Abstract
Here we describe the diversity and activity of sulfate reducing bacteria along a salinity gradient in four different soda lakes from the Kulunda Steppe (South East Siberia, Russia). For this purpose, a combination of culture-dependent and independent techniques was applied. The general bacterial and SRB diversity were analyzed by denaturing gradient gel electrophoresis (DGGE) targeting the 16S rDNA gene. DNA was used to detect the microbial populations that were present in the soda lake sediments, whereas ribosomal RNA was used as a template to obtain information on those that were active. Individual DGGE bands were sequenced and a phylogenetic analysis was performed. In addition, the overall activity of SRB was obtained by measuring the sulfate reduction rates (SRR) and their abundance was estimated by serial dilution. Our results showed the presence of minor, but highly active microbial populations, mostly represented by members of the Proteobacteria. Remarkably high SRR were measured at hypersaline conditions (200 g L−1). A relatively high viable count indicated that sulfate reducing bacteria could be highly active in hypersaline soda lakes. Furthermore, the increase of sodium carbonate/bicarbonate seemed to affect the composition of the microbial community in soda lakes, but not the rate of sulfate reduction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soda lakes are a specific type of salt lake with high to extremely high carbonate alkalinity, a pH from 9 to 11, and a moderate to extremely high salinity. They are spread all over the world, but located, as most inland salt lakes, in arid and semi-arid areas where the evaporative climate favors accumulation of salts in local depressions. The major ions in soda lake brines are sodium, carbonate/bicarbonate, chloride and sulfate, whereas calcium is virtually absent and magnesium only present at very low concentrations. In contrast to other alkaline environments, such as low-salt alkaline (“cement”) springs, soda lakes maintain a stable alkaline pH due to the high buffering capacity of the soluble carbonates. These double extreme conditions (i.e. high pH and high salinity) make soda lakes a unique ecosystem.
In the last decade, special attention has been given to the investigation of the microbial communities in soda lakes using both traditional isolation methods (Duckworth et al. 1996; Sorokin et al. 2004; Sorokin and Kuenen 2005b) and molecular biology techniques (Humayoun et al. 2003; Rees et al. 2004; Scholten et al. 2005; Foti et al. 2007). Few reviews (Zavarzin et al. 1999; Jones et al. 1998) summarize these results, showing that soda lakes contain representatives of the major trophic and phylogenetic groups of prokaryotes, and that they can be considered as autonomous systems, in which cycling of nutrients is close to complete. The most well-studied soda lakes are those located in the East African Rift Valley (Duckworth et al. 1996; Rees et al. 2004), in the Libyan desert (Wadi Natrun)(Imhoff et al. 1979) and in North America, i.e. Mono Lake (California) (Humayoun et al. 2003; Scholten et al. 2005a) and Soap Lake (Washington) (Sorokin et al. 2007a). There are also data available on soda lakes in India (Wani et al. 2006) and Central Asia (Sorokin et al. 2004; Gorlenko et al. 2004; Ma et al. 2004). This last region accommodates a great number of steppe soda lakes, especially in the Inner Mongolia (North east China) and south Siberia (Transbaikal region, Tuva Republic and Kulunda Steppe).
Except for the study conducted by Issancheko et al. (1951) between 1933 and 1935, almost nothing is known about the microbial communities in soda lakes of the Kulunda Steppe. In contrast to tropical African lakes and deep stratified North American lakes, the Central Asian lakes are very shallow and usually very small, in which the total concentration of salts and chemistry may differ considerably among nearby lakes as well as during the year. Furthermore, lakes are subjected to unstable water and temperatures regimens between −40°C and +40°C, causing frequent fluctuations of the water level and salinity.
It has been hypothesized that the diversity of metabolic types of prokaryotes decreases with salinity for thermodynamic reasons (Oren, 1999). Several studies have been performed on systems with saline gradients (Sörensen et al. 2004; Nübel et al. 2000; Casamayor et al. 2002), especially estuaries (Crump et al. 2004; Schultz and Ducklow 2001), to investigate the influence of salinity on the composition of the microbial community. However, estuaries have very low salinities compared to salt lakes or salterns (Benlloch et al. 2002; Sörensen et al. 2004). Commonly, thalassohaline ecosystems are object of investigation, in which sodium, magnesium, chloride and sulfate are the major components of the brines. In soda lakes sodium carbonate/bicarbonate is the major salt in solution, which has different physical and chemical properties, such as being a two times weaker electrolyte than sodium chloride. For this, a less energetically expensive osmotic adaptation is needed than demanded for life in NaCl brines (Sorokin and Kuenen 2005b). Nevertheless, organisms inhabiting soda lakes have to deal with high alkaline conditions, which represent an extra stress factor. Therefore, it was of interest to study the microbial community composition in soda lakes, and how this was affected by an increase in sodium carbonate/bicarbonate concentration.
Since the sulfur cycle is one of the most active cycles in soda lakes, special attention was given to the sulfate-reducing bacterial community (SRB). It has been hypothesized (Oren 1999; Ollivier et al. 1994) that specific groups of SRB, i.e. complete oxidizers, stop growing at salt concentrations above 130 g L−1. So, it was of interest to investigate the presence of these different phylogenetic groups of sulfate-reducers along a salinity gradient.
