrate in MSM), and cultured for 3 weeks at 37 °C with shaking of 180 rpm. After enrichment the cultures were diluted and plated on the solid medium with hexadecane as carbon source. The strains were purified by plate streaking twice. One strain named IM1011 growing faster on solid medium was selected for further study. The single colonies of IM1011 could be visualized by the eye after being cultured for about 6–10 days. For preparation of mother culture for hexadecane degradation, two loopful of IM1011 culture grown on plate was inoculated in liquid enriched MSM medium supplemented with 10 g/L yeast extract (Oxoid, Hampshire, UK) and 7.5 g/L casamino acids (BD Difco, New Jersey, USA). Beside hexadecane, other aliphatic hydrocarbons such as dodecane (C12) and eicosane (C20) were also used as carbon source to culture the strain IM1011, which was later named Halorientalis hydrocarbonoclasticus strain IM1011 (= CGMCC 13754) for further study in this article.
Hexadecane biodegradation
Potential of H. hydrocarbonoclasticus IM1011 for hexadecane degradation was carried out by growing it in 50 mL MSM medium supplemented with hexadecane (5 g/L). Medium was seeded with 4% inoculums from mother culture prepared in enriched MSM medium (supplemented with yeast extract and casamino acids). Degradation experiment was set in triplicate flasks and incubated on orbital shaker, 180 rpm at 37 °C for 24 days. Control (without cells) was prepared using same method and incubated under similar conditions. After 24 days of incubation, hexadecane was extracted thrice with 10 mL each of hexane. Extracts were combined and after suitable dilution with hexane, 1 μL sample was analyzed using Agilent Technology 6820 Gas Chromatograph (Agilent Technologies, California, USA) equipped with a flame ionization detector (FID) and a DB-1 capillary column (15 m × 0.53 mm i.d. × 0.5 μm film thickness; J & W Scientific). Oven temperature was programmed at 40 °C for 1 min and increased 5 °C/min from 40 to 140 °C, temperature was further increased from 140 to 280 °C at 10 °C/min. Injector and detector temperature was set at 280 °C. Quantitative estimation of hexadecane degradation was done by comparing the peak area of test samples with that of control and calculating the decrease percentage.
DNA extraction, genome sequencing and annotation
The H. hydrocarbonoclasticus IM1011 was cultured for 7 days on the solid medium with hexadecane as carbon source. For genomic DNA extraction, cells collected from the plates were resuspended in MSM, transferred to a clean Eppendorf tube, and washed twice with MSM. The total DNA was extracted using QIAGEN Blood & Cell Culture DNA Kit (catalog no. 13343) (QIAGEN, Duesseldorf, Germany). The quality and quantity of genomic DNA were ascertained by running 0.75% agarose gel, as well as by NanoDrop 1000 spectrophotometer and Qubit fluorometer.
Genome sequencing, assembling, annotation and basic analysis were performed by Nextomics Biosciences, Wuhan, China. Briefly, after quality control, shearing, library construction and library size selection, genome DNA was sequenced using single molecule real-time (SMRT) technology on a PacBio RS II sequencer (Pacific Biosciences, Nextomics Biosciences Co., Ltd., Wuhan). Sequencing generated an overall of 586,042,960 bp, and 55,180 post-filter reads with a mean length of 10,620 bp. After removing adapter, error and low-quality nucleotides, the clean data were used for the following assembling. The Hierarchical Genome Assembly Processing 2.3.0 was used to assemble the genome (Miller et al. 2008; Chin et al. 2013). The reads were preassembled and corrected by BLASTR (Chaisson and Tesler 2012). Celera Assembler program was used for full assembly, which was corrected using Quiver software. Finally, a total of 3,778,989 bp clean data with 122× genome coverage was obtained.
Gene Locator and Interpolated Markov ModelER (Glimmer) version 3.02 software was used to predict coding sequences (CDSs) (Delcher et al. 1999). tRNAs and rRNA genes were recognized by tRNAscan-SE (Lowe and Eddy 1997) and RNAmmer databases (Lagesen et al. 2007). The gene ontology (GO) (Ashburner et al. 2000), Kyoto encyclopedia of genes and genomes (KEGG) (Kanehisa et al. 2004), non-redundant protein (NR) (Pruitt et al. 2012), InterPro (Jones et al. 2014), Swiss-Prot (Magrane and Consortium 2011), and cluster of orthologous groups (COG) (Tatusov et al. 2001) databases were exercised to annotate the predicted genes.
