Abstract
Objectives
This study includes the direct sequencing of cornulin (CRNN) gene to elucidate the possible mechanism of CRNN downregulation and explore the genetic imbalances at 1q21.3 across oral squamous cell carcinoma (OSCC) samples.
Materials and methods
In mutation screening of CRNN gene, gDNA from OSCC tissues were extracted, amplified, and followed by direct sequencing. OSCC samples were also subjected to fragment analysis on CRNN gene to investigate its microsatellite instability (MSI) and loss of heterozygosity (LOH). Immunohistochemistry was performed to validate CRNN downregulation in OSCC samples.
Results
No pathogenic mutation was found in CRNN gene, while high frequency of allelic imbalances was found at 1q21.3 region. MSI was found more frequent (25.3 %) than LOH (9.3 %). Approximately 22.6 % of cases had high MSI which reflects higher probability of inactivation of DNA mismatch repair genes. MSI showed significant association with no betel quid chewing (p = 0.003) and tongue subsite (p = 0.026). LOH was associated with ethnicity (p = 0.008) and advanced staging (p = 0.039). The LOH at 1q21.3 was identified to be as an independent prognostic marker in OSCC (HRR = 7.15 (95 % CI, 1.41–36.25), p = 0.018). Downregulation of CRNN was found among MSI-positive OSCCs and was associated with poor prognosis (p = 0.044).
Conclusion
This study showed a significant correlation between LOH/MSI at 1q21.3 with clinical outcomes and that downregulation of CRNN gene could be considered as a prognostic marker of OSCC.
Clinical relevance
Insights of the downregulation mode of CRNN gene lays the basis of drug development on this gene as well as revealing its prognostic value.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oral cancer is the 6th common cancer worldwide in which more than 90 % of oral cancers are squamous cell carcinoma [1–3]. Both genetic and environmental risk factors contribute to disease development. Oral cavity epithelium forms the first shield of protection during exposure to environmental risk factors. Hence, environmental risk factors might be able to induce some genetic alterations on these epithelial cells.
Genomic instability is one of the hallmarks of cancer that is associated with disease progression [4]. Chromosome 1q21.3 is one of the cancer-related regions that showed such instability across different types of cancers such as esophageal adenocarcinoma [5], gastrinomas [6], and sporadic insulinomas [7]. More recently, we detected genomic instability at the 1q21.3 chromosomal region in oral squamous cell carcinoma (OSCC) cases [8]. This region harbors the epidermal differentiation complex (EDC), a gene cluster that plays a critical role in late epithelial differentiation [9]. Downregulation of EDC genes is reported in the progression of normal esophageal epithelium to Barrett’s esophagus [10] and esophageal adenocarcinoma [11]. Frequent loss of heterozygosity (LOH) of 1q21.3 in cases with esophageal adenocarcinoma [5] could be one of the possible explanations for downregulation of EDC genes.
Environmental stresses such as smoking, alcohol drinking, and betel-quid chewing play a critical role in causing oral lesions and reflux of acidic gastric fluid in causing oesophageal lesions [12–15]. Thus, heat shock proteins such as HSP70 and CRNN play a critical role in controlling these unusual environmental pressures placed on the squamous epithelial cells [12]. CRNN as a member of EDC genes on 1q21.3 is a squamous epithelial heat shock protein which is also known as SEP53. CRNN gene expression is confined to squamous cells, especially in esophageal squamous epithelial cells [16]. Upregulation of CRNN in response to squamous epithelial cell injury in porcine [12] and in the buccal mucosa of smokers reflects the stress respondent role of this gene [17]. High-level expression of CRNN induced by environmental pressures would arrest the cell cycle at G1/S checkpoint [12, 18, 19].
Thus, it seems that upregulation of CRNN plays a critical role to control environmental pressures and to prevent formation of lesions on the epithelial tissues. Significant downregulation of CRNN in lesional area of the skin compared with non-lesional area in patients with eczema provides evidence in agreement with this conclusion [20].
Oral epithelial cells are subject to damage from habitual risk factors with CRNN responding to DNA damages induced by environmental pressures [12]. CRNN gene expression has been shown to be significantly reduced or absent in cancer cell lines, esophageal tumors, and OSCC [11, 12, 16, 19]. Downregulation in OSCC tumors reflects that CRNN might not be able to respond to DNA damages that might be induced by habitual risk factors. Hence, we hypothesized that CRNN mutation or LOH at the relevant locus might be the possible mechanism of downregulation of CRNN gene expression in OSCC. Thus, we directly sequenced parts of the promoter region at 5′-UTR, coding regions, and 3′-UTR of CRNN and further explored the possibility of LOH at 1q21.3 using polymorphic microsatellites.