In this study we investigated the bacterial diversity and the major actively growing groups, based on ribosomal RNA determination, in four soda lakes with increasing salinity. For the first time the dominant active populations were detected in these extreme environments by targeting the 16S rRNA gene from the lake sediments. A combination of culture-dependent and -independent techniques was applied to investigate the SRB community. Identification of sulfate-reducers was done by DGGE analysis of the 16S rRNA genes, while the activity was determined by measuring the sulfate reduction rates (SRR).
Our results showed the presence of less abundant, but highly active microbial populations, mostly represented by members of the Proteobacteria. The detection of active SRB from sediment samples by targeting the 16S rRNA gene was not completely successful. However the remarkably high sulfate reduction rates (SRR) measured at hypersaline conditions (200 g L−1) and the relatively high viable counts indicated that this group of organisms was active in these hypersaline soda lakes. Furthermore, an increase of salinity seemed to affect the composition of the microbial community in soda lakes, but did not affect the rate of active processes, such as sulfate reduction.
Materials and methods
Site description
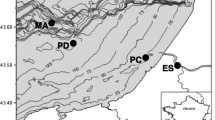
The sampled lakes are located in the southern part of the Kulunda Steppe, in south-eastern Siberia (Altai, Russia), along the north-eastern border with Kazachstan, about 320 km south–west of Barnaul (Altai capital), near the village Mikhaylovskiy (Fig. 1). The only information on the geology and chemistry of this area is available from the results of the Kulunda Expedition of the Russian Academy of Sciences taken place between 1933 and 1935 (Issachenko 1951). Lakes present in this region are generally very small and shallow, except for the large chloride–sulfate lakes with neutral pH in the north, such as Kulunda Lake, Lake Burlinskoe, and Lake Big Yarovoe. In the south of the Kulunda Steppe, the lakes are much smaller and some of them belong to the soda lake type. The concentration and nature of the salts present as well as their sediment structure may differ even between closely situated lakes. Except for Cock Lake, all investigated lakes (see Table 1) are located close to each other, i.e. within a diameter of 25 km. They belong to two systems: the Tanatar and the Bitter Lakes System, and both are part of a larger system, which is locally known as the “Salt Lake Steppe”. Although Cock Lake does not belong to the above-mentioned systems of lakes being situated ca 50 km north–west, it is very similar for its characteristics to the lakes in the Bitter Lake System. Characteristic feature of all the investigated lakes is the mineral deposition of ancient trona (i.e., sodium carbonate/bicarbonate), which confer the high salinity and alkalinity. Cock Lake was the first source of commercial soda in the Soviet Union since 1923, while Tanatar system is still actively mined for sodium carbonate. Cock Lake and Tanatar-3 presented a sandy-clay type of sediment, whereas lakes Tanatar-1 and Bitter-1 presented a black-clay type of sediment, with a strong sulfide smell. Sediment samples were taken from the mentioned lakes with increasing salinity in July 2007. The physical and chemical parameters of the lakes are presented in Table 1.
Location of the investigated soda lakes in the Kulunda Steppe (Altai, Russia). a Large scale map including Cock Lake. The lakes within the square are shown in detail in Fig. 1b. b Detailed map showing lakes Tanatar-1 (T1), Tanatar-3 (T3), and Bitter-1
Chemical analysis and sampling
Field measurements included pH, conductivity, and carbonate alkalinity in brine water. pH and conductivity were measured using a field meter supplemented with pH and conductivity probes (WTW, model pH/cond340, Weilheim, Germany). The pH probe was calibrated in saline buffers containing 0.5–2 M total Na+ (Sorokin 2005). The conductivity probe was calibrated in NaCl and carbonate buffer with pH 10 from 10 to 80 g L−1. At higher values, the samples were diluted with distilled water. Alkalinity was measured by titration with 1 M HCl using phenolphthalein (carbonate alkalinity) and methyl orange (bicarbonate alkalinity) as indicators. The salt concentration inferred from the conductivity measurements was verified by gravimetry in the laboratory. Samples were taken for nucleic acid extraction, sulfate reduction rate measurements and viable counts. Sediment cores (top 20 cm) and overlaying water were sampled manually along the littoral, using a corer. Sediment cores were divided into different layers, based on color, and stored in 50 ml Falcon tubes. Samples for nucleic acid extraction were mixed with RNAlater (Ambion, UK) in a 1:1 volume ratio. All tubes were kept at 4°C during transportation to the laboratory, and stored at −20°C until further analysis.
Sulfate reduction rates
Sulfate-reduction rate (SRR) experiments were conducted in situ and measured in 5-ml syringes capped with butyl rubber stoppers using the 35S-SO4 2− methodology (Lein et al. 2002). After 1–2 days of incubation at ambient temperature, the sediments were fixed with 10 M KOH and further processed in the laboratory. Additional analysis of total SO4 2− and CH4 content in the sediment pore water was performed in the laboratory after centrifugation of 2 cm3 sample using ion-exchange chromatography (Biotronic) for sulfate and by gas chromatography for methane.
Additional experiments to study the influence of pH/salt and nutrient additions on indigenous SRB populations were performed in the laboratory with sediment slurries (top 5 cm layer) from Cock Lake and Tanatar-1. Nutrient addition experiments were performed with native sediment slurries after addition of nitrogen (100 μM NH4Cl), phosphorus (10 μM K2HPO4) and electron donors such as hydrogen, lactate and acetate (1 mM) as substrates. The influence of pH and salt was examined after separation of the sediment solids from the pore brines by centrifugation. The influence of pH was investigated by suspending the sediments in buffers containing 5 mM sodium sulfate, varying the pH between 7 and 11 and keeping the original salinity. The influence of salt was investigated at pH 10 with increasing concentration of sodium carbonates. The SRR rates were measured in the same way as for the in situ measurements.