Bioinformatics analysis
The taxonomic relationship was analyzed by 16S rRNA gene sequence similarity in the EzTaxon database (http://www.ezbiocloud.net/) (Yoon et al. 2016). The phylogenetic tree was constructed using the software MEGA 6.0 (Tamura et al. 2013) with the neighbor-joining method (Saitou and Nei 1987). The topologies of the phylogenetic tree were assessed by bootstrap analysis (Felsenstein 1985) based on 1000 replications. Evolutionary relationship at the genomic level was analyzed based on average nucleotide identity (ANI) algorithm that mimics DNA–DNA hybridization using OrthoANI software (Lee et al. 2016).
Nucleotide sequence accession number
The complete genome sequence of Halorientalis hydrocarbonoclasticus IM1011 (= CGMCC 13754) was deposited in GenBank with accession number CP019067. The accession numbers of pHOS300 and pHOS100 plasmid sequences were CP019068 and CP019069, respectively.
Results and discussion
Isolation of a haloarchaeal strain growing on hexadecane
Bioremediation of saline/hypersaline samples are intricate because of the constraint of salinity. Halophiles are suitable microbes to overcome this constraint since they grow and carry out normal metabolism under saline conditions (Edbeib et al. 2016). Haloarchaea among halophiles are better suited for remediation of hypersaline environments for the reason that they endure and thrive at high salt concentrations. The present study was undertaken to isolate hydrocarbon-degrading haloarchaea for cleaning up oil pollution in hypersaline environments. A haloarchaeal strain IM1011 was isolated by hexadecane and salt enrichment culture from saline samples collected near Da Gang Oilfield in Tianjin. Strain IM1011 grew well on MSM plate with hexadecane as carbon source (Fig. 1a). Colonies on MSM plate were circular, red, opaque, smooth and convex (Fig. 1b). Scanning electron microscope was used to decipher the cell morphology of IM1011 cells grown in the presence of hexadecane. Cells were spherical in shape and occurred either singly or in cluster (Fig. 1c, d). Strain IM1011 was also able to grow on MSM plates supplemented with dodecane and eicosane, however, cell growth was much weaker than with hexadecane.
The growth, colony and cellular morphology of H. hydrocarbonoclasticus IM1011. The growth (a) and colony morphology (b) of H. hydrocarbonoclasticus IM1011 cultured on hexadecane plate. The scanning electron microscope photograph of cluster of cells (c) or a single cell (d) using field emission scanning electron microscopy (Hitachi SU8010, Japan). Scale bar 1 μm
Determination of hexadecane degradation
To identify whether strain IM1011 could degrade hexadecane, it was cultured in liquid medium with hexadecane as carbon source. Liquid medium culture of strain IM1011 was established in MSM containing 21% (w/v) NaCl and hexadecane (5 g/L) as carbon source. It was found to degrade hexadecane while growing on it as carbon source. Gas chromatography profile of hexadecane extracted from control and test samples demonstrated hydrocarbon biodegradation potential of IM1011 (Fig. 2). Hexadecane degradation estimation by gas chromatography revealed that 57 ± 5.2% hexadecane degradation was achieved after 24 days. In one of the earliest studies, strain EH4 (later classified as Haloarcula vallismortis) was found to degrade 66% hexadecane (0.5 g/L) in 30 days under 3.5 M NaCl milieu (Bertrand et al. 1990). Haloarcula sp. and Haloferax spp. have been found to degrade 32–95% heptadecane (0.5 g/L) in 30 days (Tapilatu et al. 2010). Hexadecane degradation of 57% in the present study is quite comparable to previous studies and is noteworthy considering that the degradation efficiency could be increased after optimization (Table 1). Indeed, the strain IM1011 stands out in the higher concentration of hydrocarbon used and relatively shorter time period required for degradation.
