Abstract
Sulfolobus acidocaldarius DSM639 produced an acid-resistant membrane-bound amylopullulanase (Apu) during growth on starch as a sole carbon and energy source. The physiological role of Apu in starch metabolism was investigated by the growth and starch degradation pattern of apu disruption mutant as well as biochemical properties of recombinant Apu. The Δapu mutant lost the ability to grow in minimal medium in the presence of starch, and the amylolytic activity observed in the membrane fraction of the wild-type strain was not detected in the Δapu mutant when the cells were grown in YT medium. The purified membrane-bound Apu initially hydrolyzed starch, amylopectin, and pullulan into various sizes of maltooligosaccharides, and then produced glucose, maltose, and maltotriose in the end, indicating Apu is a typical endo-acting glycoside hydrolase family 57 (GH57) amylopullulanase. The maltose and maltotriose observed in the culture medium during the exponential and stationary phase growth indicates that Apu is the essential enzyme to initially hydrolyze the starch into small maltooligosaccharides to be transported into the cell.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sulfolobales are considered as a model archaea because of their global abundance and relatively easy cultivation on a variety of carbon sources as well as their possession of various genetic elements such as plasmids, insertion sequences, and viruses (Grogan 1989; Zillig et al. 1998). Carbon source utilization profiles previously reported for Sulfolobus show that these species prefer starch or long-chain maltooligosaccharides to maltose and have both α-amylase and α-glucosidase activities (Bragger et al. 1989; Haseltine et al. 1996; Rolfsmeier and Blum 1995; Rolfsmeier et al. 1998). S. acidocaldarius grows faster than any other Sulfolobus species on yeast extracts, but this species have been reported to show relatively narrow ranges of temperature tolerance and carbon source utilization (Grogan 1989). We recently reported the characterization of a soluble α-glucosidase (MalA) from S. acidocaldarius DSM639 (Choi et al. 2013). The transcriptional analysis of malA gene in various sugars revealed that MalA was induced on starch medium, suggesting that this enzyme is involved in the starch utilization. The annotation of the genome sequence of S. acidocaldarius indicates that the gene encoding MalA is positioned with the gene encoding amylolytic enzyme, Saci_1162, within the putative maltose/maltodextrin ABC transporter operon (Saci_1160 to Saci_1166). Therefore, we assume that Saci_1162 degrades starch or α-linked polymers into small maltooligosaccharides. These hydrolysis products are then transported into the cell by maltose/maltodextrin transporter, where they are acted upon by an intracellular α-glucosidase, producing glucose which feeds into a modified version of the Entner–Doudoroff pathway (De Rosa et al. 1984).
Previously, major secreted α-amylase from S. solfataricus was purified and the repression of its synthesis by glucose has been observed (Haseltine et al. 1996). Later, the disruption study of amyA gene (Sso1172) in S. solfataricus proved that AmyA was a major secreted α-amylase which was essential for growth on starch, glycogen, and pullulan (Worthington et al. 2003). However, the substrate specificity of this enzyme using various substrates and the growth kinetics on various carbon sources by the mutant strain have not been studied in detail to understand how this enzyme is involved in starch metabolism.
The study reported here was carried out to identify the amylolytic enzyme of S. acidocaldarius and elucidate its role in starch utilization. The apu gene (Saci_1162) which is a homolog of Sso1172 in S. solfataricus was cloned and homologously expressed in S. acidocaldarius. Apu was localized in the membrane fraction and isolated using non-ionic surfactant. The substrate specificity of the recombinant enzyme and the gene inactivation study indicate that Apu is essential for starch utilization in this strain.
Materials and methods
Strains and cultivation
Sulfolobus acidocaldarius strain DSM639 and the Δapu mutant strain were grown aerobically at 77 °C in YT medium, comprised Brock’s mineral salts, 0.1 % tryptone, and 0.005 % yeast extract, and the medium was adjusted to pH 3.0 with 12 N H2SO4 at 25 °C. NYT medium contained only mineral salts without tryptone and yeast extract. The uracil auxotroph S. acidocaldarius MR31 was grown in the same medium containing uracil (50 µg/ml) and used as a host for transformation of Sulfolobus-Escherichia coli shuttle vector and the disruption vector for the target gene (Reilly and Grogan 2001). The solid medium was prepared using 0.8 % Gelrite (Sigma) and 10 mM CaCl2. In this case, tryptone was replaced by NZAmine (Sigma) and 0.2 % xylose was added to the medium as a carbon and energy source. E. coli strain DH5α was used for the propagation of plasmids, and was incubated in Luria–Bertani medium at 37 °C. When necessary, 100 μg/ml ampicillin was added to the medium.