Quantification of sulfate-reducing bacteria by serial dilutions
Quantification of haloalkaliphilic SRB was performed by serial decimal dilutions in Hungate tubes. The top 5 cm sediment layers were used as inoculum. Low-salt (0.6 M total Na+) and high-salt (4 M total Na+) carbonate medium at pH 10 (Sorokin and Kuenen 2005a) was used with either H2 or acetate (20 mM plus 0.5 g L−1 yeast extract) as an energy source. The inoculation was done directly in the field. Growth was monitored within 4–8 weeks incubation at room temperature by estimating the sulfide production (Trüper and Schlegel 1964). Tubes with the highest positive dilutions were then subjected to DGGE analysis.
Nucleic acids extraction
Prior to the nucleic acid extraction, RNAlater was removed by 1 min centrifugation at 5,000 rpm. The sediments were then washed three times with 1 M NaCl and 10 mM Tris (pH 7.5) to lower the pH and the salt concentration. Genomic DNA was extracted from circa 2 g of sediment using the UltraClean Soil DNA Extraction Kit (MoBio Laboratories, USA), following the manufacturer’s instructions with minor modification. Two hundred microlitres of 0.1 M AlNH4(SO4)2 was added to the sediment to remove potential PCR inhibitors, such as humic acids (Braid et al. 2003). For RNA extraction, 7 g of sediments were divided in 2-ml screw cap tubes and washed as described above. After the washing step, bead beating and the addition of 120 μl of preheated MBER® (Max Bacterial Enhancement Reagent, Invitrogen, The Netherlands) were added to each tube and incubated for 4 min at 95°C. Cells were lysed by adding 0.7 ml TRIzol® (Invitrogen, The Netherlands), a mono-phasic solution of phenol and guanidine isothiocyanate and homogenized for 45 s, followed by 5 min incubation at room temperature to allow the complete dissociation of nucleo-proteins complexes. The TRIzol® reagent maintains the integrity of the RNA during sample homogenization, while disrupting cells and dissolving cell components. After homogenization cold chloroform was added (0.1 ml/0.5 ml TRIzol®). Subsequently, the samples were vigorously shaken manually for 15 s and incubated at room temperature for 3 min followed by 15 min centrifugation at 10,000 rpm and 4°C. The aqueous upper phase was then transferred to a new tube, 0.3 ml cold isopropanol was added and mixed to precipitate the RNA. After 10 min incubation at room temperature, the pellet was collected by centrifugation at 10,000 rpm for 10 min and washed with 75% (v/v) ethanol. Samples were then vortexed and centrifuged for 5 min at 8,000 rpm. The supernatant was discarded, while the pellet was air dried and dissolved in 40 μl RNA-free water. Subsequently, DNA traces were removed using the Turbo DNA free kit (Ambion, UK) and the resulting DNA-free RNA was purified with the Qiagen RNAeasy MiniEluteKit (Qiagen, the Netherlands), following manufacturer’s instructions. All reagents, plastic and glassware were RNAase free.
RT-PCR, PCR and DGGE
RNA was converted to cDNA by reverse transcriptase using the iScript cDNA Synthesis Kit® (BioRad, The Netherlands), following the manufacture’s instructions. The purity of the extracted RNA was checked by 16S rRNA gene amplification using RNA as template; no PCR product was obtained indicating the absence of contaminating DNA. From the genomic DNA and cDNA. a fragment of the 16S rRNA gene suitable for DGGE, was amplified. A nested PCR approach was used for the amplification of the 16S rRNA gene. First the whole gene was amplified using the primer GM3F/GM4R (Brinkhoff et al. 1998). The product was then excised from the agarose gel, purified using the Qiagen Gel Extraction Kit (Qiagen, the Netherlands) and used as a template for the amplification of the a gene fragment suitable for DGGE using primer combination 341F + GC/907R (Schäfer and Muyzer 2001). To detect sulfate-reducing bacteria, six primers pairs specific for the 16S rRNA gene of different SRB groups (Table 2; Daly et al. 2000) were used as described by Dar et al. (2005). PCR products of each amplification step were excised from the gel and purified before the following PCR step to avoid non-specific amplification. The obtained PCR products were then subjected to DGGE analysis. DGGE was performed as described by Muyzer et al. (1993), using a denaturing gradient of 35 to 60% denaturants in 8% polyacrylamide gel. Individual bands were excised, reamplified, and run again on a denaturing gradient gel to check their purity. PCR products for sequencing were purified using the Qiaquick PCR purification kit (QIAGEN).
Comparative sequence analysis
The sequences were first compared to sequences stored in GenBank using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/BLAST). Subsequently, the sequences were imported into the ARB software program (Ludwig et al. 2004), automatically aligned, and added to a phylogenetic tree using the Quick-add tool. Sub-trees were then built using the Neighbor Joining algorithm with automatic selected correction settings.
Nucleotide sequence accession number
The sequences determined in this study were deposited in GenBank under the following accession numbers: EF622422–EF622488.