Comparison of residual hexadecane by gas chromatography. Hexadecane (the peak at 15.8 min) was extracted by hexane from medium with (IM1011) and without (control) inoculation of H. hydrocarbonoclasticus IM1011. Dodecane (the peak at 6.2 min) was used as internal standard. The medium used was MSM with hexadecane as carbon source. The cultures were incubated at 180 rpm, 37 °C for 24 days. The peaks time were confirmed by pure hexadecane and dodecane (data not shown)
Genome properties of strain IM1011
Strain IM1011 was selected for genome sequencing considering its ability to degrade hexadecane, as the availability of total genome is desirable to analyze the metabolic pathway of hexadecane degradation in haloarchaea. The complete genome sequence of IM1011 was of 3,778,989 bp with a GC content of 65.58%. The genome consists of one main chromosome (3,381,613 bp; GC content, 65.96%) and two megaplasmids pHOS300 (276,813 bp; GC content 61.19%) and pHOS100 (120,563 bp; GC content 65.17%) (Fig. 3). The genome information is summarized in Table 2. The genome encodes 3720 putative proteins, 75 tRNAs and two 16S–23S–5S rRNA operons. The genome information indicates that IM1011 belongs to Halorientalis genus (see below), and so far this is the first complete genome sequence determined for this genus, as previously only draft genome sequences are available for Halorientalis persicus strain IBRC-M 10043 and Halorientalis regularis strain IBRC-M 10760 in NCBI database. Draft genome sequence of H. persicus (4.87 Mb) has 4684 protein coding sequences with GC content of 63.5%, while that of H. regularis (4.03 Mb) has 3893 protein coding sequences with GC content of 65%. The genome size and the number of protein coding sequence of IM1011 and H. regularis are comparable, but the genome size of H. persicus is larger, and the number of protein coding sequence of H. persicus is higher. The annotation of protein coding sequence was predicted by NCBI Reference Sequence Database (RefSeq) in which redundant genes and pseudogenes have been removed. The average lengths of per protein coding sequence in IM1011, H. regularis and H. persicus were 1015, 1035 and 1039 bp (genome size divided by the number of protein coding sequence). The average length per protein coding sequence in IM1011 was a little lower than the other two species, H. regularis and H. persicus, of which the genomes were draft. The availability of the complete genome sequence of Halorientalis strain IM1011 is not only important for basic research of the Halorientalis genus, but also important for understanding the molecular mechanism of hydrocarbon degradation by haloarchaea.
Schematic circular illustration of complete genome of H. hydrocarbonoclasticus IM1011. From inner to outer: (1) GC skew (GC skew is calculated using a sliding window, as (G − C)/(G + C). The value is plotted as the deviation from the average GC skew of the entire sequence, (2) GC content (the GC content is plotted using a sliding window, as the deviation from the average GC content of the entire sequence), (3) tRNA and rRNA, (4 and 5) CDS (colored according to COG function categories, 4 is backward strand, 5 is forward strand), (6 and 7) m4C and m6A sites in CDS/rRNA/tRNA (6 is backward strand, 7 is forward strand), (8) m4C and m6A sites in intergenic regions
Phylogenetic analysis of strain IM1011
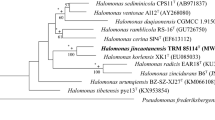
The phylogenetic classification of strain IM1011 was analyzed based on the evolutionary trees of conserved genes and ANI at the genomic level to mimic DNA–DNA hybridization. Two 16S rRNA gene sequences were found in the IM1011 genome and were named rrnA and rrnB. In the phylogenetic tree of 16S rRNA gene sequence, the evolutionary distance between two 16S rRNA genes was long, but both clustered in the branches closed to H. persicus and H. regularis (Fig. 4a). The rrnA gene was closed to H. persicus D108 (98.40%) and H. regularis TNN28 (98.12%), while rrnB gene was closed to H. persicus IBRC-M 10043T (= D108T) (98.43%) and H. regularis IBRC-M 10760T (= TNN28T) (96.93%). To confirm further, the phylogenetic tree based on the RNA polymerase beta-subunit (rpoB’) gene with one copy located in the chromosome was constructed (Fig. 4b). In this tree, strain IM1011 clustered in the Halorientalis clade. The phylogenetically closest strains were also H. persicus (94%) and H. regularis (93%).
Phylogenetic tree analysis of conserved genes. The phylogenetic tree was constructed based on the sequence of 16S rRNA gene (a) and rpoB’ gene (b). Each organism is preceded by its NCBI accession number. Bootstrap values are shown as percentages of 1000 replicates. Horizontal scale bar represents number of substitutions per nucleotide
Finally, the phylogenetic classification of IM1011 at the genomic level was deduced based on output from OrthoANI software, which includes phylogenetic tree and heatmap with OrthoANI values (Fig. 5). In the phylogenetic tree, strain IM1011 also clustered in one clade with other two Halorientalis species. In the heatmap, the OrthoANI values between species of Halorientalis were more than 80.0, while it was less than 80.0 between different genuses. It was concluded that IM1011 belongs to Halorientalis genus. The OrthoANI value between IM1011 and H. regularis was 86.0, while that between IM1011 and H. persicus was 86.2. Comparing it with the OrthoANI value of 89.1 between two different classified species, H. regularis and H. persicus; strain IM1011 could be considered as a new species. Taking into account IM1011 capability to devour and degrade aliphatic hydrocarbon hexadecane, we proposed to name it as Halorientalis hydrocarbonoclasticus sp. nov. (type strain IM1011 = CGMCC 13754).