Construction of the apu overexpression vector
The plasmid pKHapuA modified from the pKHmalA (Choi et al. 2013) was constructed for homologous expression of the apu gene in S. acidocaldarius. For the construction of pKHapuA, malA gene controlled by the promoter of gdhA in pKHmalA was replaced by apu gene. Using the genomic DNA of S. acidocaldarius as a template, the apu gene encoded by saci_1162 was amplified with the primers KH45 and KH46. PCR was carried out using 2.5 U of PrimeSTAR HS DNA polymerase (Takara, Japan) using the following conditions: 30 cycles of 98 °C for 10 s, 55 °C for 5 s, and 72 °C for 2 min. The obtained 2663 bp PCR product was purified and digested with NcoI. The digested DNA fragment was cloned into pKHmalA which was digested with same enzyme, resulted in pKHapuA. The oligonucleotide primers used for construction of pKHapuA were listed in Table S1 in the supplemental material.
Strain construction
To construct a deletion mutant strain of apu, a fragment of 792 bp containing a portion of the upstream region of apu gene was amplified with the primers KH89 and KH90, and a fragment of 761 bp containing a part of the downstream region of apu was amplified with primers KH91 and KH92 using genomic DNA from S. acidocaldarius DSM639 as a template. Two PCR fragments were fused by overlap extension PCR using the primers KH89 and KH92, as described (Choi et al. 2013). As a result, a BamHI restriction site was inserted in the internal region of the fusion fragment and a 2116-bp region of the apu gene was deleted in the fusion fragment. The 1559-bp fusion PCR product was cloned into pGEM-T (Promega) to generate pKHd1162. To disrupt the apu gene by replacement of pyrE gene in S. acidocaldarius genome, a DNA region from 280 nt upstream to 71 nt downstream of the pyrE gene from S. solfataricus P2 was amplified using the primers KH71 and KH72. The 770-bp PCR product was digested with BamHI and cloned into pKHd1162, which was digested with same enzyme, to generate apu gene targeting vector pKHΔapu. The plasmid was methylated to prevent restriction by the SuaI restriction enzyme. For that purpose E. coli ER1821 (New England Biolabs) bearing the additional plasmid pM.EsaBC4I was transformed with plasmid DNAs. The resulting methylated plasmid was transformed into parental strain of S. acidocaldarius MR31. The deletion mutant was isolated and verified by PCR and DNA sequencing as described (Choi et al. 2013).
Expression and purification of recombinant Apu from S. acidocaldarius
Sulfolobus acidocaldarius MR31 cells harboring pKHapuA were grown in YT medium containing 0.2 % sucrose at 77 °C for 48 h. The cells were harvested by centrifugation at 8000×g for 30 min and resuspended in 20 mM sodium phosphate buffer (pH 7.4). The cells were lysed by French pressure on ice, and centrifuged at 3000×g for 10 min to remove cell debris. For isolation of membrane proteins, the supernatant was centrifuged at 15,000×g for 50 min. The pellet was resuspended in 20 mM sodium phosphate containing 40 mM imidazole and solubilized with 2 % n-dodecyl β-d-maltoside (DDM) at a protein concentration of 2–3 mg/ml at room temperature. After 2 h at room temperature, nonsolubilized proteins were removed by centrifugation at 15,000×g for 50 min, and the supernatant was diluted using 20 mM sodium phosphate containing 40 mM imidazole and 0.1 % DDM. The supernatant was applied onto a Ni–NTA affinity column (GE healthcare, Freiburg, Germany) equilibrated with 20 mM sodium phosphate containing 40 mM imidazole and 0.1 % DDM. The column was washed with wash buffer [20 mM sodium phosphate buffer (pH 7.4), 0.1 % DDM, 40 mM imidazole], and then the bound proteins were eluted using the elution buffer [20 mM sodium phosphate buffer (pH 7.4), 0.1 % DDM, 1 M imidazole]. All fractions were collected and dialyzed against the 20 mM sodium phosphate buffer (pH 7.4) containing 0.01 % DDM without imidazole, and then concentrated using Centricon-10 microfilter (Millipore, Darmstadt, Germany). The protein concentration was determined according to the Bradford method with bovine serum albumin as a standard (Bradford 1976). The molecular mass and purity of the purified Apu were estimated by 12 % SDS-PAGE. To determine the native molecular mass of the purified protein, analytical size exclusion chromatography was performed using a HiLoad 16/600 Superdex 200 pg column (GE Healthcare). An isocratic gradient of 20 mM sodium phosphate (pH 7.4) with 0.5 M NaCl was used to detect the protein.
Enzyme activity assay
The α-amylase and pullulanase activities of Apu were assayed by measuring the amount of reducing sugar released during hydrolysis of starch and pullulan, respectively. The amount of reducing sugar produced was measured by the 3, 5-dinitrosalicylic acid (DNS) method (Miller 1959). All reactions were performed in triplicate. The reaction mixtures (500 μl) containing 0.1 % substrate in 20 mM sodium citrate buffer (pH 3.0), overlaid with mineral oil to minimize evaporation, were incubated with 15 μg of enzyme at 95 °C for 90 min. Five hundred μl of DNS reagent were added to the mixture and incubated at 100 °C for 5 min. The reaction was stopped by quench on ice, and aliquots of 200 μl were measured at 575 nm. One unit of the enzyme activity was defined as the amount of the enzyme that hydrolyzes the substrate to release 1 μmol of reducing sugar per minute with an appropriate standard (glucose for amylase activity and maltotriose for pullulanase activity) under the assay conditions.
