Abstract
Great Salt Lake (GSL) represents one of the world’s most hypersaline environments. In this study, the archaeal and bacterial communities at the North and South arms of the lake were surveyed by cloning and sequencing the 16S rRNA gene. The sampling locations were chosen for high salt concentration and the presence of unique environmental gradients, such as petroleum seeps and high sulfur content. Molecular techniques have not been systematically applied to this extreme environment, and thus the composition and the genetic diversity of microbial communities at GSL remain mostly unknown. This study led to the identification of 58 archaeal and 42 bacterial operational taxonomic units. Our phylogenetic and statistical analyses displayed a high biodiversity of the microbial communities in this environment. In this survey, we also showed that the majority of the 16S rRNA gene sequences within the clone library were distantly related to previously described environmental halophilic archaeal and bacterial taxa and represent novel phylotypes.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Molecular evidence has greatly increased the appreciation of microbial biodiversity and has contributed to a general consensus that the microbial world is much more diverse and complex than anyone had anticipated (Pace 1997; Harris et al. 2013). Such studies are particularly important in extreme environments where the microbial biodiversity is little explored and particularly unique.
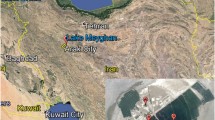
Great Salt Lake (GSL) represents one of the world’s most extreme environments, it is the fourth largest terminal lake and the second most saline lake in the world (Hassibe and Keck 1978). In the late 1950s, a railroad causeway was built, which subsequently separated the lake into a North Arm and a South Arm and induced an environmental evolution of the habitat (Cannon and Cannon 2002). In historical time, the lake’s salinity has ranged from a little less than 5 % to nearly 27 % (Utah geological survey, online publications). This lake has indeed unique characteristics when compared to other hypersaline environments. Whereas the South Arm maintains a salinity level of about 9 %, the North Arm is characterized by high levels of salinity (as high as 30 % w/v). The North Arm, near Rozel Point, is also influenced by numerous petroleum seeps that introduce into the lake nitrogen (0.5 %) and high sulfur (14 %) containing asphaltic oil (Gwynn 1980; Sinninghe Damsté et al. 1987). Moreover, the North Arm of the lake has no significant freshwater input, which prevents important fluctuations in salinity. The petroleum seeps at Rozel Point were discovered in the late 1800s, and oil production attempts began in 1904 (Eardley 1963).
Extensive studies of hypersaline environments in various geographical locations have led to the isolation and characterization of the microbial communities found in these environments (e.g., Litchfield and Gillevet 2002; Burns et al. 2004; Maturrano et al. 2006; Mesbah et al. 2007; Clementino et al. 2008; Pagaling et al. 2009; Boutaiba et al. 2011; Ghai et al. 2011; Oren 2012; Podell et al. 2013). Classical biological and microbiological studies have found in GSL, only a limited variety of identifiable halophiles that have adapted to the various levels of salinity (Post 1977, 1981; Tsai et al. 1995; Wainø et al. 2000; Weimer et al. 2009). However, molecular characterization of the microbial biodiversity in GSL still remains very limited (Baxter et al. 2005; Parnell et al. 2011; Meuser et al. 2013). This type of study is warranted to shed the light on the microbial structure in GSL and will provide greater insights into the microbial diversity by sampling genomes from organisms not amenable to classical approaches.
In this study, we performed a survey of the archaeal and bacterial communities present in the North Arm and the South Arm of GSL by analyzing 16S rRNA gene sequences using phylogenetic approaches. We assessed the biodiversity detected in the lake and compared it with the microbial communities previously characterized in other hypersaline environments. We also examined the biogeographical patterns displayed by the microbial communities in this ecosystem. GSL represents a unique environment and it is clearly understudied in terms of its microbial ecology. Such examination of the microbial life in GSL provides a new perspective on the structure and composition of the microbial communities found in this extreme environment.
