Abstract
Squamous epithelium in mammals has evolved an atypical stress response involving down-regulation of the classic HSP70 protein and induction of sets of proteins including one named SEP53. This atypical stress response might be due to the unusual environmental pressures placed on squamous tissue. In fact, SEP53 plays a role as an anti-apoptotic factor in response to DNA damage induced by deoxycholic acid stresses implicated in oesophageal reflux disease. SEP53 also has a genetic signature characteristic of an adaptively and rapidly evolving gene, and this observation has been used to imply a role for SEP53 in immunity. Physiological models of squamous tissue are required to further define the regulation and function of SEP53. We examined whether porcine squamous epithelium would be a good model to study SEP53, since this animal suffers from a bile-reflux disease in squamous oesophageal tissue. We have (1) cloned and sequenced the porcine SEP53 locus from porcine bacterial artificial chromosome genomic DNA, (2) confirmed the strikingly divergent nature of the C-terminal portion of the SEP53 gene amongst mammals, (3) discovered that a function of the conserved N-terminal domain of the gene is to maintain cytoplasmic localisation, and (4) examined SEP53 expression in normal and diseased porcine pars oesophagea. SEP53 expression in porcine tissue was relatively confined to gastric squamous epithelium, consistent with its expression in normal human squamous epithelium. Immunohistochemical staining for SEP53 protein in normal and damaged pars oesophagea demonstrated significant stabilisation of SEP53 protein in the injured tissue. These results suggest that porcine squamous epithelium would be a robust physiological model to examine the evolution and function of the SEP53 stress pathway in modulating stress-induced responses in squamous tissue.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Human cancers evolve through a series of morphological changes from normal epithelium through to cancer that are driven in part by mutations and environmental cues that together give rise to enhanced cell survival in an aberrant microenvironment (Vogelstein and Kinzler 2004). These localised environmental signals include hypoxia, DNA damage, acidification, and altered autocrine responses (Hupp 2000). In developing relevant models to understand the link between stress protein responses and cancer development, a key clinical model that is giving novel molecular mechanistic insight is adenocarcinoma of the oesophagus. This cancer is one of the fastest rising cancers in the west and is associated with stresses induced by environmental damaging agents including acid and bile reflux (Fitzgerald and Farthing 2003; Reid et al. 2003). In particular, bile acids such as deoxycholic acid can stimulate cell proliferation, migration, DNA damage, and apoptosis in gut epithelial cells (Olliver et al. 2003). Understanding fundamental aspects of how cells respond to deoxycholic acid stresses is an important part in understanding tissue-specific repair factors that control cancer development.
Stress protein responses are classically maintained by heat shock proteins or molecular chaperones that play an important role in repairing protein damage after cell injury, thus maintaining tissue integrity (Mosser and Morimoto 2004). As such, the integrity of the chaperone system can alter progression of diseases associated with ageing, DNA damage, protein damage, chronic injury, and cancer (Ciocca and Calderwood 2005). Although molecular chaperone proteins are some of the most evolutionarily conserved proteins known, there is a high degree of tissue specificity in chaperone induction. Many cells exhibit uncoupled chaperone gene expression from protein induction after stresses including hyperthermia or endotoxin exposure (Blake et al. 1991; Blake et al. 1990; Metzler et al. 2000). In Drosophila melanogaster, an atypical stress response operates in Malpighian tubules that recruits a novel HSP60 family member after heat shock (Lakhotia and Singh 1996), further highlighting that some cells in metazoans have evolved unique stress responses due to unique microenvironmental pressures during evolution.