The dependence of enzyme activity on pH and temperature was determined over a pH range of 3.0 to 8.0 and a temperature range of 60 to 120 °C. The buffers used were sodium citrate (pH 2.0–4.0), sodium acetate (pH 4.0–5.5), and MES buffer (pH 5.5–6.5). Thermostability was determined by incubating the enzyme solution (0.03 mg/ml in 20 mM sodium citrate buffer, pH 3.0) at different temperatures. After various time intervals, samples (100 μl) were withdrawn and the residual activity was measured using 0.1 % starch as described above. For the effect of metal ions, the enzyme was EDTA treated. It was dialyzed extensively first against the 20 mM sodium phosphate buffer (pH 7.4) containing 2 mM EDTA and then twice against the same buffer without EDTA. The enzyme activity was examined after preincubation of the enzyme with metal ions and chemical reagents at a 1 mM concentration, except for SDS (1 %) at 95 °C for 1 h. The residual activity was determined with 0.1 % starch as a substrate, as described above.
To analyze the membrane-bound Apu activity in S. acidocaldarius DSM639 and the Δapu mutant, the strains were incubated in YT medium containing 0.4 % starch (first grade, Shinyo, Osaka, Japan) at an OD600 of 0.5. The cells were harvested and resuspended in 20 mM sodium phosphate buffer (pH 7.4). The membrane proteins from the cells were prepared as described above. The membrane proteins (30 μg) were incubated with 0.1 % starch in 20 mM sodium citrate buffer (pH 5.0). After various time intervals, 10 μl of the sample was withdrawn and the hydrolysis patterns were confirmed by thin-layer chromatography (TLC) analysis. TLC was done on K5F silica gel plates (Whatman, Madistone, UK) with 1-butanol/ethanol/H2O (5:5:3, v/v/v) as the solvent system. After irrigating twice, the TLC plate was dried and dipped into a solution containing 0.3 % N-(1-naphthyl)-ethylenediamine and 5 % H2SO4 in methanol and then heating for 10 min at 110 °C to visualize the reaction spots.
Substrate specificity of Apu
To examine the hydrolytic patterns of Apu, the purified Apu (90 mU/ml) was incubated with 0.5 ml of 1 % soluble starch, amylose, amylopectin, glycogen, pullulan, and β-cyclodextrin in 20 mM sodium citrate buffer (pH 3.0). For maltooligosaccharides (G2 to G7) as a substrate, 1 mM of concentration was used. Each reaction was incubated at 95 °C for 90 min, and then placed immediately in an ice-water bath to stop the reaction. The resulting hydrolyzed products were identified by TLC and quantified by densitometry using ImageJ computer software (ver. 1.46, National Institute of Mental Health, Bethesda, MD) for comparison.
Zymogram
Starch zymography gel was prepared using polyacrylamide gel (12 %) containing 0.1 % starch. The samples (15 μg) were diluted five-fold in a zymogram sample buffer [0.5 M Tris–HCl (pH 6.8), 10 % SDS, 20 % glycerol, and 0.5 % bromophenol blue]. After electrophoresis at 120 V for 90 min, the gel was washed with distilled water for 30 min to eliminate SDS. And then, the gel was incubated in 20 mM sodium citrate buffer (pH 3.0) at 77 °C for 12 h. The gel was stained with 10 % Lugol’s solution, which consisted of 1.5 % iodine and 10 % potassium iodide, and the bands with amylolytic activity were visualized as non-stained regions on the gel.
Results
Sequence analysis of the putative Apu
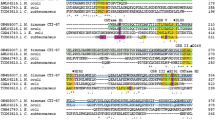
The putative apu (saci_1162) gene has been identified within the maltose/maltodextrin ABC transporter operon of S. acidocaldarius. The open reading frame of this gene is composed of 2655 nucleotides corresponding to 884 amino acids. A signal peptide was found using the SignalP 4.0 prediction program for Gram-positive bacteria mode (Bagos et al. 2009) and the potential cleavage site was predicted between 21th alanine and 22th glutamine. The conserved domain search from NCBI revealed that Apu contained the N-terminal catalytic domain of thermoactive amylopullulanases which belonged to glycoside hydrolase family 57 (GH57N-Apu) positioned 331 to 647 and a starch binding domain (CBM25) positioned 38 to 121 (Fig. 1). Apu also included the regions which are suspected to be localized to the membrane at the N-terminal and C-terminal ends as well as the GH57 N-Apu and CBM25. The protein sequence translated from the apu gene was aligned with sequences available in databanks using the BLAST algorithm. The highest identity score (59 %) was observed with S. tokodaii α-amylase (ST1102) and S. solfataricus P2 α-amylase (Sso1172) (Fig. 2). Apu sequence also shows some similarities with the sequences in the euryarchaeal Pyrococcus furiosus amylopullulanase and Thermococcus litoralis amylopullulanase within the maltodextrin ABC transporter (Imamura et al. 2004; Koning et al. 2002). The size of euryarchaeal enzymes was bigger than those of Sulfolobus enzymes and they contained an extra DOMON_glucodextranase-like domain which is known to interact with carbohydrates (Fig. 1). Apu homologs conserved the five consensus sequence motif which is the characteristic of the GH57 enzymes (Zona et al. 2004). In the Apu from S. acidocaldarius, these motifs correspond to HQP (region I), GKVEVL (region II), WTPEQA (region III), AFDGENPLIF (region IV) in GH57N-Apu domain, and AEGSDWTWQ (region V) in following α-helical region (Fig. 2). The third and fourth regions contain the catalytic nucleophile and the acid/base catalyst, respectively. It has been known that amylopullulanase is one of the GH57 family thermostable enzymes from extremophiles, which exhibit α-amylase, 4-α-glucanotransferase, amylopullulanase, α-galactosidase, and branching enzyme depending on the enzyme specificities (Blesák and Janeček 2012). Recently, the crystal structure of GH57-type branching enzyme from Thermus thermophilus revealed that the structural characteristics which contained a (β/α)7-barrel and the succeeding α-helical region was essential for correct functioning of this family (Palomo et al. 2011). Based on the amino acid sequence analysis of Apu and its comparison with other homologs, Apu is thought to be an amylopullulanase which can degrade both the α-1,4 and α-1,6-linkages in α-glucans such as starch and glycogen.