Materials and methods
Sample collection and extraction of genomic DNA
Surface water samples (a total volume of 100 ml for each sample) were collected in September and November 2004 from 20 different sampling sites located at GSL North Arm (Rozel Point) and GSL South Arm (Antelope Island State Park). The samples were collected from 17 various locations at Rozel Point, near the Spiral Jetty (41°25′N 112°39′W 1,281 m), in the immediate vicinity of oil seeps and from three different locations at Antelope Island State Park, near the shore of the lake (40°57′N 112°36′W 1,280 m). We received permission to collect in GSL from the Utah Division of Natural Resources (DNR) and this permit is on file at Brigham Young University. Our activities were not regulated by any other agency. In this study, our field research had no impact on vertebrate or protected species. During this sampling period, the water level in the lake was very low and the salinity had reached its maximum value in GSL North Arm (30 % w/v). By comparison to Antelope Island, Rozel Point is an oil seep environment; indeed the production rates of 5–10 barrels of oil per day were reported by Eardley (1963). This site is also known for its unusually high sulfur content up to 14 % per weight (Sinninghe Damsté et al. 1987). All water samples were stored in sterile Whirl–Pak plastic bags (Fisher Scientific) and kept at −20 °C until further analysis.
In order to obtain microbial DNA for sequencing, water samples (30 ml each) were centrifuged at 20,000 rpm for 1 h (Sorvall RC 5 Plus Superspeed Centrifuge, GMI Inc., Ramsey, MN, USA). After centrifugation, the supernatant was discarded and the rest of the water sample was added to the corresponding pellet for another round of centrifugation. Total genomic DNA was extracted from the pellets by the methods described in Crandall et al. (1999). The quantity of DNA from each sample was evaluated by measuring the optical density at 260 nm, and its quality was checked by electrophoresis in an agarose gel (1 %). The DNA (100 ng/μl) was stored at −20 °C in TE buffer (pH 8.0) prior to analysis.
16S rRNA clone library construction
In order to characterize the microbial communities present in this ecosystem, we generated 16S rRNA sequences from all 20 water samples collected in this study. PCR amplification of the 16S rRNA gene was performed using the primers 6F (5′-CGGTTGATCCYGCCGGM-3′) and 741R (5′-GACTACCSGGGTATCTAATCC-3′) for Archaea, and the primers 38F (5′-CAGGCCTAACACATGCAAGTC-3′) and 882R (5′-GTTTTAACCTTGCGGCCGTACTCC-3′) for Bacteria. These primers amplify a segment of the 16S rRNA gene (5′-end) with an average size of 850 bp. The size of the fragment analyzed is sufficiently discriminating to assess the microbial diversity in GSL, and is comparable to that used in other studies and therefore allows for a broad comparison (Cytryn et al. 2000; de Souza et al. 2001; Souza et al. 2006). PCR conditions were 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 60 °C (for Archaea) or 65 °C (for Bacteria) for 1 min, and 72 °C for 2 min 30 s, with a final 20-min extension at 72 °C.
The cloning and transformation procedure for all the samples under study were performed using the TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer recommendations. The successfully cloned fragments were sequenced with included M13 vector primers (forward and reverse primers) using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) on an ABI 3730XL DNA Analyzer.
16S rRNA gene library analysis
The 16S sequences were analyzed using Sequence Navigator Software (PE Applied Biosystems) and were checked for chimera formation using DECIPHER (Wright et al. 2012). Sequences were compared with each other and those with >97 % identity were clustered as operational taxonomic units (OTUs) using MOTHUR software v.1.31.2 (Schloss et al. 2009). This cut-off is a commonly used level for comparative analysis in microbial communities (Nam et al. 2011). For the grouping into OTUs, we used 175 archaeal clone sequences (149 sequences from GSL North Arm and 26 sequences from GSL South Arm) and 212 bacterial clone sequences (172 sequences from GSL North Arm and 40 sequences from GSL South Arm). We then compared our 16S rRNA sequences to the NCBI GenBank database (December 2013) and to the ribosomal database project (RDP, Release 11, Update 1, December 2013) (Cole et al. 2009). Since RDP is known to be more informative than GenBank for taxonomic assignment (Cole et al. 2007), we chose to use the comparison results from RDP to identify environmental sequences with the highest sequence identity for inclusion in our analyses. In addition, we included sequences from microbial species found in other hypersaline environments and/or those already characterized from GSL. Taxa found in other archaeal and bacterial phyla and in petroleum environments were also added to this collection for comparison purposes.