Results
Physical–chemical characterization of the sediments
The investigated lakes were less than 1 km in diameter and had a maximum depth of 1 m. The salinities varied from 60 to 200 g L−1 and the pH from 10 to 10.3 (see Table 1). The dominant salt in solution was sodium carbonate/bicarbonate and the total alkalinity was in the range of 0.9–2.82 M. Sulfate concentrations in sediment pore waters correlated with total salt and in general were very high except for Bitter Lake in which, despite the extremely high salinity, the sulfate concentration was nearly depleted (Table 1). Free sulfide (up to 0.5 mM) was also detectable in the pore waters of the top 5 cm sediment layer in Tanatar-1 and Bitter Lake. In Bitter Lake the highest methane content was measured in the sediments and even in the brines, suggesting that both sulfate reduction and methanogenesis are active.
Nucleic acids extraction and DGGE analysis of 16S rRNA genes
Nucleic acids extraction and PCR amplification of the 16S rRNA gene was successful for all analyzed samples. PCR amplification using specific primers for SRB groups was successful with most of the samples when targeting group 1 (Desulfotomaculum, DFM), group 5 (Desulfosarcina, DCC) and group 6 (Desulfovibrio–Desulfomicrobium, DVV). One, four and no positive amplifications were obtained targeting group 2 (Desulfobulbus, DBB), group 4 (Desulfobacter DSB), and group 3 (Desulfobacterium, DBM), respectively. In some cases amplification products were identified as non-specific, resulting in the detection of different phylogenetic groups than the targeted ones (see Fig. 4).
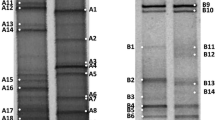
Amplicons obtained from genomic DNA and from cDNA (obtained after reverse transcription of the RNA) were then analyzed by DGGE. An average of 4 bands was observed for both DNA- and RNA-derived profiles (Fig. 2) using primers for the 16S rRNA genes of Bacteria. The DNA- and RNA-derived profiles were found to be highly different when analyzing the same samples (Fig. 2).
DGGE analysis of the bacterial 16S rRNA gene from sediment samples of the investigated soda lakes from the Kulunda Steppe. Lanes 1–6 Cock Lake; lanes 7–10 lake Tanatar-3; lanes 11–14 lake Tanatar-1; lanes 15–18 lake Bitter-1. Sediment depths have been analyzed for each lake (see Table 1). DNA (open square)- and RNA (filled circle)-derived profiles of the same samples are situated close to each other. Bands indicated by a white dot have been excised from the gel and sequenced
DGGE analysis of PCR products obtained with the primer pairs specific for the 16S rRNA genes of different SRB groups (results not shown) gave complex patterns for both group 1 and 6, whereas for group 2 and 4 just one dominant band was detected, corresponding to Desulfobulbus propionicus and Desulfosalina propionicus respectively. From the highest positive tube of the dilution series for the SRB enrichment, 1 and 5 bands were obtained, using hydrogen or acetate as electron donor, respectively. Almost all sequences grouped with members of the low G + C gram-positive bacteria and not to SRB of the Delta-proteobacteria.
The DGGE analysis of the different soda lakes did not show a decrease in the number of bands at different sediment depths or along the salinity gradient, except for the RNA-derived profiles obtained for Lake Tanatar-1 and Bitter-1 (Fig. 2). Rather, the composition of bacterial taxa decreases at higher salinities (Fig. 3). In Cock Lake, the lowest saline lake (60 g L−1), ten phylogenetic groups were detected, in Tanatar-3 (112 g L−1), eight groups and in Tanatar-1 (200 g L−1) and Bitter Lake (200g L−1), 3 and 5, respectively (see Fig. 3).
Microbial diversity along a salinity gradient
From the different DGGE gels obtained targeting the 16S rRNA gene, a total of 72 partial sequences have been analyzed phylogenetically. An overview of the obtained results is shown in Table 2 and Fig. 4.
Phylogenetic trees based on the 16S rRNA gene. The open squares and the closed circles represent sequences retrieved from DNA- and RNA-derived DGGE profiles, respectively. The stars indicate sequences or organisms originated from (hyper) saline/alkaline habitats. The initial of the lake is followed by the sediment layer, and eventually by the SRB-specific group set of primers used (see Table 2). a Proteobacteria, b Clostridia, c Actinobacteria, d Bacilli, e Bacteroidetes–Chlorobi
The less saline Cock Lake showed the highest bacterial taxa richness (Fig. 3). The top 10 cm sediments were characterized by the presence of sequences affiliated to the halo (alkaliphilic) Delta-proteobacterial SRB, such as D. propionicus and Desulfonatronum thiodismutans, to the Gamma-proteobacterial Acinetobacter junii and to the actinobacterium Propionibacterium acnes. The sequences retrieved from the RNA-derived DGGE profiles, thus presumed to correspond to the active population, belonged to the Cyanobacteria (i.e. Oscillatoria neglecta), Deinococcus, Actinobacteria and Beta-proteobacteria (i.e. Tepidimonas aquatica). The deepest sediment layer (10–20 cm) was mostly characterized by sequences related to uncultured low G + C organisms. From RNA-based DGGE’s, sequences related to the alphaproteobacterium Brevundimonas and to uncultured green sulfur bacteria (Fig. 2; lane 6; band a) were observed.
In lake Tanatar-3 (112 g L−1) members of the Alphaproteobacteria and Deinococcus–Thermus were detected. Sequences retrieved from DNA-based profiles were affiliated to the Delta-proteobacteria (D. propionicus and D. lacustre), Actinobacteria, Clostridia and Cyanobacteria (Leptolyngbya sp., i.e., Fig. 2, lane 7, band a)). The sequences presumed to correspond to the active population were related to the actinobacterium P. acnes, the gammaproteobacterium M. osloensis and Stenotrophomonas sp., the betaproteobacterium Massilia sp. (Fig. 2) and to the Bacillus group.