Comparison of the domain structure of Apu with other homologs of hyperthermophilic archaea. CBM25, carbohydrate binding module 25; GH57N-Apu, N-terminal catalytic domain of thermoactive amylopullulanases; Domon_glucodextranase-like domain, DOMON_glucodextranase-like domain of various glycoside hydrolases. S. acidocaldarius, Sulfolobus acidocaldarius; S. solfataricus, Sulfolobus solfataricus (BAA11010.1); T. acidophilum, Thermoplasma acidophilum (CAC11276.1); P. furiosus, Pyrococcus furiosus (ABA33719.1); P. abyssi, Pyrococcus abyssi (CCE69556.1); T. litoralis, Thermococcus litoralis (BAC10983.1); Thermococcus sp. CL1 (AFL95093.1); T. hydrothermalis, Thermococcus hydrothermalis (AAD28552.1); P. aerophilum, Pyrobaculum aerophilum str. IM2 (AAL64927.1)
Sequence alignment of Apu (Saci_1162) with S. tokodaii α-amylase (ST1102) and S. solfataricus α-amylase (Sso1172). Multiple alignments were carried out with the Clustal W2 algorithm. Region I to V represent conserved motifs in the GH57 enzymes. The conserved sequences are signified by asterisks. The colon and period indicate conservation between groups of strongly and weakly similar properties, respectively
Identification of Apu critical for starch degradation in S. acidocaldarius
The growth characteristics of the Δapu mutant in the NYT and YT medium supplemented with starch were examined and compared with that of the uracil auxotrophic parental strain MR31. At first, a nutrient-rich YT medium was used to examine the growth rate. The growth rate of MR31 was severely affected by the disruption of apu gene (Fig. 3a). The growth phenotype of Δapu mutant was very similar to ΔmalEFG mutant which the maltose/maltodextrin transporter genes were disrupted in our previous study (Choi et al. 2013). The growth pattern on YT medium with starch was similar to that on YT medium without sugar, indicating that the Δapu mutant cannot utilize the starch as a carbon source. The wild-type DSM639 and MR31 strains failed to grow in the NYT medium without starch, but they started to grow by the addition of starch and reached the final cell density up to 0.6 at OD600 indicating that starch can be used as a sole carbon and energy source. The specific growth rates of the DSM639 and MR31 strains on NYT medium were three-four times slower than that of the strains on YT medium (K. H. Choi, unpublished data). The Δapu mutant strain failed to grow on NYT medium by the addition of starch. These results imply that Apu is essential for the growth in the presence of starch.
Growth (a) and the hydrolyzed products of starch by membrane fraction (b) of S. acidocaldarius DSM639 (solid circle) and Δapu mutant (open circle) in YT medium supplemented with starch. The cultivation of each strain was carried out at 77 °C and 0.4 % soluble starch was used as a carbon source. The membrane proteins were incubated with 0.1 % soluble starch in 20 mM sodium citrate (pH 5.0) at 95 °C and the hydrolysis patterns of the starch were analyzed by TLC at the indicated times. G1–G7 oligosaccharide standard is glucose through maltoheptaose
The starch degradation patterns of the MR31 and Δapu mutant strain grown on YT medium in the presence of starch were examined. The membrane fractions were isolated when the cell density was reached OD600 of 0.5, and incubated with starch up to 12 h. The sugar composition of the reaction products was analyzed by TLC to identify the degradation products produced by the amylolytic activities of membrane-bound enzymes. In an early stage, starch was degraded into maltooligosaccharides, and later glucose and maltose were gradually increased (Fig. 3b). In the Δapu mutant, the hydrolyzed products were not detected, indicating that Apu would be a key enzyme which initially attacks the starch to degrade into smaller maltooligosaccharides.