Materials and methods
Samples
Sixty-two tumor samples from patients with OSCC that consisted of 41 Indians, 3 Indigenous, 12 Malays, and 6 Chinese who had a history of risk habits were included for direct sequencing of CRNN gene. If any variation was detected in the tumor, the matched normal sample from peripheral blood of the same patient was used for excluding the germline variations. In addition, 75 OSCC samples comprised 35 from the initial set of 62 OSCC cases with the addition of 40 independent OSCCs with their matched DNA samples from peripheral blood were recruited for microsatellite instability (MSI) and LOH analysis. Samples were taken from OSCC of the gum, floor of mouth, lip, tongue, palate, and buccal mucosa.
All samples were obtained from the Malaysian Oral Cancer Database and Tissues Bank System (MOCDTBS) coordinated by the Oral Cancer Research and Coordinating Centre, University of Malaya [21]. The American Joint Committee on cancer staging criteria was used for tumor staging [22]. Written informed consent has been taken from all patients and normal healthy controls before collecting the samples [21]. This study was approved by Medical Ethics Committee, Faculty of Dentistry, University of Malaya (MEC No. DFOP1108/0083(L)).
Direct sequencing of CRNN gene
Specific primers were designed using Primer3 program V0.4.0 [23], covering the coding regions, untranslated regions (5′- and 3′-UTR) and splicing sites (Table 1). The primers were designed approximately 100 bp beyond the splicing sites into the intronic region. PCR amplification was performed in a total volume of 25 μl, including 40 ng genomic DNA, 0.4 μm of each primer, and 12.5 μl GoTaq Mater Mix, 2× (Promega, USA). The PCR conditions were as follows: initial denaturation for 5 min at 95 °C, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 45 s and elongation at 72 °C for 30 s, and final elongation at 72 °C for 10 min.
The purified PCR products were directly sequenced using the BigDye terminator V3.1 sequencing standard kit (Applied Biosystems), as recommended by the manufacturer, and run on an automated Applied Biosystems 3730XL Genetic Analyzer. DNA sequences were analyzed using the Bioedit sequence alignment editor software, V5.0.9 [24], and then were BLASTed against reference sequences for CRNN. DNA sequence variants were confirmed by the sequencing of both forward and reverse strands. Reference sequences for CRNN coding DNA and protein were NM_016190.2, NP_057274.1, and NG_007081.1, respectively, according to the human genome assembly GRCh37.p10/hg19.
The PolyPhen-2 program was further used to predict the functional effects of non-synonymous variants. PolyPhen score is ranged from 0 to 1, where 1 indicates probable damage and 0 indicates that mutation is benign.
LOH and MSI analysis
To elucidate the LOH and MSI events on the 1q21.3 region, we have selected four highly polymorphic CA dinucleotide short tandem repeat (STR) markers with a heterozygosity of ≥8, covering the 1q21.3 chromosomal region (Table 2). Very highly heterozygous markers increase the probability of LOH detection. However, CRNN is located at this region, but there was no documented polymorphic STR marker inside the gene.
Specific primers were designed using Primer3 program, and the forward primers were labeled with fluorescent dyes. Multiplex PCR amplification was performed in a total volume of 50 μl, including 100 ng genomic DNA, 0.4 μm of each primer and 25 μl GoTaq Mater Mix, 2× (Promega, USA). The PCR conditions were as follows: initial denaturation for 5 min at 95 °C, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 2 min and elongation at 72 °C for 30 s, and final elongation at 60 °C for 45 min. The PCR products were capillary electrophoresed using Applied Biosystems 3730XL Genetic Analyzer and analyzed by Peak Scanner V2.0 and GeneMapper V4.0 software (Applied Biosystems).
Total loss or significantly reduced signal intensity of one allele in tumor sample was considered as LOH. LOH was estimated based on the previously reported formula as below, ratios >50 % was considered as LOH [25]. Presence of a new fragment in the tumor samples compared with matched normal was considered as MSI. Detecting a new allele in more than one STR marker was considered as high MSI (MSI-high).
Immunohistochemistry analysis of CRNN protein
Of the 75 OSCC samples that were used for MSI/LOH analysis, 43 samples were recruited for immunohistochemical (IHC) analysis. However, only 31 samples were deemed as interpretable for analysis in this study. Non-interpretable data was the consequence of damaged sections. Lack of fresh tissue impeded further sectioning and analysis. Frozen tissues were sectioned at 5 μm thickness and placed onto SuperfrostExcell™ microscope slides (Fisher Scientific, Pittsburgh, PA) for IHC study. Prior to performing IHC using the Envision technique, DAKO REAL EnVision Detection System and Peroxidase/DAB+ (Dako, USA), all sections were immersed in precooled acetone (−20 °C) for 10 min at room temperature for tissue fixation. The sections were immersed in blocking solution (Dako Corporation, Carpinteria, CA, USA) for 10 min at room temperature followed by incubation with primary antibody (1:200, anti-CRNN rabbit polyclonal antibody, 11799-1-AP, Proteintech Group, Chicago, IL, USA) for 1 h at room temperature. Visualization was achieved by incubation with the peroxidase-labeled secondary antibody from the Envision kit (Dako Corporation, Carpinteria, CA, USA) for one hour at room temperature followed by staining with 3’3 diaminobenzidine substrate chromogen (Dako Corporation) and counterstaining with Mayer’s hematoxylin, dehydration, and mounting. Normal oral mucosal tissues were used as positive controls because CRNN is highly expressed in oral mucosa. For negative control, the primary antibody was replaced with phosphate-buffered saline. The scoring assessment was done by two oral pathologists independently based on the semi-quantitative scoring system. The intensity scores were quantified using the following scores: negative = 0, weak = 1, moderate = 2, and strong = 3. The proportion of immune-positive cells was quantified as follows: 0 = negative, 1 = 0–5 % 2 = 6–50 %, and 3 = 51–100 % of positive cells. The final immunoreactive score (IRS) was determined by multiplying the positive intensity and the positive proportion scores to obtain an immunoreactive score ranging from 0 to 9. The consolidated immunoreactive scores for each case were recorded. Protein expression of CRNN was classified into two groups; high and low with a cutoff value based on the median of the respective immunoreactive score, by which IRS scores of ≥6 and <6 were used as high and low levels of CRNN expression, respectively.