The lakes with highest salinity, i.e. Lake Tanatar-1 and Bitter-1 (200 g L−1), showed the poorest bacterial diversity (Fig. 3). Both were mostly represented by low G + C organisms and Delta-proteobacterial SRB related to D. propionicus. The latter were predominant in the RNA-based DGGE`s obtained by targeting the SRB groups specific 16S rRNA gene (data not shown). The top 4 cm sediment of Lake Bitter-1 was found to be particularly rich in sequences belonging to the Proteobacteria (see Fig. 2, lane 17 and Table 2) and all of them were retrieved from the RNA-derived profiles.
Remarkably, almost all identified sequences were related to organisms isolated from halo (alkaliphilic) environments (Fig. 4).
Sulfate-reducing activity
High sulfate reducing-activity was observed in the near bottom water and in the sediment layers of all four investigated soda lakes (Table 1). The rates were comparable with previous measurements (Foti et al. 2007) and, in general, corresponded to characteristic values of eutrophic marine sediments (Lein et al. 2002). Salinity seems to have no effect on the sulfate reduction process in the studied soda lakes. In fact, the highest SRR (1.426 103 μmol dm−3 day−1) were observed in Bitter Lake, at highest salinity conditions (200 g L−1).
Results of the sediment slurry experiments to investigate the influence of pH and salt concentration suggested that the SRB populations in low-salt Cock Lake were limited by electron donors, and not by inorganic nutrients, i.e. nitrogen and phosphor, while in high-salt Tanatar-1 lake both inorganic nutrients, and electron donors were moderately stimulating (Fig. 5a). Furthermore, both populations were active between pH 8 and 10 (Fig. 5b), but differed in salt optimum-tolerance (Fig. 5c), although not as much as might be expected from the actual difference in salt content in the two lakes.
Influence of nutrients (a), pH (b), and salt (c) on SRR in the top sediment layer of Cock Lake (gray columns or open circles) and lake Tanatar-1 (black columns or closed circles), incubated for 48 h at 30°C. pH profiles were measured at 0.6 M total Na+ for the Cock Lake sediments, and at 2 M Na+ for the lake Tanatar-1 sediments. Salt profiles were measured at pH 10 with sodium carbonate/bicarbonate
The presence of active SRB populations in sediments of the investigated lakes was confirmed by relatively high viable counts obtained both at high and low-salinities (Table 1). However, the heterotrophic cultures with acetate and yeast extract proved to be unstable during further transfers. In contrast, autotrophic enrichments with H2 were active and transferable even at 4 M of total Na+ with prominent development of relatively large vibrio’s resembling the extremely natronophilic sulfite-disproportionating isolate ASO3-1, that previously was isolated from Kulunda soda lakes (Foti et al. 2007).
Discussions
Microbial diversity along a salinity gradient
Here we describe, for the first time, the diversity and activity of microbial communities in general and SRB communities in particular along a salinity gradient in different soda lakes. For this purpose we used DGGE analysis of 16S rRNA gene fragments obtained from genomic DNA (as a measure for diversity) and from rRNA (as a measure for activity). In addition, sulfate reduction rates were measured. Salinity did affect the general microbial composition, but not the sulfate reducing activity. As reported in other studies (Wu et al. 2006; Bennloch et al. 2002), the number of DGGE bands did not show a negative correlation with increasing salinity, rather a decrease in the richness of bacterial genera was observed. This suggests an increase of the microdiversity, within the remaining phylogenetic group, at higher salinities.
At the highest salinities, the dominant actively growing populations were belonging to the Delta and Gamma-proteobacteria. The latter were also detected previously in hypersaline lakes (Wu et al. 2006; Bennloch et al. 2002), together with organisms belonging to the Alphaproteobacteria. In the literature, little information can be found about the microbial diversity along salt gradients in athalassophilic habitats; most of the studies have been conducted either on estuaries, with overall salinities 10–100 times lower than in hypersaline lakes, or on sea solar salterns. Furthermore, in most of these studies water and brines were the target and not the sediments, making comparison difficult or impossible.
The highest bacterial diversity was observed in Cock Lake (60 g L−1) and Tanatar-3 (120 g L−1) with actively growing populations affiliated to members of the Proteobacteria and bacilli. At the lowest salinity, a sequence corresponding to the cyanobacterium Oscillatoria neglecta was detected, which is probably responsible of the bulk of primary production. Interesting was the detection of one sequence closely related the N 2 -fixing cyanobacterium Leptolyngbia sp. PCC7104. So far, no cyanobacteria capable in N 2 -fixation have been detected or isolated from soda lakes, and so it is still not clear which organisms are responsible for this process. Hydrolytic (bacilli, Bacteroidetes), lipolytic (Stenotrophomons sp.), and fermentative (Propionibacteria, Clostridia, lactobacilli) bacteria were also detected as actively growing. Interesting was the identification in Cock Lake of two sequences related to the Beta-proteobacteria (Tepidimonas aquatica) and to an uncultured Chlorobium-like organism, both of which might be involved in the sulfur and carbon cycle. Tepidimonas aquatica is a slightly thermophilic, facultatively chemolithoheterotrophic bacterium able to oxidize thiosulfate and tetrathionate, and able to grow up to pH 8.5 (Freitas et al. 2003). Since the phylum green sulfur bacteria (GSB) are known to be a homogeneous physiological group, it can be assumed that members of this group inhabiting soda lakes might be involved in the sulfur cycle. This shows the necessity of looking more closely at the anaerobic phototrophic green sulfur bacteria in soda lakes. This Chlorobium-like sequence was detected in the deepest sediment layer, in which low light intensity and high sulfide are favorable for GSB. Unexpectedly the chemolithoautrophic sulfur-oxidizing bacteria of the genera Thioalkalivibrio and Thioalkalimicrobium were not detected in this study despite the fact of high viable numbers (up to 106 cell cm−3) and the isolation of 12 extremely natronophilic Thioalkalivibrio strains in pure culture (D. Sorokin, unpublished results). This clearly shows the limitation of the DGGE analysis method, which detects major components, but is less suitable than cloning when microbial diversity is analyzed.