Cells of the normal human oesophageal squamous epithelium are under relatively unique environmental stresses, including bacterial infestation, viruses, thermal stresses, refluxed acid, and bile adducts (Wild and Hardie 2003). Thus, by this rationale, we had defined previously the stress-responsive pathways in normal human squamous oesophageal epithelial cells using a “functional proteomics” approach. Ex vivo organ culture in conjunction with specific stresses, including ethanol and heat shock, identified a novel class of stress proteins in normal squamous epithelium; these include SEP70, SEP53, and glutamine–glutamyl transferase (Hopwood et al. 1997; Yagui-Beltran et al. 2001). The SEP53 gene is located on chromosome 1q21 within a group of proteins named the epidermal differentiation complex fused-gene complex that is silenced as part of a general mechanism that suppresses genes from this locus in cancer cells (Xu et al. 2000; Imai et al. 2005). Molecular characterisation of SEP53 has demonstrated that its role as a stress-responsive protein is linked to its activity as an anti-apoptotic factor; death induced by exposure of cells to normally lethal levels of deoxycholic acid can be attenuated by SEP53 (Darragh et al. 2006). Subsequent studies have also identified a strong signature of adaptive evolution on SEP53 DNA sequence of a type that is commonly associated with a coevolutionary arms race with a pathogen (Little et al. 2007), suggesting SEP53 as a candidate component of the mucosal/epithelial innate immune response engaged in an ongoing interaction with a pathogen.
There are no current physiological models to analyse SEP53 function, since its expression has not been observed in cell lines in vitro. As such, mammalian squamous tissue models are needed to study the SEP53 pathway, and we have evaluated whether SEP53 expression in diseased porcine tissue where environmental damage plays a role in tissue injury. Porcine oesophageal squamous epithelium has been reported to be a much better model for oesophageal squamous cell responses than the mouse (Christie et al. 1995). Furthermore, hyperplastic and erosive damage of the porcine pars oesophagea is a documented physiological disease where lesions are confined to the squamous epithelium adjacent to the glandular gastric mucosa (Roosendaal et al. 2000), and approximately 80% of finishing pigs have a bile-induced lesion in this area (Friendship 1999, 2003). There is a relationship between small food particle size and increased incidence of lesions that allows bile acids to continuously bathe the mucosa (Ayles et al. 1996). As this mammal may provide a correlate with human reflux disease at the cellular and tissue level and a robust model to understand squamous mucosal tissue injury, we focused on cloning and sequencing the porcine SEP53 gene, evaluated its adaptive evolution, and examined whether the SEP53 protein is dysregulated in porcine squamous bile-reflux disease.
2 Materials and methods
2.1 Porcine BAC library
2.1.1 In silico identification of porcine SEP53 BAC clones
Bioinformatic analysis of porcine SEP53 gene was initiated using human SEP53 sequence (GenBank accession no. AF077831), by searching the non-redundant, expressed sequence tag, high throughput genomic sequences in EMBL/GenBank or the NCBI/Ensembl trace repository. These searches did not reveal homologous sequences for porcine SEP53. However, by inspection of the alignment of pig bacterial artificial chromosome (BAC) end sequences with the human genome sequence at the SEP53 (CRNN) locus, as visualised in the ‘Pig BAC ends’ DAS track in human Ensembl contig view (http://www.ensembl.org/Homo_sapiens), we identified PigE BAC clones from the Roslin BAC library (Anderson et al. 2000) that potentially harboured the porcine SEP53 locus. Further candidate PigE BAC clones were identified from the pig BAC contig physical map (Humphray et al. 2007; http://www.sanger.ac.uk/Projects/S_scrofa/ and http://pre.ensembl.org/Sus_scrofa_map/index.html). These candidate BAC clones were screened by Southern hybridisation and polymerases chain reaction (PCR) for the presence of SEP53 sequences. Since our current in-house porcine complementary DNA (cDNA) libraries did not harbour porcine SEP53, we designed primers based on the human SEP53 sequence and used these to amplify porcine SEP53 exonic sequences from the BAC clone.
2.2 BAC recovery and purification
For Southern blot hybridisation, PigEBAC clones were grown (from Luria–Bertani (LB) agar stab cultures) on LB-agar/12.5 μg ml−1 chloramphenicol (CAMP) plates for 16 h, single colonies picked, and BAC DNA purified using a modification of the Qiagen Plasmid Mini Kits (i.e. performed steps 1–4, omitting the spin column step, followed by a standard ethanol-precipitation and resuspension in Tris–EDTA buffer). Ten micrograms of each genomic BAC clone (PigEBAC 67g6 and 67g7, initially identified as potentially harbouring genomic SEP53) was digested for 16 h with EcoRI/BamHI (NEB); after the addition of (5×) gel-loading buffer, samples were heated to 56°C for 2 min (to disrupt potential base-pairing between protruding cohesive ends) and run on a 0.8% agarose gel for 4 h at 50 V. The gel was then subjected to Southern hybridisation, followed by drying and covalent cross-linking of DNA to the membrane using a Spectronics Corporation XL-1500 UV Crosslinker.