Statistical analysis
The association between the LOH, MSI, CRNN protein expression, and the clinico-pathological parameters were analyzed by Chi-square test (or Fisher’s exact test where appropriate). Survival curves were plotted using the Kaplan–Meier analysis and compared using the log rank tests. Of 75 OSCC samples that have been used for MSI and LOH analysis, 10 samples with no survival data and 7 samples that were diagnosed after 2012 were excluded for survival analysis. Multivariate Cox regression analysis was conducted to evaluate the LOH and MSI at 1q21.3 as an independent prognostic factor. The protein expression of CRNN was compared between tumor and normal tissues using the Mann–Whitney U test. All statistical analyses were performed using the SPSS statistical package (SPSS version 12.0, Chicago, IL, USA), and the p values <0.05 was considered significant.
Results
LOH/MSI at 1q21.3 chromosomal region
Revisiting aCGH data revealed that 64.3 % of OSCC samples had a chromosomal gain in the 1q21.3 region in contrast to 35.7 % that had losses [8]. Two hotspots for alteration in copy numbers were detected in 1q21.3 region, where the CRNN gene is located in the 2nd hotspot (Fig. 1a). MSI/LOH analysis was performed on 75 OSCC samples using highly polymorphic STR markers. Most of the cases were informative (≥82 %) and remained into analysis (Fig. 1b). LOH at single STR marker was ranged from 1.3 to 5.3 %, while MSI was ranged from 18.6 to 22.6 %. LOH plus MSI were detected in 29.3 % of all cases. When all loci were considered, the overall MSI was more frequent (25.3 %) than LOH (9.3 %). Most of the cases had instability in more than one marker (22.6 %) than single marker. LOH at D1S2345 and MSI at D1S2346 markers, closest markers to the CRNN, were more frequent.
Association of LOH and MSI at 1q21.3 with socio-demographic and clinico-pathologic parameters
A significant statistical association was found with LOH at 1q21.3 among OSCC cases (Table 3). As, LOH was significantly associated with Malay ethnicity (Malay, 71.4 % vs. non-Malay, 28.6 %; p value = 0.008) and advanced staging (advanced staging, 100 % vs. early stage, 0 %, p value = 0.039). Of 18 Malay patients, 15 patients were diagnosed as having advanced stage disease. MSI was found to be significantly associated with non-betel quid chewing (non-chewer, 78.9 % vs. chewer, 21.1 %, p value = 0.003) and tongue SCC (tongue, 52.6 % vs. other subsites, 47.4 %, p value = 0.026) (Table 3).
Association of LOH and MSI of at 1q21.3 with survival outcome as prognostic indicators
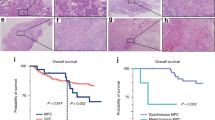
In the present study, the follow-up time for patients ranged from 1 to 88 months (mean, 23.78 months; median, 20.0 months). Two-year survival rates for negative and positive LOH at 1q21.3 chromosomal region were 61.43 and 25.0 %, respectively. Results of Kaplan–Meier analysis revealed a significant association between LOH at 1q21.3 and poor prognosis (p = 0.029) (Fig. 2a). In multivariate Cox regression analysis, LOH at 1q21.3 remained as a significant prognostic factor for survival after adjustment for age, gender, habitual risk factors, and lymph node metastasis (LNM) which are the common confounding factors in OSCC (HRR = 7.15 (95 % CI, 1.41–36.25), p = 0.018).
Three-year survival rates for negative and positive MSI at 1q21.3 chromosome region were 59.1 and 37.3 %, respectively. Results of Kaplan–Meier analysis showed no significant association between MSI at 1q21.3 and poor prognosis (p = 0.177) (Fig. 2b).