Average nucleotide identity matrix analysis at the genome level. The ANI phylogenetic tree was constructed based on OrthoANI values. GenBank no.: H. persicus IBRC-M 10043, FOCX00000000.1; H. regularis IBRC-M 10760, FNBK00000000.1; Halorientalis sp. IM1011, NZ_CP019067.1, NZ_CP019068.1 (plasmid pHOS300), NZ_CP019069.1 (plasmid pHOS100); Halomicrobium mukohataei DSM 12286, CP001688.1, CP001689.1 (plasmid pHmuk01); Halosimplex carlsbadense 2-9-1, NZ_AOIU00000000.1; Halorhabdus tiamatea SARL4B, NC_021921.1, NC_021913.1 (plasmid pHTIA); Halovenus araenesis IBRC-M 10015, FNFC00000000.1
Currently, only eight cultivable isolates (including IM1011) of the Halorientalis genus are available in the NCBI database, of which the three previously identified species are, H. regularis isolated from marine solar salterns in eastern China (Cui et al. 2011), H. persicus isolated from Aran–Bidgol salt lake in Iran (Amoozegar et al. 2014) and Halorientalis brevis isolated from Yuncheng salt lake in Shanxi, China (Yuan et al. 2015). Amongst this genus, none of the isolates have been documented for future biotechnological application. H. hydrocarbonoclasticus stands out from the perspective of its intended biotechnological relevance in hydrocarbon bioremediation in hypersaline environments.
Putative pathway involved in the hexadecane degradation
To understand the molecular basis of hexadecane degradation, insights from IM1011 genome sequence was explored (Table 3). Pathway for aerobic degradation of aliphatic alkanes is elucidated in bacteria only wherein the first step involves conversion of alkanes to corresponding alcohols by addition of oxygen. A set of enzymes called alkane hydroxylase catalyzes the addition of oxygen for initial activation of alkane molecules. Among the various types of this set of enzymes, soluble cytochrome P450 and integral membrane non-heme alkane hydroxylases (AlkB) are involved in oxidation of medium-chain length alkanes (C5–C17). Flavin-binding monooxygenase (AlmA) and luciferase-like monooxygenase (LadA) are involved in oxidation of long-chain length alkanes (C15–C36) (Rojo 2009; Wang and Shao 2013). Among these group of alkane hydroxylase enzymes, four and seven gene copies of cytochrome P450 and luciferase-like monooxygenase/LLM class oxidoreductase (LadA), respectively, were found in IM1011 genome (Table 3). Interestingly, a cyclohexanone monooxygenase (WP_077205775) was found, but the ability of cyclohexanone degradation need to be identified. The alkB gene was not found in IM1011 genome. A study on alkane hydroxylases genes has reported that alkB gene was not found in archaeal genomes (Nie et al. 2014).
Terminal oxidation of alkane involves additional three steps before alkane is completely metabolized by β-oxidation pathway (Fig. 6). Genes for these three steps enzymes, alcohol dehydrogenase (4 copies), aldehyde dehydrogenase/aldehyde ferredoxin oxidoreductase (8 copies) and acyl-CoA synthetase/long-chain fatty acid-CoA ligase/AMP-dependent synthetase (15 copies) are also present in IM1011 genome (Table 3). Factually, many copies of putative short-chain alcohol dehydrogenase were annotated (not shown); however, considering that alkanes in hypersaline environments were usually long-chain or medium-chain, only four copies which might be related with alkane degradation are summarized in Table 3. For the similar reason, long-chain fatty acid-CoA ligase was the most possible enzyme catalyzing the biosynthesis of acyl-CoA, but some proteins named acyl-CoA synthetase or AMP-dependent synthetase catalyzes the same reaction were also listed, in case of the omission of some enzymes that could bind and catalyze both short-chain and long-chain or medium-chain fatty acids.