The following 5′-biotinylated primer pair (Sigma-GenoSys Custom Oligos) was employed to PCR-amplify exon II of human SEP53 (GenBank accession no. AF077831; synonyms: C1orf10; cornulin, CRNN): HuSEP53ExII F ATGCCTCAGTTACTGCAAAACA and HuSEP53ExII R-CACAATCACATCGGCAAACTCT, from the pcDNA3.0/Human SEP53 vector construct (Darragh et al. 2006). The resulting ∼140 bp amplicon was gel-purified (QIAquick Gel Extraction Kit 28704), quantified, and subsequently used as probe for hybridisation using the North2South Hybridisation and Detection Kit (Pierce N2S; kit 17097) according to the manufacturer’s instructions. Briefly, 300 ng of HuSEP53-biotinylated probe in 10 ml hybridisation buffer was incubated with the Hybond-N+ membrane/immobilised BAC DNA overnight at 58°C in 50 ml Falcon tubes. After three times stringency washes at 55°C, probe was detected by subsequent formation of streptavidin-horseradish peroxidase conjugates and addition of Luminol/Enhancer solution; the probe signal was identified after 1-min exposure to photographic film (Kodak).
2.3 Porcine BAC purification and sequencing strategy
The NucleoBond® BAC 100 Kit (Macherey-Nagel, GmbH, Germany) was used to obtain ultrapure DNA from the PigEBAC clones (which have low copy number). First, ElectroMAX DH10B Cells (Invitrogen) were transformed by electroporation (Bio-Rad MicroPulser with 1-cm cuvettes, 2.5 kV; 5 ms). Cells were recovered in 1 ml SOC medium (Gibco-BRL) at 37°C for 1 h before plating on LB agar (as above) and incubated at 37°C for 24 h. Single colonies containing PigEBACs were individually picked into sterile 15-ml Greiner tube containing 5 ml LB medium/12.5 μg ml−1 CAMP and cultured 8 h at 37°C in an orbital shaker. Two-liter flasks were then inoculated in LB/CAMP with 200 μl of the above culture and incubated 16–18 h at 37°C in the orbital shaker. Next, each culture was transferred to a sterile Sorvall Dri-spin 250 ml centrifuge bottle; centrifuged 5 min at 10,000 rpm in a Sorvall RC5B with an F16-250 rotor at 4°C. Media was discarded and pellets resuspended using 12 ml Buffer S1+RnaseA per pellet and transferred to sterile Nalgene 50-ml tubes. The NucleoBond kit manufacturers’ protocol was subsequently followed to obtain ultrapure BAC DNA, which was quantified using the NanoDrop® ND-1000 Spectrophotometer (both PigEBAC 67g6 and 67g7 clones had λ 260/280 ratios > 2.0).