At the highest salinities a decrease of the bacterial genera richness was observed, with the actively growing groups in these lakes represented by the Delta and Gamma-proteobacteria, together with a minority of Clostridia (Natronoanaerobium halophilum). Saprophytic (Acinetobacter sp.), lipolytic (Stenotrophomonas sp.) and saccharolytic (Eubacterium sp., Bacteroidetes) bacteria were the dominant active groups in the Bitter Lake sediments.
Only few of the identified sequences were related to culturable organisms. Unexpectedly, none of those have been described as tolerant halo-alkaliphilic or halo-alkalitolerant (e.g. Stenotrophomonas sp., Massilia sp., Brevundimonas sp.). Still, the majority (90%) of the identified sequences were related to representatives, i.e. uncultured organisms, from alkaline and saline environments (Fig. 4), indicating that many haloalkaliphilic phylotypes remain unknown. One such example is a new cluster of Actinobacteria, from which the first culturable representative recently has been described. Strain ANL-iso2 was enriched and isolated from Kulunda Steppe soda lake sediments using isobutyronitrile as the only substrate. It is specialized in the degradation of C3-C6 aliphatic nitriles (Sorokin et al. 2007b).
Sulfate reduction rates (SRR) and sulfate reducing community
Remarkably high SRR values have been observed in all four investigated soda lakes, especially in the top organic-rich sediment layer, with the exception of the sandy sediments of Tanatar-3, in which we measured the highest SRR in the deepest layer. The highest SRR were found in Bitter Lake-1, an organic-rich hypersaline lake. In contrast to other studies (Sörensen et al. 2004; Brandt et al. 2001), in which a remarkable decrease of the sulfate-reducing activity at salt concentrations greater than 12% was documented, we did not observe inhibition of SRR by increasing salinity. Brandt et al. investigated three different stations located in the moderate and hypersaline arms of the Great Salt Lake, Utah. A difference of factor 10 was observed between the stations at concentrations 120 and 274 g L−1 of NaCl. Sörensen (2004) studied the salinity responses of the benthic microbial communities in the Solar salterns (Israel) and they also observed a strong inhibition of the sulfate reduction activity at salinity of 215 g L−1. In case of the soda lakes investigated in this study, the highest SRR were found at the highest salinity. To the best of our knowledge, these high SRR have never been measured before in hypersaline soda lakes. Only Sorokin et al. (2004) reported a higher measurement in a Mongolian soda lake, but at lower salinity (60 g L−1). Life at high salt concentrations is energetically costly and organisms with a dissimilatory metabolism, which yield low energy, e.g. autotrophic nitrification, methanogenesis and complete-oxidizer SRB, may not be able to deal with high salinities for thermodynamic reasons (Oren 1999). However, soda lakes are a specific type of saline lake in which NaHCO3/Na2CO3 is the major salt in solution, a two times weaker electrolyte than sodium chloride, demanding approximately two times less energy to adapt to osmotic stress (Sorokin and Kuenen 2005b). Furthermore, at concentrations higher than 2 M total Na+, sodium carbonates are present only in their undissociated form, causing less stress than sodium chloride, which is fully dissociate up to saturation (5 M Na+). This might be one of the reasons of the observed unexpectedly high SRR in the investigated hypersaline soda lakes.
In a recent work conducted on two different soda lakes, i.e. Mono Lake and Searles Lake, Kulp et al. (2007) found different results. They observed a reduction of the SRR in Mono Lake with increasing salinity and no SRR at all was measured in Searles Lake, in which the sulfate reduction activity seemed to be inhibited by the high concentrations of borate. However, they did impose the salinity gradients in the laboratory, without analyzing similar lakes with increasing salinities, as we did.
Even if we measured the highest SRR at the highest salt conditions, SRR incubation experiment conducted at different salt concentration (see Fig. 5c) showed the optimum at 1 M total Na+, which was two times lower than the actual salt concentration. Furthermore, the optimum pH measured in the similar experiment was also lower (pH 8.5–9) than the natural pH, suggesting that the indigenous SRB population is living under stress in these extreme environments. Spiking experiments demonstrated that the SRB populations are limited by availability of electron donors at both salinities, but especially in low-salt lakes, whereas nitrogen and phosphate were required only in high-salt sediments. These results might be explained by a higher microbial activity and nutrient turnover in the low-salt Cock Lake. Most likely acetate is utilized only by a small, but highly active population, which might correspond to the detected D. propionicus–like organisms. In fact, at the highest salt concentrations, at which the highest SRR were measured, this group of organisms has been persistently detected by molecular analysis, both by DNA- and RNA-based approaches. D. propionicus, a complete oxidizing halophilic SRB (Kjeldsen et al. 2007), can grow at salinity up to 190 g L−1, using propionate as a carbon source and electron donor. Since propionibacteria have also been detected in the investigated lakes, there might be a trophic link between these groups of primary and secondary anaerobes in soda lake sediments. In our previous study (Foti et al. 2007), the predominance of the Desulfosalina-like sequences was also observed when targeting the dsrB gene, indicating the relevance of these bacteria in soda lakes of the Kulunda Steppe. So far, complete oxidizing SRB have never been isolated from soda lakes with extreme salinity, therefore it is important to identify these organisms and to infer their functional role in saline lakes.