Mutation screening of CRNN gene
In direct sequencing of CRNN gene in 62 OSCC samples, five known and two novel variants were found (Fig. 1c). The novel variant of c.*158G>C was found as heterozygous at 3′-UTR in an Indian patient with a history of betel-quid chewing. Sample was taken from the gum of a patient who was at the advanced stage. The p.Glu205Asp was found as heterozygous non-synonymous variant in which glutamic acid in the position site of 205 was substituted with an aspartic acid. This missense variation was detected in a Malay OSCC patient who was at advanced stage and with lymph node metastasis. Sample was taken from tongue.
With the exception of three Indian patients who were heterozygous for rs3215709 variant, an intronic insertion in intron 2, almost all other samples were homozygous for rs3215709 variant (Table 4). However, the wild-type allele G of rs72477395 is reported ≥98 % in the HAPMAP population, but in contrast, the mutant allele was more frequent in all four ethnicities in the present study (Table 4).
CRNN protein expression among OSCCs
Immunohistochemical analyses showed high (IRS >6) CRNN expression in 28/31 (90.3 %) cases. Strong and extensive staining in the nuclei and the cytoplasm of the tumor cells was observed in these cases. Weak (IRS <6) staining of the tumor cells was seen in 3/31 (9.7 %) cases (Fig. 3). The socio-demographical and clinico-pathological parameters of the 31 OSCC patients recruited for IHC study is shown in Table 5. In all normal oral mucosal tissues (positive control), the epithelial cells showed strong (IRS = 9) cytoplasmic and nuclear staining with anti-CRNN antibody in the spinous and keratinized layer of the epithelium (Fig. 3).
Immunohistochemistry of CRNN. Normal oral mucosal tissue. a H&E stain (magnification × 400 and × 1600); d anti-CRNN antibody immunostain was strongly positive in normal oral mucosa (magnification × 400 and × 1600); OSCC (b, c) H&E stain (magnification × 400 and × 1600); e anti-CRNN antibody immunostaining showed low expression and f high expression in the cytoplasm and nuclei of the epithelial tumor cells (magnification × 400 and × 1600)
Correlation of CRNN protein expression with LOH, MSI, and clinico-pathological parameters
Expression of CRNN protein was significantly higher in normal oral mucosa tissues compared with OSCC samples (p < 0.05), but it showed no significant association with clinico-pathologic factors. In contrast, low expression of CRNN protein was significantly correlated with MSI (MSI yes, 100 % vs. MSI no, 0 %, p value = 0.037). We were able to recruit only two samples with LOH for IHC analysis. Therefore, lack of correlation between LOH and CRNN protein expression could be attributed to the small sample size of cases with LOH.
Significance of CRNN protein expression as a prognostic indicator in OSCC
The follow-up time of patients analyzed for CRNN expression ranged from 1 to 88 months (mean, 26.27 months; median, 22.5 months). Two-year survival rates for low and high CRNN expressions were 33.3 and 68.4 %, respectively. The low expression of CRNN protein was significantly correlated with poor prognosis (p = 0.044) in Kaplan–Meier analysis (Fig. 4).
Discussion
Genomic instability as the hallmark of cancer refers to increased tendency of alteration such as copy number changes, MSI/LOH, single gene mutations, and epigenetic changes in the genome [26]. EDC genes are clustered at 1q21, and the mechanisms that regulate the normal balanced expression of these genes are still unknown [27]. CpG methylation appeared to play a role in this event but chromatin remodeling and other epigenetic modifications most likely are contributing predominantly [28]. In addition, pool of transcription factors that are acting in a gene- and cell-specific manner to regulate the normal balance of gene expression play pivotal role [27, 29].
It is however controversial that genomic instability is the consequence of cancer progression or the event that drives tumorigenesis, but these alterations can be indicative of a cancer-related gene. Detection of genetic instabilities from clinically benign appearing lesions to potentially malignant disorders (OPMD) to invasive oral tumors provide evidence that genomic instabilities play a role in both genesis and progression of cancer [30–34]. We more recently detected high frequency of chromosomal instabilities such as gains or losses among OSCC cases [8]. Revisiting our aCGH data revealed that 64.3 and 35.7 % of OSCC samples had gain and loss at 1q21.3, respectively. In addition, revisiting our unpublished gene expression microarray data showed that CRNN is downregulated among OSCCs. However, genes in a region with copy number changes might be amplified partially which are either non-functional or act as a gene expression silencer [35]. Here, we hypothesized that downregulation of CRNN gene might be the consequence of mutation or LOH at 1q21.3. Hence, we further explored this region using polymorphic STR markers and the CRNN gene was directly sequenced.