2.4 Porcine BAC sequencing
PigEBAC clones 67g6 and 67g7 were initially sequenced using human SEP53 ExonII primer set (HuExIIF1_ ATGCCTCAGTTACTGCAAAACA; and HuExIIR2_ CTCAATCACATCGGCAAACTCT). From these sequence data, subsequent porcine primer sets were designed to facilitate a ‘primer-walking’ sequencing strategy, involving sequencing of overlapping genomic fragments. After sample preparation and automated sequencing on an ABI 3700 sequencer (according to the vendor; PE-Applied Biosystems), contiguous consensus sequences were constructed using the IFOM CAP EST Assembler (http://bio.ifom-firc.it/ASSEMBLY/assemble.html) spanning the entire porcine SEP53 gene. Any sequence disparity was affirmed using BioEdit Sequence Alignment Editor (version 7.0.5.3: http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
2.5 Porcine tissue panel RT-PCR
Porcine tissue isolated from 5 kg Large White piglets was rinsed in sterile, ice-cold phosphate-buffered saline (PBS; pH 7.4) and immediately placed in 10× volumes RNA later solution (Ambion; no. 7020) to inactivate RNase and stabilise total RNA. Total RNA was isolated from tissues using Trizol (Invitrogen) according to manufacturers’ instructions. Homogenisation of each tissue type in Trizol was carried out using a Fast-Prep FP120 cell disrupter (BIO101) in Lysing matrix D beads at speed 5 for 30 s. Total RNA was then treated to remove contaminating DNA with the DNA-free kit as per protocol (Ambion; no. 7020). Primers were designed from porcine SEP53 genomic sequence, which would amplify a product ∼600 bp spanning both coding exons’ II and III: PigExIIF_ATGCCTCAGTTACTGCGAAACA and PigExIIR_TGTCCTTGCACACTCTCTGG. First-strand cDNA was synthesised from 0.5 μg of total RNA from each porcine tissue type using Omniscript Reverse Transcription (ORT) Kit as described (Qiagen; no. 205111), with poly(dT)15 Primer (Promega), RNaseI, and 100 mM dithiothreitol; controls: RT mix; minus ORT (C1) or minus total RNA (C2). PCR was performed using HotStarTaq DNA Polymerase kit (Qiagen; no. 203203) with the above primers (400 nM) and 10% (v/v) of cDNA, and PCR products run on a 1% agarose gel.
2.6 Cell transfection and laser scanning confocal microscopy
HCT116 human colon carcinoma cells were transfected at 70% confluency onto four-chamber slide flasks (BD Biosciences) at 5 × 104/cm2, with 1 μg DNA of either wild type (pEYFP::SEP53), N-terminal Ca domain-truncated construct (pEYFP::ΔCa:SEP53), or control pEYFP-N1 vector, as described (Darragh et al. 2006). After transfection, cells were washed in PBS and fixed with ice-cold methanol for 10 min at −20°C, aspirated, and air-dried 10 min before storage in foil at −20°C until analysis. Cell nuclei were stained with TO-PRO3 (Molecular Probes; 1:1000) for 10 min, washed briefly in dH2O before mounting in one drop DakoCytomation Fluorescent Mounting Medium (Dako, no. 3023), coverslips sealed with clear nail varnish, and left overnight at 4°C to set. To detect yellow fluorescent protein expression in transfected cells, we used the λ 514 nm line of the Argon laser to excite and a long-pass λ530 nm filter to collect the emission, on a Zeiss LSM 510 confocal system, using a 63× oil immersion lens. Images were processed and viewed as a projected focus image from a stack of optical sections using the Zeiss LSM Image Browser (http://www.zeiss.co.uk/uk/micro/home.nsf/).
2.7 SEP53 protein expression: immunostaining of SEP53 protein in normal and diseased porcine pars oesophagea
Porcine tissues were obtained from a local abattoir and defined morphologically as being normal or diseased, as indicated by a veterinary pathologist (JRT). Human cancer tissues were obtained with approval of the ethics committee from the Masaryk Memorial Cancer Institute, Brno, Czech Republic. The tissue was fixed in 10% phosphate-buffered formalin before processing and embedding in paraffin wax. Tissue samples were cut at 4 μm onto Biobond-coated microscope slides (British Biocell). Sections were allowed to dry overnight at 37°C before staining. Fixed and processed tissue sections were de-waxed and rehydrated; endogenous peroxidase activity was blocked using 3% H2O2 in distilled H2O for 20 min. Antigen retrieval was then carried out using 0.01 M citrate buffer (pH 6.0) within a plastic pressure cooker (A Menarini Diagnostics) using a microwave oven as the heat source (800 W) for 15 min at full pressure followed by 15 min at 30% power. Non-specific protein blocking was carried out using a Vectastain Elite Universal ABC kit (Vector Laboratories) as per manufacturer’s instructions. The primary antibody to SEP53 was used at a dilution of 1/500 and applied for 1 h at room temperature followed by the secondary biotinylated reagent for 30 min. Visualisation was performed using 3,3′-diaminobenzidine tetrahydrochlorideDAB (Vector Laboratories) and viewed with an Olympus BX40 microscope. Negative controls were incorporated for each procedure using negative control serum (Dako).