One of the goals of this research was to determine the diversity and activity of SRB communities along a salinity gradient. In general, this question was only partly answered due to non-specific PCR amplification, resulting mainly in the detection of non-SRB low G + C Gram-positive bacteria. This was also observed in another study conducted on Mono Lake (Scholten et al. 2005), suggesting that the specific amplification of the 16S rRNA gene from six SRB-groups is not recommended in environments rich in low-G + C organisms. It is possible that preferential amplification of the DNA template from low G + C content occurred due to the easier dissociation into single DNA strand than higher G + C templates, resulting in a bias in the favor of the former group (Suzuki and Giovannoni 1996). On the other hand, it could also be that low G + C bacteria, like acetogenic clostridia, out-number SRB at hypersaline alkaline conditions, since it is known that the acetogenic haloalkaliphiles benefit from sodium-based energetics (Häse et al. 2001). This was also observed by DGGE analysis and microscopic analysis of the highest positive tubes of the serial dilutions in SRB media, which revealed the predominance of Clostridia even at the highest dilutions. The discrepancy between observed high SRR rates/SRB viable count and the detection of SRB by molecular techniques might in fact imply that sulfate-reducers are represented by very active, but numerically minor populations in soda lake sediments.
Concluding, in this work we investigated the bacterial diversity and activity in sediments along a salinity gradient in different soda lakes of the Kulunda Steppe in southeastern Siberia. For the first time the actively growing bacterial groups, based on ribosomal RNA determination, have been detected by culture-independent techniques with special attention to SRB representing one of the most active bacterial functional groups in this extreme habitat. A new cluster of complete oxidizing SRB seems to be active in hypersaline soda lakes. In agreement with previous studies a decrease of the general bacterial diversity has been observed with increasing salinity, in contrast to the sulfate reduction activity, which seems to be well adapted even to saturating concentrations of sodium carbonates.
References
Benlloch S, Lopez-Lopez A, Casamayor EO, Ovreas L, Goddard V, Daae FL, Smerdon G, Massana R, Joint I, Thingstad F, Pedros-Alio C, Rodriguez-Valera F (2002) Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol 4:349–360
Braid MD, Daniels ML, Kitts CL (2003) Removal of PCR inhibitors from soil DNA by chemical flocculation. J Microbiol Methods 52:389–393
Brandt KK, Vester F, Jensen AN, Ingvorsen K (2001) Sulfate reduction dynamics and enumeration of sulfate-reducing bacteria in hypersaline sediments of the Great Salt Lake (Utah, USA). Microb Ecol 41:1–11
Brinkhoff T, Santegoeds CM, Sahm K, Kuever J, Muyzer G (1998) Polyphasic approach to study the diversity and vertical distribution of sulfur-oxidizing Thiomicrospira species in coastal sediments of the German Wadden Sea. Appl Environ Microbiol 64:4650–4657
Casamayor EO, Massana R, Benlloch S, Ovreas L, Diez B, Goddard VJ, Gasol JM, Joint I, Rodriguez-Valera F, Pedros-Alio C (2002) Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solern saltern. Environ Microbiol 4:338–348
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE (2004) Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol 70:1494–1505
Daly K, Sharp RJ, McCarthy AJ (2000) Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiology 146:1693–1705
Dar SA, Kuenen JG, Muyzer G (2005) Nested PCR-denaturing gradient gel electrophoresis approach to determine the diversity of sulfate-reducing bacteria in complex microbial communities. Appl Environ Microbiol 71:2325–2330
Duckworth AW, Grant WD, Jones BE, van Steenbergen R (1996) Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol Ecol 19:181–191
Foti M, Sorokin DY, Lomans B, Mussmann M, Zacharova EE, Pimenov NV, Kuenen JK, Muyzer G (2007) Diversity, activity and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl Environ Microbiol 73:2093–2100
Freitas M, Rainey FA, Nobre MF, Silvestre AJD, da Costa MS (2003) Tepidimonas aquatica sp. nov., a new slightly thermophilic β-proteobacterium isolated from a hot water tank. Syst Appl Microbiol 26:376–381
Gorlenko V, Tsapin A, Namsaraev Z, Teal T, Tourova T, Engler D, Mielke R, Nealson K (2004) Anaerobranca californiensis sp.nov., an anaerobic, alkalithermophilic , fermentative bacterium isolated froma hot spring on Mono lake. Int J Syst Evol Micriobiol 54:739–743
Häse CC, Fedorova ND, Galperin MY, Dibrov PA (2001) Sodium ion cycle in bacterial pathogens: evidences from cross-genome comparison. Microbiol Mol Biol Rev 65:353–370
Humayoun SB, Bano N, Hollibaugh JT (2003) Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl Environ Microbiol 69:1030–1042
Imhoff JF, Sahl HG, Soliman GSH, Truper HG (1979) The Wadi Natrun: chemical composition and microbial mass developments in alkaline brines of eutrophic desert lakes. Geomicrobiol J 1:219–234