Homologous expression and purification of membrane-bound Apu
The analysis of the secondary structure prediction program of membrane proteins (SOSUI) indicates that Apu is a membrane-bound protein since it is predicted to possess a putative N- and C-terminal transmembrane segment (TMS) that may function as a membrane anchor. In order to identify the localization of Apu expression, the amylolytic activity of the cells grown in starch medium was examined in an extracellular or membrane-bound fractions. The activity was confirmed by quantifying the amount of reducing sugar released during enzyme assay. The amylolytic activity was detected in all the fractions, but strong activity was only observed in membrane fraction, implying that Apu is a membrane-bound protein (data not shown). The expression of Apu was tried in E. coli to obtain high and reproducible production of the enzyme compared to that of the natural host, S. acidocaldarius. However, the amylolytic activity was undetectable in the transformed cells and the presence of an Apu was not observed. It was assumed that the putative membrane-anchoring N-terminal peptide could impair the correct folding of Apu and hence favors rapid degradation of the misfolded protein.
In order to produce functional enzyme, a homologous expression in S. acidocaldarius was tried. The coding sequence of apu containing downstream 6× histidine tag was located under the gdhA promoter of pKHmalA. The recombinant enzyme was expressed and the enzyme activity was detected in the insoluble cell pellet suspension after the cell breakage indicating that the enzyme should be localized in the cell membrane. A membrane-bound enzyme was solubilized using a non-ionic detergent DDM, and then purified by one-step Ni–NTA affinity chromatography. The apparent molecular mass of the purified Apu was estimated as 100 kDa by SDS-PAGE, which was in good agreement with the size calculated from the primary sequence (Fig. 4a). The molecular mass of Apu by gel filtration chromatography was approximately 435 kDa, which indicated that Apu is a tetramer in its native form (Fig. 4b). The dimeric conformation of the protein was observed with α-amylase from S. solfataricus (Haseltine et al. 1996). Approximately 0.28 mg Apu with a specific activity of 2.2 U/mg, corresponding to a yield of 0.03 % of the total activity in cell-free extracts, was obtained. The hydrolysis of starch was visualized on a native PAGE by zymographic assay, indicating that Apu was successfully expressed in S. acidocaldarius (Fig. 4c). The pI value of Apu was estimated to 5.31 by ExPASy ProtParam tool program (http://web.expasy.org/protparam).
SDS-PAGE analysis (a), determination of molecular mass (b), and zymogram (c) of purified recombinant Apu. During purification step, the amylolytic activity was revealed by zymography on a gel containing 0.1 % starch stained with iodine solution. Lane M molecular weight standard (Dokdo Mark, broad range, Elpis Biotech, Daejeon, Korea), lane 1 cell-free extract, lane 2 membrane proteins, lane 3 2 % DDM-solubilized proteins, lane 4 purified enzyme after Ni–NTA chromatography. The molecular mass of the Apu was determined by analyzing the molecular mass of standard proteins eluted during gel filtration chromatography performed using a HiLoad 16/600 Superdex 200 pg column. The column was calibrated with blue dextran (2000 kDa); thyroglobulin (669 kDa); apoferritin (443 kDa); β-amylase (200 kDa); alcohol dehydrogenase (150 kDa); Albumin, bovine (66 kDa); Carbonic anhydrase (29 kDa)
Catalytic properties of membrane-bound Apu
The amylolytic activity of Apu toward starch was most active at pHs ranging from 3.0 to 3.5 (Fig. 5a). The enzyme activity decreased sharply at pHs over 4.5 or below 2.5. The optimum pH of Apu is consistent with the pH in growth medium of S. acidocaldarius supporting that Apu is a membrane-bound enzyme. The optimal temperature of Apu was at least 20 °C higher than maximum growth temperature of S. acidocaldarius, and the activity was rapidly decreased below 80 °C (Fig. 5b). Moreover, the recombinant Apu possessed remarkable thermostability, retaining its full activity after 40-h incubation at 90 and 95 °C (Fig. 5c). The half-life of Apu was 2.5 h at 100 °C. These results imply that Apu was successfully expressed in S. acidocaldarius. The effects of metal ions and chemical reagents on enzyme activity were also examined (Table S2 in the supplemental material). Most of the metal ions did not affect enzyme activity or stability. Co2+ and Mg2+, as chloride salts at a concentration of 1 mM, slightly enhanced the enzyme activity. The addition of CaCl2 to the enzyme did not enhance the activity. EDTA was not found to be inhibitory for Apu. Even after preincubation for 1 h, the enzyme activity was not reduced. Apu exhibited significant tolerance against the chemical reagents except for SDS. The addition of 1 % SDS completely inactivated the enzyme.