3 Results
3.1 Cloning of the SEP53 gene from porcine genomic DNA
The availability of a high quality clone-based (BAC contig) physical map of the pig genome and the alignment of BAC end sequences with the human genome facilitate the rapid in silico identification of BAC clones potentially containing the porcine homologues of genes of interest. We identified BAC clones, including PigE-146b11 and PigE-78i12, from the Roslin PigEBAC library (Anderson et al. 2000) that potentially contained the porcine SEP53 locus from the pig–human BAC end sequence alignments. Further candidate PigE BAC clones, including PigE-67g6 and PigE-67g7, were identified from the homologous region of the pig physical map. DNA was prepared from the candidate BAC clones and screened for the presence of SEP53 sequences by Southern blot analyses using a 5′-biotinylated primer pair to PCR-amplify exon II of human SEP53 (GenBank accession no. AF077831; C1orf10). The hybridisation confirming the presence of the SEP53 orthologue in PigEBAC 67g6 and 67g7 is shown in Fig. 1, lanes 3–6. PigEBAC clones 67g6 and 67g7 were initially sequenced using human SEP53 ExonII primer set, and from this sequence data, subsequent porcine primer sets were designed to facilitate a ‘primer-walking’ sequencing strategy, involving sequencing of overlapping genomic fragments. The complete sequence of the porcine SEP53 locus (as present in the PigE-67g6 and PigE-67g7 clones) was deposited with GenBank (accession no. DQ910536).
Identification of PigEBAC clones harbouring the SEP53 locus. BAC clones from the putative region harbouring SEP53 were isolated from our in-house pig BAC genomic library (Anderson et al. 2000). A Restricted and B analysed by Southern blotting with a human SEP53 exon II biotinylated probe. The BAC clones number (lanes 1–6) are listed below B, and the arrow highlights the position of the positive signal in two BAC clones, 67g6 and 67g7
Subsequent comparisons of the PigE-67g7 clone sequence (DQ910536) revealed that the SEP53 locus is unstable in the PigE-67g7 clone and prone to deletion of sequences in the repeat region described below. The pig SEP53 locus present in the CH242-126j8 BAC clone (CU076058) is currently being sequenced as part of the pig genome sequencing project led by the Swine Genome Sequencing Consortium (Schook et al. 2005). The SEP53 locus found in the PigE-67g7 clone sequence (DQ910536) is missing 168 bp present in the CH242-126j8 clone (CU076058). We have determined the sequence of the SEP53 locus in the Meishan–Large White F1 male pig (95-0283), from which the PigEBAC library was constructed, by direct sequencing of genomic DNA. The SEP53 locus in this pig contains the missing 168 bp. We sequenced a new isolate of the PigE-67g7 BAC clone and found a similar, but non-identical deletion in the same region of the SEP53 locus confirming that these sequences are unstable in the PigEBAC library. Subsequent analyses of the SEP53 gene is based on the CH242-126j8 clone sequence (CU076058) and the sequence data from direct sequencing of the SEP53 locus in the Meishan–Large White F1 male pig (95-0283).