In the present study, MSI was found more frequent than LOH, which was consistent with the results of aCGH by which gains were more frequent than losses at 1q21.3. In addition, MSI showed a significant association with no betel-quid chewing and tongue SCC. In a study on Japanese cases with OSCC, a high frequency of MSI was detected among tongue SCC which is in agreement with our finding [36]. Lack of association between MSI and habitual risk factors and identifying MSI-H with a high frequency reflects the higher probability of inactivation in DNA mismatch repair (MMR) genes. MSI phenotype is strongly associated with mutation in genome, especially in MMR genes [37]. Inactivation of MMR genes either by mutation or methylation has been previously detected in oral cancer [38, 39]. MSI in various loci is reported in a range of 7 % up to 60 % among oral cancers with a higher frequency among Asian countries [33, 34, 40, 41]. In this study, LOH showed significant association with Malay ethnicity than other ethnicities in Malaysia. However, the sample size of Malays was not that much large in comparison with Indians and Indians had more habitual risk factors than Malays. Thus, these observations reflect that LOH/MSI at 1q21.3 might not be affected by habitual risk factors such as betel-quid chewing. This statement was in line with that from the significant association between no betel-quid chewing and MSI in the current study. In addition, detecting higher frequency of LOH among Malays could be attributed to similar predisposing hereditary and dietary factors that are different in other ethnicities in Malaysia. However, we were unable to exclude the possible role of these habitual risk factors in mutagenesis of MMR genes by which subsequently will cause genetic instability. In agreement with the current study, LOH has been detected in OSCC cases with advanced stage [42], as all cases with LOH had LNM as well. In this study, a significant difference was found among those with and without LOH in survival analysis. Poor prognosis in OSCC cases with LOH at 1q21.3 could be explained by the fact that over-expression of CRNN gene suppresses cell proliferation by arresting the cell cycle progression at the G1/S phase and downregulation of cyclin D1 in oral cancer [19].
Downregulation of CRNN gene has been well documented in tumoregenesis of esophageal squamous cell carcinoma (ESCC) [18, 43, 44]. They found that significant loss of CRNN expression to be associated with advanced stage, invasive behavior of tumor, LNM, and poor survival [18, 43, 44]. This gene has shown downregulation among OSCCs as well [19], which is consistent with our findings. CRNN expression was downregulated in OSCCs compared with normal oral mucosal samples, and it was significantly correlated with poor prognosis. Loss of CRNN expression has been detected consistently across these studies with no further investigation on the possible mechanisms. Consistent with the current study, evidence has shown that single nucleotide changes might not have a remarkable impact on CRNN downregulation among ESCCs [45]. While LOH/MSI was frequent in this study, hence we suggested that genetic instabilities could be one of the possible mechanisms that diminish the expression of the genes that are located at 1q21.3.
To the best of our knowledge, mutation screening of CRNN in patients with ESCC is the only documented report, so that except for several known polymorphic variants, no novel variation was detected [45]. In direct sequencing of CRNN among OSCC patients with a history of habitual risk factors, five known and two novel germline variations were detected in the current research. Nucleotide substitutions were more frequently observed in exon three. All known variants have been previously reported in the National Center for Biotechnology Information (NCBI) SNP database. The novel variant of c.*158G>C was found at 3′-UTR of the gene in an Indian patient who had a history of betel-quid chewing. However, this position site is conserved among primates, but it is not conserved among placental mammals. The 3′-UTR of protein coding genes harbors the binding site of microRNAs (miRNAs) [46]. Disruption of these binding sites tends to be damaging and might be involved in disease development via regulation of gene expression [47, 48]. Somatic mutations that created or disrupted the miRNA binding sites have been reported in many cancer-related genes [49]. Thus, the potential impact of variants at 3′-UTR should be kept in mind.
The p.Glu205Asp was found in a Malay patient who was smoker. The p.Glu205Asp was predicted to be benign (PolyPhen score = 0.001), and this position site is not conserved among different species. Both novel variants were detected in matched normal tissues as well, excluding somatic mutation. Hence, we did not further explore these variants among normal healthy controls. However, patients with novel variants were at advanced stage, but they had no LOH/MSI at 1q21.3. In addition, the p.Gly480Ser which is a synonymous variant was predicted as benign as well (PolyPhen score = 0.18), and this position site is not conserved.
With the exception of several known and two benign novel germline variants, no pathogenic mutation was found in mutation screening of CRNN gene among OSCC cases. Thus, it appears that habitual risk factors might not be associated with increased risk of mutation in CRNN and mutation in the coding regions might not be the mechanism of downregulation in gene expression. However, promoter plays a central role in regulation of gene expression, but no statistical difference in the expression of CRNN gene has been detected among patients with variations in the promoter region [45].
Identifying LOH in 9.3 % of our samples reflects that small part of CRNN downregulation could result from LOH, which is consistent with results of aCGH. Despite significant difference in CRNN expression between normal oral mucosal samples and OSCCs, we were unable to draw a clear pattern between LOH and CRNN expression due to insufficient sample size. Evidence has shown that MSI would be able to alter the transcription factor binding and gene expression [50]. Consistent with this evidence, our MSI-positive OSCC samples showed low expression of CRNN reflecting the impact of MSI on CRNN expression. Thus, the MSI-H phenotype that was found in the current research could be associated with a higher mutation rate in the genome which in turn could alter the CRNN expression profile in OSCC. Downregulation of CRNN is detected to be significantly associated with lymph node metastases, advanced clinical stage, and overall survival rate, and it thus remained as an independent prognostic marker for ESCC [18]. Taken together, a part of downregulation of CRNN among OSCC cases could be resulted from LOH/MSI at 1q21.3 than mutation in the CRNN gene. However, epigenetic changes should be kept in mind as another possibility of downregulation for this gene.