Each data set was aligned using MUSCLE, a multiple sequence alignment software (Edgar 2004), and the resulting alignments were checked using MacClade (Maddison and Maddison 2005). Because many environmental sequences used for the analyses are extremely divergent from the archaeal and bacterial ingroup taxa, and therefore difficult to align reliably, Gblocks v0.91b was used to select the conserved blocks in the alignments prior to phylogenetic analyses (Castresana 2000).
Phylogenetic analyses
The phylogenetic relationships from the combined data set were estimated using maximum likelihood analysis (Felsenstein 1981) with nodal support assessed via bootstrapping (100 pseudo-replicates) (Felsenstein 1985) as implemented in PhyML v3.0 (Guindon et al. 2010). Different sister groups to the archaeal and bacterial communities were chosen as outgroups for rooting our phylogenetic trees. For the archaeal sequence analysis, we selected the phyla Crenarchaeota, Korarchaeota, and Nanoarchaeota to serve as outgroups. For the bacterial sequence analysis, we selected Actinobacteria, Bacteroidetes, Firmicutes, Cyanobacteria, Spirochaetes, Chlamydiae, Deinococcus-Thermus, and Aquificae to serve as outgroups. These outgroups confirmed the stability of the topology of the trees. Model selection for these analyses followed the procedure outlined by Posada and Buckley (2004) as implemented in ModelTest v3.7 (Posada and Crandall 1998). The model chosen for both communities was GTR+I+G selected by the Akaike information criterion (AIC). Theoretically, AIC reduces the number of unnecessary parameters that contribute little to describing the data by penalizing more complex models.
Statistical analyses
The MOTHUR software was used to calculate archaeal and bacterial richness and diversity (Schloss et al. 2009). The diversity indices included the non-parametric richness estimators ACE, Chao1, the Simpson diversity index and the Shannon–Weaver diversity index. The library coverage was also calculated using MOTHUR software (Schloss et al. 2009). For all these estimates, we used the default setting with a 3 % cut-off in MOTHUR. Species evenness (species’ relative abundance) was calculated using Pielou’s evenness index (Pielou 1975). A value of evenness close to 1 means that the community is diverse, whereas a value close to 0 means that the community is not diverse.
Nucleotide sequence accession numbers
All the archaeal and bacterial sequences of the 16S rRNA gene clone library generated in this study were deposited in NCBI GenBank database under accession numbers KF160960 to KF161059 and KF585215 to KF585501.
Results
Biodiversity of the archaeal and bacterial clone libraries
In this study, we used a total of 175 archaeal and 212 bacterial clones. 58 OTUs were identified from the archaeal library and 42 from the bacterial library (Table 1). A comparison with environmental 16S sequences deposited in RDP website showed a different distribution in the archaeal and bacterial OTUs in GSL (Fig. 1). Of the total archaeal OTUs, 5 % showed less than 60 % identity with formally described lineages referenced in RDP and the majority of archaeal OTUs (57 %) displayed identity levels ranging from 60 to 90 % (Table 1; Fig. 1). Only 38 % of the total archaeal OTUs showed higher sequence identities (≥90 % identity). A different scenario was observed in the bacterial communities in GSL. The bacterial OTUs included a higher proportion of sequences (62 % of the total OTUs analyzed) displaying a high sequence identity in RDP (≥90 % identity). However, only 38 % of the total bacterial OTUs sequences showed in RDP a sequence identity ranging from 60 to 90 %, and none of the bacterial clone sequences showed a sequence identity lower than 60 % (Table 1; Fig. 1). In comparison with the archaeal OTUs in GSL, the majority of the bacterial clone sequences (62 % in comparison with only 38 % in archaeal OTUs) were more closely related to previously described environmental lineages. Furthermore, the comparison of our clone library with the sequences deposited in RDP showed that 78 % of the total archaeal OTUs (45 archaeal OTUs) and 43 % of the total bacterial OTUs (18 bacterial OTUs) did not associate with any of the previously known taxa and might represent novel environmental species yet to be determined (Table 1).