Issachenko BL (1951) Chloride, sulfate and soda lakes of Kulunda steppe and its biogenic processes. In: Selected works, vol 2. Academy of Sciences USSR, Leningrad (in Russian), pp 143-162
Jones BE, Grant WD, Duckworth AW, Owenson GG (1998) Microbial diversity of soda lakes. Extremophiles 2:191–200
Kjeldsen KU, Loy A, Jakobsen TF, Thomsen TR, Wagner M, Ingvorsen K (2007) Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol Ecol 60:287–298
Kulp TR, Han S, Saltikov XW, Lanoil BD, Zargar K, Oremland RS (2007) Effects of imposed salinity gradient on dissimilatory arsenate reduction, sulphate reduction and other microbial processed in sediments from two California soda lakes. Appl Environ Microbiol 73:5130–5137
Lein A, Pimenov N, Guillou C, Martin J-M, Lancelot C, Rusanov I, Yusupov S, Miller Y, Ivanov M (2002) Seasonal dynamics of the sulfate reduction rate on the north-western Black Sea shelf. Estuar Coast Shelf Sci 54:385–401
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart W, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lûmann R, May M, Nonhoff B,Reichel B, Strehlow R, Stamatakis A ,Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Ma Y, Zhang W, Xue Y, Zhou P, Ventosa A, Grant WD (2004) Bacterial diversity of the Inner Mongolian Baer Soda Lake as revealed by 16S rRNA gene sequence analysis. Extremophiles 8:45–51
Muyzer G, deWaal EC, Uitterlinden A (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction—amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nübel U, Garcia-Pichel F, Clavero E, Muyzer G (2000) Matching molecular diversity and ecophysiology of benthic cyanobacteria and diatoms in communities along a salinity gradient. Environ Microbiol 2:217–226
Ollivier B, Caumette P, Garcia J, Mah RA (1994) Anaerobic bacteria from hypersaline environments. Microbiol Rev 58:27–38
Oren A (1999) Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev 63:334–348
Rees HC, Grant WD, Jones BE, Heaphy S (2004) Diversity of Kenyan soda lake alkaliphiles assesses by molecular methods. Extremophiles 8:63–71
Schafer H, Muyzer G (2001) Denaturing gradient gel electrophoresis in marine microbial ecology. In: Paul JH (ed) Methods in microbiology. Academic, New York
Scholten JCM, Joye SB, Hollibaugh JT, Murrell JC (2005) Molecular analysis of the sulfate-reducing and archaeal community in a meromictic soda lake (mono Lake, California) by targeting 16SrRNA, mcrA, apsA and dsrAB genes. Microb Ecol 50:29–39
Schultz GE, Ducklow H (2001) Changes in bacterioplankton metabolic capabilities along a salinity gradient in the York River estuary, Virginia, USA. Aquat Microb Ecol 22:167–174
Sörensen KB, Canfield DE, Oren A (2004) Salinity responses of benthic microbial communities in a solar saltern (Eilat, Israel). Appl Environ Microbiol 70:1608–1616
Sorokin DY, Kuenen JK (2005a) Haloalkaliphilic sulphur-oxidizing bacteria in soda lakes. FEMS Microbiol Rev 29:685–702
Sorokin DY, Kuenen JG (2005b) Chemolithotrophic haloalkaliphiles from soda lakes. FEMS Microbiol Ecol 52:287–295
Sorokin DY, Gorlenko VM, Namsaraev BB, Namsaraev ZB, Lysenko AM, Eshinimaev BT, Khmelenina VN, Trotsenko YA, Kuenen JG (2004) Prokaryotic communities of the north-eastern Mongolian soda lakes. Hydrobiologia 522:235–248
Sorokin DY, Foti M, Pinkart HC, Muyzer G (2007a) Sulfur-oxidizing bacteria in Soap Lake (Washington State), a meromictic, halosaline lake with an unprecedented high sulphide content. Appl Environ Microbiol 73:451–455
Sorokin DY, van Pelt S, Tourova TP, Muyzer G (2007b) Microbial isobutyronitrile utilization at haloalkaline conditions. Appl Environ Microbiol 73:5574–5579
Suzuki MT, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62:625–630
Trüper HG, Schlegel HG (1964) Sulphur metabolism in Thiorhodaceae. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenh 30:225–238
Wani AA, Surakasi VP, Siddharth J, Raghavan RG, Patole MS, Ranade D, Shouche YS (2006) Molecular analysis of microbial diversity associated with the Lonar soda lake in India: an impact crater in a basalt area. Res Microbiol 157:928–937
Wu QL, Zwart G, Schauer M, Kamst-van Agterveld MP, Hahn MW (2006) Bacterioplankton community composition along a salinity gradient of sixteen high-mountain lakes on the Tibetan Plateau, China. Appl Environ Microbiol 72:5478–5485
Zavarzin GA, Zhilina TN, Kevbrin VV (1999) The alkaliphilic microbial community and its functional diversity. Microbiology 63:503–521
Acknowledgments
This work was supported by the Dutch Science Foundation for Applied Research (STW) and by NWO-RFBR (grant 047.011.2004.010).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Horikoshi.
Rights and permissions
About this article
Cite this article
Foti, M.J., Sorokin, D.Y., Zacharova, E.E. et al. Bacterial diversity and activity along a salinity gradient in soda lakes of the Kulunda Steppe (Altai, Russia). Extremophiles 12, 133–145 (2008). https://doi.org/10.1007/s00792-007-0117-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-007-0117-7