In addition, this study showed a significant correlation between LOH/MSI at 1q21.3 with clinical outcomes. The LOH at this region was identified to be as an independent prognostic marker in OSCC. These findings provide further evidences on the roles of CRNN in tumor progression of OSCC and as a prognostic marker to predict the disease outcome. However, further investigation on the interaction between CRNN and other potential genes or environmental risk factors would shed light on the pathogenesis of OSCC.
References
Warnakulasuriya S (2009) Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 45(4–5):309–316. doi:10.1016/j.oraloncology.2008.06.002
Neville BW, Day TA (2002) Oral cancer and precancerous lesions. CA Cancer J Clin 52(4):195–215
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90. doi:10.3322/caac.20107
de Martel C, Franceschi S (2009) Infections and cancer: established associations and new hypotheses. Crit Rev Oncol Hematol 70(3):183–194. doi:10.1016/j.critrevonc.2008.07.021
Maru DM, Luthra R, Correa AM, White-Cross J, Anandasabapathy S, Krishnan S, Guha S, Komaki R, Swisher SG, Ajani JA, Hofstetter WL, Rashid A (2009) Frequent loss of heterozygosity of chromosome 1q in esophageal adenocarcinoma: loss of chromosome 1q21.3 is associated with shorter overall survival. Cancer 115(7):1576–1585. doi:10.1002/cncr.24122
Chen YJ, Vortmeyer A, Zhuang Z, Huang S, Jensen RT (2003) Loss of heterozygosity of chromosome 1q in gastrinomas: occurrence and prognostic significance. Cancer Res 63(4):817–823
Yang YM, Liu TH, Chen YJ, Jiang WJ, Qian JM, Lu X, Gao J, Wu SF, Sang XT, Chen J (2005) Chromosome 1q loss of heterozygosity frequently occurs in sporadic insulinomas and is associated with tumor malignancy. Int J Cancer J 117(2):234–240. doi:10.1002/ijc.21175
Vincent-Chong VK, Anwar A, Karen-Ng LP, Cheong SC, Yang YH, Pradeep PJ, Rahman ZA, Ismail SM, Zaini ZM, Prepageran N, Kallarakkal TG, Ramanathan A, Mohayadi NA, Rosli NS, Mustafa WM, Abraham MT, Tay KK, Zain RB (2013) Genome wide analysis of chromosomal alterations in oral squamous cell carcinomas revealed over expression of MGAM and ADAM9. PLoS One 8(2):e54705. doi:10.1371/journal.pone.0054705
Marenholz I, Zirra M, Fischer DF, Backendorf C, Ziegler A, Mischke D (2001) Identification of human epidermal differentiation complex (EDC)-encoded genes by subtractive hybridization of entire YACs to a gridded keratinocyte cDNA library. Genome Res 11(3):341–355. doi:10.1101/gr.114801
Kimchi ET, Posner MC, Park JO, Darga TE, Kocherginsky M, Karrison T, Hart J, Smith KD, Mezhir JJ, Weichselbaum RR, Khodarev NN (2005) Progression of Barrett’s metaplasia to adenocarcinoma is associated with the suppression of the transcriptional programs of epidermal differentiation. Cancer Res 65(8):3146–3154. doi:10.1158/0008-5472.CAN-04-2490
Luthra MG, Ajani JA, Izzo J, Ensor J, Wu TT, Rashid A, Zhang L, Phan A, Fukami N, Luthra R (2007) Decreased expression of gene cluster at chromosome 1q21 defines molecular subgroups of chemoradiotherapy response in esophageal cancers. Clin Cancer Res Off J Am Assoc Cancer Res 13(3):912–919. doi:10.1158/1078-0432.CCR-06-1577
Nelson L, Anderson S, Archibald AL, Rhind S, Lu ZH, Condie A, McIntyre N, Thompson J, Nenutil R, Vojtesek B, Whitelaw CB, Little TJ, Hupp T (2008) An animal model to evaluate the function and regulation of the adaptively evolving stress protein SEP53 in oesophageal bile damage responses. Cell Stress Chaperones 13(3):375–385. doi:10.1007/s12192-008-0037-1
Jaber MA, Porter SR, Gilthorpe MS, Bedi R, Scully C (1999) Risk factors for oral epithelial dysplasia—the role of smoking and alcohol. Oral Oncol 35(2):151–156
Pentenero M, Broccoletti R, Carbone M, Conrotto D, Gandolfo S (2008) The prevalence of oral mucosal lesions in adults from the Turin area. Oral Dis 14(4):356–366. doi:10.1111/j.1601-0825.2007.01391.x
Thomas G, Hashibe M, Jacob BJ, Ramadas K, Mathew B, Sankaranarayanan R, Zhang ZF (2003) Risk factors for multiple oral premalignant lesions. Int J Cancer 107(2):285–291. doi:10.1002/ijc.11383