Effect of pH and temperature on the activity and stability of Apu. a For optimal pH determination, the following buffers were used: pH 2.0–4.0, sodium citrate (inverted triangle); pH 4.0–5.5, sodium acetate (open circle); pH 5.5–6.5, MES (solid circle). Activity was assayed at 95 °C for 5 min in various buffers (20 mM). b To determine the optimal temperature, activity was measured at the indicated temperatures under the standard conditions of the assay. c To determine the thermostability, the enzyme was preincubated at 90 °C (solid circle), 95 °C (solid square) and 100 °C (solid triangle) in 20 mM sodium citrate buffer (pH 3.0) without substrate. After various time intervals, samples were withdrawn and the residual activity was measured at 95 °C for 5 min. Error bar indicates standard deviations from 3 separate experiments
The substrate hydrolysis pattern of Apu was determined by TLC. Apu hydrolyzed a wide variety of substrates, such as soluble starch, amylose, amylopectin, glycogen, pullulan, and maltooligosaccharides (Fig. 6a and Table S3 in the supplemental material). When the amylolytic activity on pullulan was taken as standard, most of the polymeric substrates showed high activity. Starch and its constituent polysaccharides amylose and amylopectin were hydrolyzed into glucose, maltose, and maltotriose. Apu preferentially hydrolyzed starch (78 %) and amylopectin (72 %), while hydrolysis activity toward amylose (46 %) was slightly decreased. Highly branched α-glucans like glycogen which contains α-1,6 glycosidic linkages were also hydrolyzed efficiently with 53 %. Pullulan, α-glucan in which every third glycosidic linkage is an α-1,6 linkage, was hydrolyzed into maltotriose with minor amounts of glucose and maltose. The small substrates such as maltose and maltotriose were not hydrolyzed very well. The hydrolytic pattern of Apu toward various substrates is consistent with those of the reported amylopullulanases from T. litoralis and P. furiosus except those enzymes could not hydrolyze maltose through maltohexaose (Brown and Kelly 1993).
Hydrolysis patterns of Apu toward various substrates. a The substrates (1 mM or 0.1 %) were digested with recombinant Apu (0.2 U) in 20 mM sodium citrate buffer (pH 3.0) for 12 h at 95 °C. The products were separated on a silica gel TLC plate. Lane S G1–G7 (glucose through maltoheptaose) oligosaccharide standard (1 mM), lane 1 starch, lane 2 amylose, lane 3 amylopectin, lane 4 glycogen, lane 5 pullulan, lane 6 β-CD, lanes 7–12, maltose through maltoheptaose. b Time courses of the hydrolyzed products from starch. The enzyme was incubated with 0.1 % soluble starch in 20 mM sodium citrate (pH 3.0) at 95 °C and the hydrolysis patterns were analyzed by TLC at the indicated times. c Schematic diagram of starch hydrolysis by Apu. Black arrow indicates α-1,4 glycosidic linkage and red arrow indicates α-1,6 glycosidic linkage
The action mode of Apu was analyzed by the hydrolyzed products during the incubation of the enzyme with starch at different time points (Fig. 6b). Apu initially hydrolyzed starch into various sizes of maltooligosaccharides, and then produced glucose, maltose, and maltotriose in the end, indicating Apu acts as an endo-type fashion. As summarized from these results, Apu is indeed a membrane-bound amylopullulanase which can degrade both α-1,4 and α-1,6-linkages of α-glucan (Fig. 6c).
The role of Apu in starch metabolism of S. acidocaldarius
The wild-type DSM639 strain was grown on YT medium including starch and the sugar composition of the culture supernatant was analyzed by TLC to identify the degradation products produced by membrane-bound Apu from lag phase through stationary phase (Fig. 7a). In an exponential phase, starch was degraded into maltooligosaccharides, and later short-chain maltooligosaccharides such as maltose, maltotriose, and maltoteraose were gradually increased until stationary phase was reached. The medium pH was increased from 3.5 to 5.0 during this period. Unlike the purified Apu mainly produced glucose, maltose, and maltotriose from starch, no glucose was detected in the culture supernatant during the growth of the wild-type cells. It can be explained by the difference of the hydrolytic activity of Apu at pH 3.5 and pH 5.0, respectively. As shown in Fig. 7b, c, Apu showed strong activities toward various substrates at pH 3.5, but the activities were sharply decreased when the pH was shifted to 5.0. At pH 5.0, the hydrolytic activities of Apu toward pullulan and glycogen which have 5–30 % of α-1,6 glycosidic linkages were much lower than those of maltooligosaccharides which have only α-1,4 glycosidic linkages. The hydrolyzed products by the action of most α-amylases are mainly maltose and maltooligosaccharides. Therefore, the lack of glucose in the culture supernatants of the growing cells can be explained by the amylase-type activity of Apu during the growth period between exponential and stationary phases. Alternative explanation that glucose was not found in the supernatants of the growing cells can be that it was used as an energy source by the growing cells.
The growth curve and starch degradation pattern of DSM639 in YT medium with starch (a) and TLC analysis of the hydrolyzed products produced from various substrates by Apu at pH 3.5 (b) and pH 5.0 (c). Lane S G1–G7 oligosaccharide standard, lane 1 starch, lane 2 amylose, lane 3 amylopectin, lane 4 glycogen, lane 5 pullulan, lane 6 β-cyclodextrin, lanes 7 maltopentaose, lane 8 maltohexaose, lane 9 maltoheptaose
Therefore, membrane-bound Apu is the first enzyme to attack the starch to degrade into smaller maltooligosaccharides such as maltose and maltotriose, and the hydrolyzed products would then be imported into the cytoplasm by maltodextrin transporters and eventually metabolized inside the cells.