Phylogenetic analyses showed also a very high level of biodiversity among the archaeal and bacterial communities in GSL (Figs. 2, 3). All archaeal OTUs belonged to the phylum Euryarchaeota (Fig. 2). Even though most of the OTUs did not cluster with environmental sequences, representatives of the following genera: Natronomonas, Halorhabdus, Halorubrum, Haloquadratum, Haloferax, Halogeometricum, Haloarcula and Halobacterium were represented in our archaeal clone library (Fig. 2). Bacterial OTUs showed, in contrast to archaeal OTUs, a higher proportion of clones clustered with environmental sequences (Fig. 3). All bacterial OTUs grouped within the phylum Proteobacteria, except a single OTU that clustered within the phylum Firmicutes. Moreover, the majority of the bacterial OTUs clustered with environmental taxa known to degrade hydrocarbons: Shewanella, Halomonas, Idiomarina, Alcanivorax, Pseudomonas and Marinobacter (Fig. 3).
Phylogenetic analysis showing the biodiversity of the archaeal clone library in Great Salt Lake, UT, along with reference sequences belonging to different phyla. The archaeal OTUs described in this study are marked with a star symbol. Branch points with bootstrap values of >90 and 70–90 % are shown in dark and clear circles, respectively. Branch points without circles are not resolved (bootstrap values <70 %). An uncultured nanoarchaeote (AJ458436) was used as outgroup in this tree
Phylogenetic analysis showing the biodiversity of the bacterial clone library in Great Salt Lake, UT, along with reference sequences belonging to different phyla. The bacterial OTUs described in this study are marked with a star symbol. Branch points with bootstrap values of >90 and 70–90 % are shown in dark and clear circles, respectively. Branch points without circles are not resolved (bootstrap values <70 %). Chlamydophila abortus (AB001816) was used as outgroup in this tree
Diversity and richness analyses
Richness estimators (Chao1 and ACE) were significantly higher than the observed number of OTUs in both archaeal and bacterial communities (Table 2). These results indicate that the diversity observed in this study is, by far, an underestimate of the total archaeal and bacterial diversity present in GSL. A further sampling would yield a more accurate assessment of the diversity in these communities. Also, Shannon–Weaver and Simpson analyses indicated a high biodiversity within the archaeal (3.99 and 0.0018, respectively) and bacterial (3.67 and 0.0023) clone libraries (Table 2). The low level found in library coverage in both communities represents another indication for an incomplete representation of the biodiversity in these microbial communities (Table 2). A more extensive sampling in GSL is warranted. Moreover, values obtained of evenness for both archaeal and bacterial libraries (0.98) reinforce the statement of high biodiversity in GSL (Table 2).
Discussion
In this study, a culture-independent survey of the archaeal and bacterial communities was performed in the North (Rozel Point) and South (Antelope Island) Arms of GSL. Since the studies by Post in the 1970s (Post 1977), little information is available on the microbial community structure in GSL (Weimer et al. 2009; Parnell et al. 2009, 2011; Baxter et al. 2005). A very recent study (Meuser et al. 2013) examined the community assembly in the stratified water column in the South Arm of GSL, however knowledge on species composition of the archaeal and bacterial communities in this extreme environment still remains very limited. This ecosystem remains indeed understudied, especially when its large size and high number of microenvironments are taken into account. In this study, our analysis revealed a high biodiversity in this ecosystem and an abundance of yet-to-be described archaeal and bacterial taxa.
Many of our OTU clone libraries from GSL are unequivocally different and unique from other hypersaline environments, with 45 archaeal and 18 bacterial OTUs not affiliated with any of the previously described taxa. Similar findings have been observed in other extreme environments where diverse and novel microbial communities were also detected (Harris et al. 2013; Mesbah et al. 2007; Podell et al. 2013; Walker et al. 2005; Boujelben et al. 2012; Huber et al. 2002; Daffonchio et al. 2006).
The diversity indices in addition to the other statistical parameters estimated in this study also indicated that both the archaeal and bacterial communities in GSL were highly diverse and that a more extensive sampling in this environment was warranted to allow a better estimate of species richness. While this survey was modest compared to some environmental sampling efforts (Venter et al. 2004), it still represents a prerequisite step to assess the biodiversity in the microbial communities recovered from GSL.