Xu Z, Wang MR, Xu X, Cai Y, Han YL, Wu KM, Wang J, Chen BS, Wang XQ, Wu M (2000) Novel human esophagus-specific gene c1orf10: cDNA cloning, gene structure, and frequent loss of expression in esophageal cancer. Genomics 69(3):322–330. doi:10.1006/geno.2000.6344
Kupfer DM, White VL, Jenkins MC, Burian D (2010) Examining smoking-induced differential gene expression changes in buccal mucosa. BMC Med Genet 3:24. doi:10.1186/1755-8794-3-24
Chen K, Li Y, Dai Y, Li J, Qin Y, Zhu Y, Zeng T, Ban X, Fu L, Guan XY (2013) Characterization of tumor suppressive function of cornulin in esophageal squamous cell carcinoma. PLoS One 8(7):e68838. doi:10.1371/journal.pone.0068838
Imai FL, Uzawa K, Nimura Y, Moriya T, Imai MA, Shiiba M, Bukawa H, Yokoe H, Tanzawa H (2005) Chromosome 1 open reading frame 10 (C1orf10) gene is frequently down-regulated and inhibits cell proliferation in oral squamous cell carcinoma. Int J Biochem Cell Biol 37(8):1641–1655. doi:10.1016/j.biocel.2005.02.005
Lieden A, Ekelund E, Kuo IC, Kockum I, Huang CH, Mallbris L, Lee SP, Seng LK, Chin GY, Wahlgren CF, Palmer CN, Bjorksten B, Stahle M, Nordenskjold M, Bradley M, Chua KY, D’Amato M (2009) Cornulin, a marker of late epidermal differentiation, is down-regulated in eczema. Allergy 64(2):304–311. doi:10.1111/j.1398-9995.2008.01856.x
Zain RB, Athirajan V, Ghani WM, Razak IA, Raja Latifah RJ, Ismail SM, Sallam AA, Bustam AZ, Rahman ZA, Hussien A, Talib N, Cheong SC, Jallaludin A (2013) An oral cancer biobank initiative: a platform for multidisciplinary research in a developing country. Cell Tissue Bank 14(1):45–52. doi:10.1007/s10561-012-9298-0
Sobin LH, Gospodarowicz MK, Wittekind C (2009) TNM classification of malignant tumours. Wiley, Chichester, West Sussex, UK
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series, pp 95–98
Shi X, Li J, Li A, Lv S, Xu G (2006) Simultaneous analysis of microsatellite instability and loss of heterozygosity by capillary electrophoresis with a homemade kit. J Chromatogr B Anal Technol Biomed Life Sci 834(1–2):122–127. doi:10.1016/j.jchromb.2006.02.052
Shen Z (2011) Genomic instability and cancer: an introduction. J Mol Cell Biol 3(1):1–3. doi:10.1093/jmcb/mjq057
Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY (2012) Epigenetic regulation of gene expression in keratinocytes. J Investig Dermatol 132(11):2505–2521. doi:10.1038/jid.2012.182
Elder JT, Zhao X (2002) Evidence for local control of gene expression in the epidermal differentiation complex. Exp Dermatol 11(5):406–412
Kypriotou M, Huber M, Hohl D (2012) The human epidermal differentiation complex: cornified envelope precursors, S100 proteins and the ‘fused genes’ family. Exp Dermatol 21(9):643–649. doi:10.1111/j.1600-0625.2012.01472.x
Tsui IF, Poh CF, Garnis C, Rosin MP, Zhang L, Lam WL (2009) Multiple pathways in the FGF signaling network are frequently deregulated by gene amplification in oral dysplasias. Int J Cancer 125(9):2219–2228. doi:10.1002/ijc.24611
Garnis C, Chari R, Buys TP, Zhang L, Ng RT, Rosin MP, Lam WL (2009) Genomic imbalances in precancerous tissues signal oral cancer risk. Mol Cancer 8:50. doi:10.1186/1476-4598-8-50
Mao L, Lee JS, Fan YH, Ro JY, Batsakis JG, Lippman S, Hittelman W, Hong WK (1996) Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat Med 2(6):682–685
Ishwad CS, Ferrell RE, Rossie KM, Appel BN, Johnson JT, Myers EN, Law JC, Srivastava S, Gollin SM (1995) Microsatellite instability in oral cancer. Int J Cancer 64(5):332–335
Mahale A, Saranath D (2000) Microsatellite alterations on chromosome 9 in chewing tobacco-induced oral squamous cell carcinomas from India. Oral Oncol 36(2):199–206
Vazquez-Mena O, Medina-Martinez I, Juárez-Torres E, Barrón V, Espinosa A, Villegas-Sepulveda N, Gómez-Laguna L, Nieto-Martínez K, Orozco L, Roman-Basaure E (2012) Amplified genes may be overexpressed, unchanged, or downregulated in cervical cancer cell lines. PLoS One 7(3):e32667
Kakimoto Y, Numasawa H, Yamamoto N, Takeda E, Yamauchi T, Shibahara T (2007) Loss of heterozygosity and microsatellite instability on the long arm of chromosome 2 in human oral squamous cell carcinoma. Dent Jpn 43:70–73