Discussion
The hydrolysis of starch by S. solfataricus takes place in the extracellular medium and the resultant maltodextrins are taken up to the cell to be metabolized (Haseltine et al. 1996; Worthington et al. 2003). Extracellular enzyme AmyA (Sso1172) has been known to play a role for breakdown the starch into maltodextrin during this process. The purpose of our study is to find a major enzyme to initially break the exogenous starch to maltodextrin which can be transported and metabolized in the cells for starch utilization in S. acidocaldarius. Two genes (saci_1162 and saci_1200) in the S. acidocaldarius genome are annotated as encoding an α-amylase that hydrolyzes α-glucans. Saci_1162 is annotated as a membrane-bound amylopullulanase and Saci_1200 is an intracellular α-amylase, respectively. Therefore, it is assumed that Saci_1162 which is closely located to the genes coding for the putative ABC maltose/maltodextrin transporter and binding protein is actually utilized to degrade starch into maltodextrin and Saci_1200 located within the putative glycogen operon is involved in the intracellular utilization of storage carbohydrate glycogen. In this study, we identified the saci_1162 (apu) gene encoding the putative amylopullulanase of S. acidocaldarius and investigated its physiological role by the analysis of growth pattern and starch utilization of apu disruption mutant as well as the biochemical properties of recombinant Apu.
Comparison of the growth pattern and amylolytic activity of the wild-type and Δapu mutant strains grown on minimal and rich medium supplemented with starch reveals that the major enzyme responsible for the hydrolysis of starch is Apu. Whereas the wild-type DSM639 was grown in minimal medium in the presence of starch, the Δapu mutant failed to grow in the same medium. Furthermore, the amylolytic activity observed in the membrane fraction of the wild-type strain was not detected in the Δapu mutant. The amylolytic activity found in the membrane fraction indicates that Apu is localized to the cell membrane. The cellular localization of Apu is consistent with the prediction based on the secondary structure of Apu. In S. solfataricus major α-amylase essential for growth on starch was secreted in the culture supernatant, while in S. tokodaii and S. acidocaldarius a significant portion of the α-amylase activity was cell-associated (Ellen et al. 2010; Haseltine et al. 1996). The different cellular locations between two Sulfolobus enzymes were explained by the subtle difference in the structure and composition of S-layer which is known to be the unique outside structure of Sulfolobales (Ellen et al. 2010).
The membrane-associated Apu was solubilized using non-ionic detergent and purified by Ni–NTA affinity chromatography. Apu shows the highest activity over a pH range from 3.0 to 4.0, with an optimum at pH 3.5 when soluble starch was used as a substrate. As the active site of Apu is thought to be located toward outside from the cell membrane based on the SOSUI program, the enzyme might be exposed to the acidic milieu of the habitat. S. acidocaldarius grows between pH 3.0 and 3.5, a range corresponding to the optimum pH range of the enzyme. The same is true for the temperature-activity profile of this enzyme. Using a 10-min assay, at its optimal pH, Apu displayed maximum rates of substrate hydrolysis at 95–105 °C.
The substrate specificity of Apu shows that Apu can hydrolyze pullulan as well as starch and amylopectin. As shown in Fig. 6a, degradation of soluble starch and pullulan by Apu resulted in the mixture of glucose, maltose, and maltotriose. Homologous protein search of Apu reveals that the enzyme is closely related to the thermophilic archeal proteins which belong to GH57 family amylopullulanase (Zona et al. 2004). These enzymes possess both α-amylase and pullulanase activities which defined by a unique active site. This property of Apu suggests that it is an endo-type amylopullulanase rather than a typical α-amylase. Similar enzymes have been reported from T. litoralis, T. hydrothermalis, and P. furiosus (Brown and Kelly 1993; Dong et al. 1997; Erra-Pujada et al. 2001). An extracellular amylopullulanase (PF1935) has been known to be a key enzyme responsible for starch degradation in P. furiosus (Lee et al. 2006). The secreted AmyA of S. solfataricus is also known to be essential for growth on starch and pullulan although the hydrolytic characterization of this enzyme for various carbohydrates has not been examined in details (Worthington et al. 2003). Therefore, it is thought that an amylopullulanase which has bifunctional activities hydrolyzing both α-1,4 and α-1,6 glycosidic linkages plays an essential role in starch utilization instead of α-amylase in most thermophilic archaea. The genetic location of the amylopullulanase gene which is flanked by the genes encoding maltodextrin transporter in various hyperthermophilic archaea also supports that this enzyme is involved in the starch or maltodextrin metabolism (Elferink et al. 2001; Koning et al. 2002; Noll et al. 2008; Rolfsmeier et al. 1998).
In conclusion, Apu located in the putative ABC maltose/maltodextrin operon is an amylopullulanase which hydrolyzes the starch into mostly maltose and maltotriose. The binding protein MalE within ABC transporters captures the hydrolyzed products and transfers them to the cognate transporter complex MalFG and ATPase MalK for import into the cytosol. Finally, MalA (α-glucosidase) which is active on maltose and maltotriose can convert them into glucose, which is further utilized as an energy source in S. acidocaldarius.