Putative aerobic metabolism pathway of alkane degradation in H. hydrocarbonoclasticus IM1011. AH alkane hydroxylase (including luciferase-like monooxygenase/LLM class oxidoreductase and cytochrome P450), AD alcohol dehydrogenase, ALD/ALFO aldehyde dehydrogenase/aldehyde ferredoxin oxidoreductase, ACSL long-chain fatty acid-CoA ligase/acyl-CoA synthetase/AMP-dependent synthetase, ACD acyl-CoA dehydrogenase, ECH enoyl-CoA hydratase, HAD 3-hydroxyacyl-CoA dehydrogenase/3-hydroxybutyryl-CoA dehydrogenase, ACAT acetyl-CoA acetyltransferase/3-ketoacyl-CoA thiolase
Aerobic degradation of aliphatic alkanes by terminal or subterminal oxidation as well as degradation under anaerobic condition essentially involves β-oxidation pathway (Rojo 2009). Complete β-oxidation pathway with genes for enzymes acyl-CoA dehydrogenase (19 copies), enoyl-CoA hydratase (8 copies), 3-hydroxyacyl-CoA dehydrogenase/3-hydroxybutyryl-CoA dehydrogenase (4 copies), and 3-ketoacyl-CoA thiolase/acetyl-CoA acetyltransferase (8 copies) are present in IM1011 genome (Table 3). Remarkably, two of the 3-hydroxyacyl-CoA dehydrogenases/3-hydroxybutyryl-CoA dehydrogenases were fused to enoyl-CoA hydratase and also have enoyl-CoA hydratase activity. Information from IM1011 genome suggests possible involvement of alkane hydroxylase enzymes and β-oxidation pathway in hexadecane biodegradation.
The isolate H. hydrocarbonoclasticus IM1011 is the first halophilic hydrocarbon degrader from genus Halorientalis; this coincides with the finding of Halorientalis genus by culture-independent study of hydrocarbonoclastic microflora in hypersaline soil and water samples from sabkhas in Kuwait (Al-Mailem et al. 2014). Strain IM1011 capability of biodegradation of hydrocarbons under high salt concentration suggested the potential use in bioremediation of petroleum pollution. Moreover, the sequencing and analysis of first complete genome of haloarchaeal hydrocarbon degrader has yielded key information about hydrocarbon degradation by haloarchaea. This study provides leads for further investigation to decipher mechanism of hydrocarbon degradation under hypersaline condition by haloarchaea. H. hydrocarbonoclasticus strain IM1011 and its genome is vital resource for applications in industrial biotechnology and scientific research.
References
Al-Mailem DM, Sorkhoh NA, Al-Awadhi H, Eliyas M, Radwan SS (2010) Biodegradation of crude oil and pure hydrocarbons by extreme halophilic archaea from hypersaline coasts of the Arabian Gulf. Extremophiles 14:321–328. doi:10.1007/s00792-010-0312-9
Al-Mailem D, Eliyas M, Khanafer M, Radwan S (2014) Culture-dependent and culture-independent analysis of hydrocarbonoclastic microorganisms indigenous to hypersaline environments in Kuwait. Microb Ecol 67:857–865. doi:10.1007/s00248-014-0386-5
Amoozegar MA, Makhdoumi-Kakhki A, Mehrshad M, Fazeli SAS, Spröer C, Ventosa A (2014) Halorientalis persicus sp. nov., an extremely halophilic archaeon isolated from a salt lake and emended description of the genus Halorientalis. Int J Syst Evol Microbiol 64:940–944. doi:10.1099/ijs.0.058164-0
Ashburner M et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. doi:10.1038/75556
Bertrand JC, Almallah M, Acquaviva M, Mille G (1990) Biodegradation of hydrocarbons by an extremely halophilic archaebacterium. Lett Appl Microbiol 11:260–263. doi:10.1111/j.1472-765X.1990.tb00176.x
Castillo-Carvajal LC, Sanz-Martin JL, Barragan-Huerta BE (2014) Biodegradation of organic pollutants in saline wastewater by halophilic microorganisms: a review. Environ Sci Pollut Res Int 21:9578–9588. doi:10.1007/s11356-014-3036-z
Chaisson MJ, Tesler G (2012) Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinform 13:238. doi:10.1186/1471-2105-13-238
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi:10.1038/nmeth.2474
Cuadros-Orellana S, Pohlschröder M, Durrant LR (2006) Isolation and characterization of halophilic archaea able to grow in aromatic compounds. Int Biodeterior Biodegrad 57:151–154. doi:10.1016/j.ibiod.2005.04.005
Cui H-L, Yang X, Gao X, Xu X-W (2011) Halobellus clavatus gen. nov., sp. nov. and Halorientalis regularis gen. nov., sp. nov., two new members of the family Halobacteriaceae. Int J Syst Evol Microbiol 61:2682–2689. doi:10.1099/ijs.0.025841-0
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27:4636–4641