Vilar E, Gruber SB (2010) Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 7(3):153–162. doi:10.1038/nrclinonc.2009.237
Czerninski R, Krichevsky S, Ashhab Y, Gazit D, Patel V, Ben-Yehuda D (2009) Promoter hypermethylation of mismatch repair genes, hMLH1 and hMSH2 in oral squamous cell carcinoma. Oral Dis 15(3):206–213. doi:10.1111/j.1601-0825.2008.01510.x
Shin KH, Park KH, Hong HJ, Kim JM, Oh JE, Choung PH, Min BM (2002) Prevalence of microsatellite instability, inactivation of mismatch repair genes, p53 mutation, and human papillomavirus infection in Korean oral cancer patients. Int J Oncol 21(2):297–302
Piccinin S, Gasparotto D, Vukosavljevic T, Barzan L, Sulfaro S, Maestro R, Boiocchi M (1998) Microsatellite instability in squamous cell carcinomas of the head and neck related to field cancerization phenomena. Br J Cancer 78(9):1147–1151
Jin YT, Myers J, Tsai ST, Goepfert H, Batsakis JG, el-Naggar AK (1999) Genetic alterations in oral squamous cell carcinoma of young adults. Oral Oncol 35(3):251–256
Chen L, Wong MP, Cheung LK, Samaranayake LP, Baum L, Samman N (2005) Frequent allelic loss of 21q11.1 approximately q21.1 region in advanced stage oral squamous cell carcinoma. Cancer Genet Cytogenet 159(1):37–43. doi:10.1016/j.cancergencyto.2004.09.011
Hsu PK, Kao HL, Chen HY, Yen CC, Wu YC, Hsu WH, Chou TY (2014) Loss of CRNN expression is associated with advanced tumor stage and poor survival in patients with esophageal squamous cell carcinoma. J Thorac Cardiovasc Surg 147(5):1612–1618. doi:10.1016/j.jtcvs.2013.09.066, e1614
Pawar H, Maharudraiah J, Kashyap MK, Sharma J, Srikanth SM, Choudhary R, Chavan S, Sathe G, Manju HC, Kumar KV, Vijayakumar M, Sirdeshmukh R, Harsha HC, Prasad TS, Pandey A, Kumar RV (2013) Downregulation of cornulin in esophageal squamous cell carcinoma. Acta Histochem 115(2):89–99. doi:10.1016/j.acthis.2012.04.003
Zhang W, Chen X, Luo A, Lin D, Tan W, Liu Z (2009) Genetic variants of C1orf10 and risk of esophageal squamous cell carcinoma in a Chinese population. Cancer Sci 100(9):1695–1700. doi:10.1111/j.1349-7006.2009.01240.x
Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG (2012) Functional microRNA targets in protein coding sequences. Bioinformatics 28(6):771–776. doi:10.1093/bioinformatics/bts043
Wiemer EA (2007) The role of microRNAs in cancer: no small matter. Eur J Cancer 43(10):1529–1544. doi:10.1016/j.ejca.2007.04.002
Saunders MA, Liang H, Li WH (2007) Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A 104(9):3300–3305. doi:10.1073/pnas.0611347104
Ziebarth JD, Bhattacharya A, Cui Y (2012) Integrative analysis of somatic mutations altering microRNA targeting in cancer genomes. PLoS One 7(10):e47137. doi:10.1371/journal.pone.0047137
Martin P, Makepeace K, Hill SA, Hood DW, Moxon ER (2005) Microsatellite instability regulates transcription factor binding and gene expression. Proc Natl Acad Sci U S A 102(10):3800–3804. doi:10.1073/pnas.0406805102
Acknowledgments
This study was supported by the High Impact Research MoE Grant UM.C/625/1/HIR/MoE/DENT/08. Special thanks to Lee Peng Karen-Ng for providing expression data for CRNN gene and to the Oral Cancer Research and Co-ordinating Centre (OCRCC), University of Malaya (UM) for providing tissue and data from the Malaysian Oral Cancer Database and Tissue Bank System (MOCDTBS). The authors also thank the clinicians and pathologists from the Ministry of Health Malaysia for their clinical expertise required to successfully complete this study.
Conflict of interests
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Iman Salahshourifar and Vui King Vincent-Chong contributed equally to this work.
Rights and permissions
About this article
Cite this article
Salahshourifar, I., Vincent-Chong, V.K., Chang, HY. et al. Downregulation of CRNN gene and genomic instability at 1q21.3 in oral squamous cell carcinoma. Clin Oral Invest 19, 2273–2283 (2015). https://doi.org/10.1007/s00784-015-1467-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-015-1467-7