Since a large proportion of the bacterial clones were clustered with petroleum-degrading bacteria, one might hypothesize that the abundance of yet-to-be described archaeal and bacterial OTUs in GSL could be explained by their ability to benefit from the presence and metabolism of petroleum hydrocarbons, particularly at Rozel Point. These new microbial communities might have evolved in this ecosystem to use petroleum as a carbon source. Furthermore, though oil degradation by halophilic bacteria has been well documented (Riis et al. 2003; Brakstad and Lødeng 2005), such a biological process has yet to be conclusively demonstrated in archaeal communities (Head et al. 2006; Tapilatu et al. 2010) despite some studies showing that these microorganisms are able to grow in presence of hydrocarbons (Raghavan and Furtado 2000; Tischer et al. 2012).
The current survey performed on microbial communities in GSL demonstrates the intriguing abundance of novel phylotypes in archaeal and bacterial clone libraries in addition to the high diversity in the overall microbial composition in this ecosystem. Similar findings have been also documented in recent studies (Weimer et al. 2009; Parnell et al. 2009, 2011). Based on the comparison with environmental sequences deposited in RDP and the low sequence identity found with previously described taxa, these new lineages seem to be exclusively present in GSL and might represent distinct microbial communities than the ones found in other hypersaline environments (Litchfield and Gillevet 2002; Sørensen et al. 2005; Lay et al. 2012).
Microbial biodiversity studies have led to two major conflicting hypotheses (Martiny et al. 2006). The first is the “Global Dispersal” hypothesis, which suggests that microorganisms are ubiquitous and have few barriers to gene flow resulting in similar microbial communities across different spatial scales and habitats (Baas-Becking 1934; de Wit and Bouvier 2006; O’Malley 2007; Finlay 2002). The alternative is the “Barriers to Dispersal” hypothesis, which suggests microbial community differentiation is driven by ecological and/or geographic barriers, showing mostly similar patterns as those seen in plants and animals (Whitaker et al. 2003; Horner-Devine et al. 2004; Bell et al. 2005; Fierer and Jackson 2006). Since this study showed an abundance of novel phylotypes that have not yet been described in other hypersaline environments, this finding supports the “Barriers to Dispersal” hypothesis, in which local factors in an environment dictate the selection of adapted organisms from a global pool (Daffonchio et al. 2006; Fierer and Jackson 2006; Rengefors et al. 2012; Logares et al. 2013). However, the mechanism driving “Barriers to Dispersal” requires further investigation. Adaptation driven by ecological parameters and/or mere physical isolation could be responsible for driving the microbial biodiversity in GSL (Whitaker et al. 2003; Papke and Ward 2004). Therefore, both ecological and geographic barriers may be the driving forces creating and maintaining biodiversity in GSL, and thus responsible for the development of global microbial community structure in this unique ecosystem. A recent study also showed that adaptation to high salt content was associated with a specific genome signature (Paul et al. 2008), and this finding may support the “Barriers to Dispersal” hypothesis in the GSL microbial community. An extensive sampling in this ecosystem combined with a thorough comparison with similar environments in other locations throughout the world is therefore needed to elucidate the questions related to the microbial biogeography in GSL. Furthermore, the composition of microbial assemblages in GSL points out spatial and evolutionary relationships of the microbiota in extreme environments, and suggests low levels of gene flow that maintains the distinct microbial community structure in this ecosystem (Souza et al. 2006; Rengefors et al. 2012). A better understanding of the microbial diversity in GSL will also shed the light into the biogeochemical processes in this ecosystem. The findings of this study provide a glimpse into the ecological relationships among microbial communities in GSL, as well as an additional perspective on the nature of life in such extreme environments.
Given our results, we are also attempting to culture these novel species belonging to the different new phylotypes to characterize better their biological properties and to elucidate the differences between these new microbial communities found in GSL and all the previously described archaeal and bacterial communities. The characterization of these new archaeal and bacterial lineages found in GSL will allow the preparation of probes for these specific communities (FISH analysis) to estimate their relative abundance and to monitor the quantitative changes in the community composition in this unique ecosystem over seasons and multiple years (Konno et al. 2013).