3.2 Genomic structure and evolution of the SEP53 gene
In a previous study, we compared the SEP53 gene in a range of mammal species (Little et al. 2007). To further establish whether the porcine SEP53 gene might be a robust animal model with which to analyse reflux-disease-mediated injury, in this study, we briefly review the major points of the previous comparative work (Little et al. 2007) while providing additional relevant detail. Bioinformatic analysis using a BLAST search of porcine SEP53 sequence confirmed that SEP53 is confined to mammals and that some regions of the gene are well conserved across the taxa studied (human, pig, rat, mouse, chimpanzee, cow, macaque, orangutan, and gorilla). For the human, chimpanzee, and porcine SEP53, we evaluated both the genomic sequence and the deduced coding sequence. SEP53 gene structure (Fig. 3a, exons I, II, and III) is defined by a small number of introns, which is common to some heat shock proteins, presumably limiting requirements for multi-site splicing after stress and allowing for more rapid protein production. The loci are similarly divided into one non-coding 50-bp exon, a 151-bp coding exon containing 50% of the highly conserved EF-hand calcium-binding domain, and a single, variable, and larger coding exon of 1,671 bp, representing the entire C-terminal domain, which is substantially less conserved (Figs. 2 and 3a). The position of the two introns is similar, but their length is divergent. Putative exon/intron splice/acceptor sites and a 3′ polyAdenylation signal (AATAAAA) are evident in the sequenced SEP53 gene region (see GenBank accession no. DQ910536). The above domains appear to be present in all mammalian SEP53 open reading frames (Little et al. 2007; Fig. 2). A striking feature of both porcine and chimpanzee SEP53 genes is that each has a large insertion (Fig. 3a,b). The insertion of a long stretch of amino acids is particularly unusual in the Pan-Homo comparisons, given the great similarity between DNA sequences in these species. Further, SEP53 contains repeat regions (Fig. 3a,b), and the insertion sequence in Pan is comprised of two additional tandem repeats that the humans lack (Figs. 2 and 3a,b). The insertion at the porcine SEP53 locus similarly is comprised of an additional repeat that most other species lack (Figs. 2 and 3a,b).
Predicted amino acid sequence and alignment of SEP53 proteins from three mammalian species. The highly conserved N-terminal Ca+-binding domain spans amino acids 1 to 90. The broken lines designate the positions of the insertion sites of chimp and pig (LW and MS strains), see also Fig. 3a and b. The black shading highlights conserved amino acids between pig, human, and chimpanzee. Grey shading represents regions of conservation in two or three out of the four representative protein sequences
SEP53 gene structure. a Schematic of the general structure of the human, chimpanzee, and porcine SEP53 gene. The locus in all species is composed of three exons (shaded grey), a non-coding exon I, start (ATG) and stop (TGA) codons, two coding exons (II–III) and two introns (green) of divergent length (exon size in nucleotides shown). Positions of species-specific insertions (black bars) are demarcated: humans have two repeats, chimpanzee’s have three, and pigs have five. b Partial sequence alignment of predicted SEP53 amino acid sequence repeats of (1) Pan/Homo and (2) pig. b Comparison with human SEP53 reveals amino acid insertion sequences in chimpanzee and pig. The black shading highlights conserved amino acids between pig, human, and chimpanzee. Grey shading represents regions of conservation in two or three out of the four representative protein sequences
While the calcium-binding domain of SEP53 is highly conserved across taxa (Fig. 2) and can maintain the cytoplasmic localisation of SEP53 protein (Fig. 4), parts of the C-terminal domain show exceptional levels of divergence. In particular, Little et al. (2007) showed an elevated rate of replacement nucleotide substitutions (substitutions that result in an amino acid change, K A) compared to the rate of synonymous nucleotide substitutions (substitutions that do not result in an amino acid substitutions, K S, which evolve at an approximately neutral rate). As most genes are subject to purifying selection, i.e. where replacement mutations produce inferior phenotypes that are pruned from the population, K A/K S between species tends to be much less than 1.0, typically <0.2 (Hurst 2002; Schlenke and Begun 2003), but for certain pairwise comparisons, K A/K S at the SEP53, the C-terminal open reading frame was well above 1.0 (Little et al. 2007). Phylogenetic analysis of all nine mammalian species confirmed that SEP53 was subject to adaptive evolution. However, we now note that the highest K A/K S values were observed only in comparisons between two primates (average K A/K S between two primates = 0.99), and comparisons involving non-primates indicated more modest K A/K S at SEP53 (average K A/K S between two non-primates or between a primate and a non-primate = 0.42). Thus, while SEP53 does evolve rapidly through positive selection, this phenomenon may be restricted to primates.