Díaz MP, Boyd KG, Grigson SJW, Burgess JG (2002) Biodegradation of crude oil across a wide range of salinities by an extremely halotolerant bacterial consortium MPD-M, immobilized onto polypropylene fibers. Biotechnol Bioeng 79:145–153. doi:10.1002/bit.10318
Edbeib MF, Wahab RA, Huyop F (2016) Halophiles: biology, adaptation, and their role in decontamination of hypersaline environments. World J Microbiol Biotechnol 32:135. doi:10.1007/s11274-016-2081-9
Fathepure BZ (2014) Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front Microbiol 5:173. doi:10.3389/fmicb.2014.00173
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi:10.2307/2408678
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics (Oxford, England) 30:1236–1240. doi:10.1093/bioinformatics/btu031
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–D280. doi:10.1093/nar/gkh063
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi:10.1093/nar/gkm160
Le Borgne S, Paniagua D, Vazquez-Duhalt R (2008) Biodegradation of organic pollutants by halophilic bacteria and archaea. J Mol Microbiol Biotechnol 15:74–92. doi:10.1159/000121323
Lee I, Kim YO, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. doi:10.1099/ijsem.0.000760
Lefebvre O, Moletta R (2006) Treatment of organic pollution in industrial saline wastewater: a literature review. Water Res 40:3671–3682. doi:10.1016/j.watres.2006.08.027
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Magrane M, Consortium U (2011) UniProt knowledgebase: a hub of integrated protein data. Database J Biol Databases Curation 2011:bar009. doi:10.1093/database/bar009
Margesin R, Schinner F (2001) Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl Microbiol Biotechnol 56:650–663. doi:10.1007/s002530100701
Miller JR, Delcher AL, Koren S, Venter E, Walenz BP, Brownley A, Johnson J, Li K, Mobarry C, Sutton G (2008) Aggressive assembly of pyrosequencing reads with mates. Bioinformatics (Oxford, England) 24:2818–2824. doi:10.1093/bioinformatics/btn548
Nie Y, Chi CQ, Fang H, Liang JL, Lu SL, Lai GL, Tang YQ, Wu XL (2014) Diverse alkane hydroxylase genes in microorganisms and environments. Sci Rep 4:4968. doi:10.1038/srep04968
Oren A, Gurevich P, Azachi M, Henis Y (1992) Microbial degradation of pollutants at high salt concentrations. Biodegradation 3:387–398. doi:10.1007/bf00129095
Pruitt KD, Tatusova T, Brown GR, Maglott DR (2012) NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40:D130–D135. doi:10.1093/nar/gkr1079
Rojo F (2009) Degradation of alkanes by bacteria. Environ Microbiol 11:2477–2490. doi:10.1111/j.1462-2920.2009.01948.x
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi:10.1093/oxfordjournals.molbev.a040454
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tapilatu YH, Grossi V, Acquaviva M, Militon C, Bertrand JC, Cuny P (2010) Isolation of hydrocarbon-degrading extremely halophilic archaea from an uncontaminated hypersaline pond (Camargue, France). Extremophiles 14:225–231. doi:10.1007/s00792-010-0301-z
Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28
Wang W, Shao Z (2013) Enzymes and genes involved in aerobic alkane degradation. Front Microbiol 4:116. doi:10.3389/fmicb.2013.00116
Ward DM, Brock TD (1978) Hydrocarbon biodegradation in hypersaline environments. Appl Environ Microbiol 35:353–359
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2016) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol. doi:10.1099/ijsem.0.001755
Yuan P-P, Yin S, Han D, Zhang W-J, Cui H-L (2015) Halorientalis brevis sp. nov., isolated from an Inland Salt Lake of China. Curr Microbiol 71:382–386. doi:10.1007/s00284-015-0861-3
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant no. 31330001) and the Hundred Talents Program of the Chinese Academy of Sciences (to H.X.). This work was also supported by Chinese Academy of Sciences visiting professorship for postdoctoral researchers to S.K. (Grant no. 2016PB047).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by A. Driessen.
Rights and permissions
About this article
Cite this article
Zhao, D., Kumar, S., Zhou, J. et al. Isolation and complete genome sequence of Halorientalis hydrocarbonoclasticus sp. nov., a hydrocarbon-degrading haloarchaeon. Extremophiles 21, 1081–1090 (2017). https://doi.org/10.1007/s00792-017-0968-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-017-0968-5