References
Baas-Becking LGM (1934) Geobiologie of inleiding tot de milieukunde. W.P. van Stockum and Zoon, The Hague
Baxter BK, Litchfield CD, Sowers K, Griffith JD, DasSarma PA, DasSarma S (2005) Great Salt Lake microbial diversity. In: Gunde-Cimerron N, Oren A, Plemenita A (eds) Adaptation to life in high salt concentrations in Archaea, Bacteria, and Eukarya. Springer, The Netherlands
Bell T, Ager D, Song JI, Newman JA, Thompson IP, Lilley AK, van der Gast CJ (2005) Larger islands house more bacterial taxa. Science 308:1884
Boujelben I, Gomariz M, Martínez-García M, Santos F, Peña A, López C, Antón J, Maalej S (2012) Spatial and seasonal prokaryotic community dynamics in ponds of increasing salinity of Sfax solar saltern in Tunisia. Antonie Van Leeuwenhoek 101:845–857
Boutaiba S, Hacene H, Bidle KA, Maupin-Furlow JA (2011) Microbial diversity of the hypersaline Sidi Ameur and Himalatt salt lakes of the Algerian Sahara. J Arid Environ 75:909–916
Brakstad OG, Lødeng AG (2005) Microbial diversity during biodegradation of crude oil in seawater from the North Sea. Microb Ecol 49:94–103
Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML (2004) Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl Environ Microbiol 70:5258–5265
Cannon JS, Cannon MA (2002) The southern Pacific Railroad trestle: past and present. In: Gwynn JW (ed) Great Salt Lake, an overview of change. Special Publication of the Utah Department of Natural Resources, Salt Lake City pp 283–294
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Clementino MM, Vieira RP, Cardoso AM, Nascimento AP, Silveira CB, Riva TC, Gonzalez AS, Paranhos R, Albano RM, Ventosa A, Martins OB (2008) Prokaryotic diversity in one of the largest hypersaline coastal lagoons in the world. Extremophiles 12:595–604
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35:D169–D172
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Crandall KA, Fetzner JW, Lawler SH, Kinnersley M, Austin CM (1999) Phylogenetic relationships among the Australian and New Zealand genera of freshwater crayfishes (Decapoda: Parastacidae). Austral J Zool 47:199–214
Cytryn E, Minz D, Oremland RS, Cohen Y (2000) Distribution and diversity of archaea corresponding to the limnological cycle of a hypersaline stratified lake (Solar Lake, Sinai, Egypt). Appl Environ Microbiol 66:3269–3276
Daffonchio D, Borin S, Brusa T, Brusetti L, van der Wielen PW, Bolhuis H, Yakimov MM, D’Auria G, Giuliano L, Marty D, Tamburini C, McGenity TJ, Hallsworth JE, Sass AM, Timmis KN, Tselepides A, de Lange GJ, Hübner A, Thomson J, Varnavas SP, Gasparoni F, Gerber HW, Malinverno E, Corselli C, Garcin J, McKew B, Golyshin PN, Lampadariou N, Polymenakou P, Calore D, Cenedese S, Zanon F, Hoog S (2006) Stratified prokaryote network in the oxic-anoxic transition of a deep-sea halocline. Nature 440:203–207
de Souza MP, Amini A, Dojka MA, Pickering IJ, Dawson SC, Pace NR, Terry N (2001) Identification and characterization of bacteria in a selenium-contaminated hypersaline evaporation pond. Appl Environ Microbiol 67:3785–3794
de Wit R, Bouvier T (2006) ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ Microbiol 8:755–758
Eardley AJ (1963) Oil seeps at Rozel Point. Utah Geol Mineral Survey Spec Stud 5:32
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631
Finlay BJ (2002) Global dispersal of free-living microbial eukaryote species. Science 296:1061–1063
Ghai R, Pašić L, Fernández AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD, Papke RT, Stepanauskas R, Rodriguez-Brito B, Rohwer F, Sánchez-Porro C, Ventosa A, Rodríguez-Valera F (2011) New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1:135