References
Bagos PG, Tsirigos KD, Plessas SK, Liakopoulos TD, Hamodrakas SJ (2009) Prediction of signal peptides in archaea. Protein Eng Des Sel 22:27–35
Blesák K, Janeček Š (2012) Sequence fingerprints of enzyme specificities from the glycoside hydrolase family GH57. Extremophiles 16:497–506
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Bragger JM, Daniel RM, Coolbear T, Morgan HW (1989) Very stable enzymes from extremely thermophilic archaebacteria and eubacteria. Appl Microbiol Biotechnol 31:556–561
Brown SH, Kelly RM (1993) Characterization of amylolytic enzymes, having both both α-1,4 and α-1,6 hydrolytic activity, from the thermophilic archaea Pyrococcus furiosus and Thermococcus litoralis. Appl Environ Microbiol 59:2614–2621
Choi KH, Hwang SM, Cha J (2013) Identification and characterization of MalA in the maltose/maltodextrin operon of Sulfolobus acidocaldarius DSM639. J Bacteriol 195:1789–1799
De Rosa M, Gambacorta A, Nicolaus B, Giardina P, Poerio E, Buonocore V (1984) Glucose metabolism in the extreme thermoacidophilic archaebacterium Sulfolobus solfataricus. Biochem J 224:407–414
Dong G, Vieille C, Zeikus JG (1997) Cloning, sequencing, and expression of the gene encoding amylopullulanase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol 63:3577–3584
Elferink MG, Albers SV, Konings WN, Driessen AJM (2001) Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol Microbiol 39:1494–1503
Ellen AF, Albers SV, Driessen AJM (2010) Comparative study of the extracellular proteome of Sulfolobus species reveals limited secretion. Extremophiles 14:87–98
Erra-Pujada M, Chang-Pi-Hin F, Debeire P, Duchiron F, O’Donohue MJ (2001) Purification and properties of the catalytic domain of the thermostable pullulanase type II from Thermococcus hydrothermalis. Biotechnol Lett 23:1273–1277
Grogan DW (1989) Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J Bacteriol 171:6710–6719
Haseltine C, Rolfsmeier M, Blum P (1996) The glucose effect and regulation of α-amylase synthesis in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 178:945–950
Imamura H, Jeon BS, Wakagi T (2004) Molecular evolution of the ATPase subunit of three archaeal sugar ABC transporters. Biochem Biophys Res Commun 319:230–234
Koning SM, Konings WN, Driessen AJM (2002) Biochemical evidence for the presence of two α-glucoside ABC-transport systems in the hyperthermophilic archaeon Pyrococcus furiosus. Archaea 1:19–25
Lee HS, Shockley KR, Schut GJ, Conners SB, Montero CI, Johnson MR, Chou CJ, Bridger SL, Wigner N, Brehm SD, Jenney FE Jr, Comfort DA, Kelly RM, Adams MW (2006) Transcriptional and biochemical analysis of starch metabolism in the hyperthermophlic archaeon Pyrococcus furiosus. J Bacteriol 188:2115–2125
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Ann Biochem 31:426–428
Noll KM, Lapierre P, Gogarten JP, Nanavati DM (2008) Evolution of mal ABC transporter operons in the Thermococcales and Thermotogales. BMC Evol Biol. doi:10.1186/1471-2148-8-7
Palomo M, Pijning T, Booiman T, Dobruchowska JM, van der Vlist J, Kralj S, Planas A, Loos K, Kamerling JP, Dijkstra BW, van der Maarel MJEC, Dijkhuizen L, Leemhuis H (2011) Thermus thermophilus glycoside hydrolase family 57 branching enzyme: crystal structure, mechanism of action, and products formed. J Biol Chem 286:3520–3530
Reilly MS, Grogan DW (2001) Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J Bacteriol 183:2943–2946
Rolfsmeier M, Blum P (1995) Purification and characterization of a maltase from the extremely thermophilic crenarchaeote Sulfolobus solfataricus. J Bacteriol 177:482–485
Rolfsmeier M, Haseltine C, Bini E, Clark A, Blum P (1998) Molecular characterization of the alpha-glucosidase gene (malA) from the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 180:1287–1295
Worthington P, Hoang V, Perez-Pomares F, Blum P (2003) Targeted disruption of the α-amylase gerne in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol 185:482–488
Zillig W, Arnold HP, Holz I, Prangishvili D, Schweier A, Stedman K, She Q, Phan H, Garrett R, Kristjansson JK (1998) Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 2:131–140
Zona R, Chang-Pi-Hin F, O’Donohue MJ, Janeček Š (2004) Bioinformatics of the glycoside hydrolase family 57 and identification of catalytic residues in amylopullulanase from Thermococcus hydrothermalis. Eur J Biochem 271:2863–2872
Acknowledgments
This study was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2013R1A2A2A01068096).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Driessen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Choi, KH., Cha, J. Membrane-bound amylopullulanase is essential for starch metabolism of Sulfolobus acidocaldarius DSM639. Extremophiles 19, 909–920 (2015). https://doi.org/10.1007/s00792-015-0766-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-015-0766-x