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Gwynn JW (1980) Great Salt Lake: a scientific, historical and economic overview. Utah Geol Mineral Surv Bull 116:83–96
Harris JK, Caporaso JG, Walker JJ, Spear JR, Gold NJ, Robertson CE, Hugenholtz P, Goodrich J, McDonald D, Knights D, Marshall P, Tufo H, Knight R, Pace NR (2013) Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J 7:50–60
Hassibe WR, Keck WG (1978) The Great Salt Lake. US Geological Survey, Reston
Head IM, Jones DM, Röling WF (2006) Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182
Horner-Devine MC, Lage M, Hughes JB, Bohannan BJ (2004) A taxa-area relationship for bacteria. Nature 432:750–753
Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO (2002) A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63–67
Konno U, Kouduka M, Komatsu DD, Ishii K, Fukuda A, Tsunogai U, Ito K, Suzuki Y (2013) Novel microbial populations in deep granitic groundwater from Grimsel Test Site, Switzerland. Microbiol Ecol 65:626–637
Lay CY, Mykytczuk NC, Niederberger TD, Martineau C, Greer CW, Whyte LG (2012) Microbial diversity and activity in hypersaline high Arctic spring channels. Extremophiles 16:177–191
Litchfield CD, Gillevet PM (2002) Microbial diversity and complexity in hypersaline environments: a preliminary assessment. J Ind Microbiol Biotechnol 28:48–55
Logares R, Lindström ES, Langenheder S, Logue JB, Paterson H, Laybourn-Parry J, Rengefors K, Tranvik L, Bertilsson S (2013) Biogeography of bacterial communities exposed to progressive long-term environmental change. ISME J 7:937–948
Maddison DR, Maddison WP (2005) MacClade 4: analysis of phylogeny and character evolution. Version 4.08a. Sinauer Associates, Sunderland
Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreås L, Reysenbach AL, Smith VH, Staley JT (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112
Maturrano L, Santos F, Rosselló-Mora R, Antón J (2006) Microbial diversity in Maras salterns, a hypersaline environment in the Peruvian Andes. Appl Environ Microbiol 72:3887–3895
Mesbah NM, Abou-El-Ela SH, Wiegel J (2007) Novel and unexpected prokaryotic diversity in water and sediments of the alkaline, hypersaline lakes of the Wadi An Natrun, Egypt. Microbiol Ecol 54:598–617
Meuser JE, Baxter BK, Spear JR, Peters JW, Posewitz MC, Boyd ES (2013) Contrasting patterns of community assembly in the stratified water column of Great Salt Lake, Utah. Microb Ecol 66:268–280
Nam YD, Jung MJ, Roh SW, Kim MS, Bae JW (2011) Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS One 6:e22109
O’Malley MA (2007) The nineteenth century roots of ‘everything is everywhere’. Nature Rev Microbiol 5:647–651
Oren A (2012) Taxonomy of the family Halobacteriaceae: a paradigm for changing concepts in prokaryote systematics. Int J Syst Evol Microbiol 62:263–271
Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276:734–740
Pagaling E, Wang H, Venables M, Wallace A, Grant WD, Cowan DA, Jones BE, Ma Y, Ventosa A, Heaphy S (2009) Microbial biogeography of six salt lakes in Inner Mongolia, China, and a salt lake in Argentina. Appl Environ Microbiol 75:5750–5760
Papke RT, Ward DM (2004) The importance of physical isolation to microbial diversification. FEMS Microbiol Ecol 48:293–303
Parnell JJ, Crowl TA, Weimer BC, Pfrender ME (2009) Biodiversity in microbial communities: system scale patterns and mechanisms. Mol Ecol 18:1455–1462
Parnell JJ, Rompato G, Crowl TA, Weimer BC, Pfrender ME (2011) Phylogenetic distance in Great Salt Lake microbial communities. Aquat Microbiol Ecol 64:267–273
Paul S, Bag SK, Das S, Harvill ET, Dutta C (2008) Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol 9:R70
Pielou EC (1975) Ecological diversity. Wiley, New York
Podell S, Ugalde JA, Narasingarao P, Banfield JF, Heidelberg KB, Allen EE (2013) Assembly-driven community genomics of a hypersaline microbial ecosystem. PLoS One 8:e61692
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Post FJ (1977) The microbial ecology of the Great Salt Lake. Microbiol Ecol 3:143–165
Post FJ (1981) Microbiology of the Great Salt Lake North arm. Hydrobiologia 81:59–69
Raghavan TM, Furtado I (2000) Tolerance of an estuarine halophilic archaebacterium to crude oil and constituent hydrocarbons. Bull Environ Contam Toxicol 65:725–731
Rengefors K, Logares R, Laybourn-Parry J (2012) Polar lakes may act as ecological islands to aquatic protists. Mol Ecol 21:3200–3209
Riis V, Kleinsteuber S, Babel W (2003) Influence of high salinities on the degradation of diesel fuel by bacterial consortia. Can J Microbiol 49:713–721
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Sinninghe Damsté JS, de Leeuw JW, Kock-van Dalen AC, de Zeeuw MA, de Lange F, Rijpstra WIC, Schenck PA (1987) The occurrence and identification of series of organic sulphur compounds in oils and sediment extracts: I. A study of Rozel Point Oil (USA). Geochim Cosmochim Acta 51:2369–2391
Sørensen KB, Canfield DE, Teske AP, Oren A (2005) Community composition of a hypersaline endoevaporitic microbial mat. Appl Environ Microbiol 71:7352–7365
Souza V, Espinosa-Asuar L, Escalante AE, Eguiarte LE, Farmer J, Forney L, Lloret L, Rodríguez-Martínez JM, Soberón X, Dirzo R, Elser JJ (2006) An endangered oasis of aquatic microbial biodiversity in the Chihuahuan desert. Proc Natl Acad Sci USA 103:6565–6570
Tapilatu YH, Grossi V, Acquaviva M, Militon C, Bertrand JC, Cuny P (2010) Isolation of hydrocarbon-degrading extremely halophilic archaea from an uncontaminated hypersaline pond (Camargue, France). Extremophiles 14:225–231
Tischer K, Zeder M, Klug R, Pernthaler J, Schattenhofer M, Harms H, Wendeberg A (2012) Fluorescence in situ hybridization (CARD-FISH) of microorganisms in hydrocarbon contaminated aquifer sediment samples. Syst Appl Microbiol 35:526–532
Tsai CR, Garcia JL, Patel BK, Cayol JL, Baresi L, Mah RA (1995) Haloanaerobium alcaliphilum sp. nov., an anaerobic moderate halophile from the sediments of Great Salt Lake, Utah. Int J Syst Bacteriol 45:301–307
Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74
Wainø M, Tindall BJ, Ingvorsen K (2000) Halorhabdus utahensis gen. nov., sp. nov., an aerobic, extremely halophilic member of the Archaea from Great Salt Lake, Utah. Int J Syst Evol Microbiol 50:183–190
Walker JJ, Spear JR, Pace NR (2005) Geobiology of a microbial endolithic community in the Yellowstone geothermal environment. Nature 434:1011–1014
Weimer BC, Rompato G, Parnell JJ, Gann R, Ganesan B, Navas C, Gonzalez M, Clavel M, Albee-Scott S (2009) Microbial biodiversity of Great Salt Lake, Utah. In: Oren A, Naftz DL, Palacios P, Wurtsbaugh SJ (eds) Saline lakes around the world: unique systems with unique values (Natural Resources and Environmental Issues, vol. 15). S.J. and Jessie E. Quinney Natural Resources Research Library, Logan, pp 15–22
Whitaker RJ, Grogan DW, Taylor JW (2003) Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78:717–725
Acknowledgments
We gratefully thank Sunny S. Drysdale and Scott B. Dahlquist for their technical assistance in collecting samples and sequence data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Rights and permissions
About this article
Cite this article
Tazi, L., Breakwell, D.P., Harker, A.R. et al. Life in extreme environments: microbial diversity in Great Salt Lake, Utah. Extremophiles 18, 525–535 (2014). https://doi.org/10.1007/s00792-014-0637-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-014-0637-x