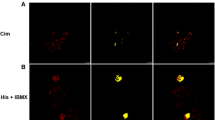
Function of the conserved N-terminal calcium-binding domain of SEP53. Laser scanning confocal immunofluorescent images of modified SEP53 expression in the human colonic carcinoma cell line HCT116. Deletion of the highly conserved EF-hand calcium-binding domain prevents cytoplasmic expression of SEP53 showing that this domain is required to maintain cytoplasmic expression of SEP53. Vectors encoding full-length SEP53 and a form of SEP53 with a deletion of the calcium-binding domain were transfected into HCT116 cells, fixed, counterstained with ToPro3 (blue), and viewed with a Zeiss 510Axioskop confocal microscope: A Full-length (wt; wild type) SEP53, B N-Terminal truncated; (DCa-SEP53), C Vector only, and D vehicle only
3.3 Over-expression of SEP53 protein in lesional porcine epithelium
Having acquired genomic sequence of the porcine SEP53 locus with a similar divergent C-terminal repeat domain to the human gene, we generated a set of PCR primers that would allow analysis of SEP53 mRNA using RT-PCR, as it was important to determine whether in fact the SEP53 gene is expressed in normal oesophageal squamous tissue. RNA was isolated from a range of tissues, and a product generated by RT-PCR to the correct length was only observed from oesophagus (Fig. 5A). These data indicate that the SEP53 gene is expressed in squamous epithelium. Similar expression of murine SEP53 has been reported from murine EST databases in skin and cervical squamous epithelium (data not shown).
Porcine SEP53 is expressed in oesophagus and differentially expressed in diseased pars oesophagea (PO) tissue. A RT-PCR analysing SEP53 expression in a porcine tissue panel; B (a–d) SEP53 immunostaining (a) normal PO—strong punctate staining (suprabasal epithelial cells, arrows), weak staining (basal layer, open arrows). (b) Hyperkeratotic PO—strong staining (basal and superficial epithelium), weak in differentiating cells (arrows). (c) Ulcerated PO—upregulation in scattered epithelial cells (arrows). (d) Healing PO—irregular staining of epithelial cells (arrows), weak staining of proliferating basal cells (open arrow). RT-PCR Tissue panel controls C1, template, C2, no RT-enzyme. Scale bars (μm): a–b 160; c–d 40. C Differential SEP53 protein expression in human oesophageal cancer tissue: The panels show SEP53 protein expression in: a adenocarcinoma of oesophagus vs b normal oesophageal squamous epithelium; scale bars (μm): a 160; b 40
The SEP53 protein is largely confined to squamous tissue, and as the protein is not expressed in commonly available cancer cell lines (data not shown), there are limited models with which to study SEP53 function and regulation. In fact, the previous characterisation required the production of stable SEP53-producing cell lines by transfection (Darragh et al. 2006). As such, animal models are required that can be subjected to transgenic technologies. Although mice are classically used as transgenic models of human gene function, the mouse does not have a bile–acid reflux tissue injury syndrome characteristic of humans and mice only have keratinised squamous oesophageal epithelium. By comparison, pathophysiological disease of the oesophagus has been reported in pig tissue: porcine gastro-oesophageal ulceration. This condition is mediated by reflux of gastric contents, presumably pepsin, bile, and acid, and has a significant impact on injury at the junction between oesophagus and stomach (Friendship 2003; Christie et al. 1995). We described porcine SEP53 expression with the goal of developing an animal model to allow, among other things, confirmation that SEP53 functions similarly in different taxa. Compared to the SEP53 protein expression patterns observed in human squamous epithelium (see Fig. 5C), SEP53 protein expression was detected in normal porcine squamous epithelium (Fig. 5B, a) with staining confined to the cytoplasm. However, a striking upregulation of SEP53 protein was observed in diseased porcine pars oesophagea. Staining in hyperplastic and hyperkeratotic lesions indicated upregulation in both the basal zone and in more mature differentiated cells with a striking band of lower intensity staining in cells in the intermediate stage of differentiation (Fig. 5B, b). In more advanced eroded and ulcerated lesions, there was irregular upregulation throughout the maturing epithelium, which was most intense in the region of ulceration (Fig. 5B, c). Healing and scarred lesions (Fig. 5B, d) showed patchy upregulation with minimal expression in the proliferating basal layer. By comparison, we observed much lowered and dysregulated SEP53 expression in the nucleus in oesophageal adenocarcinoma (Fig. 5C). Together, these changes provide evidence for differential expression and upregulation of SEP53 in response to squamous epithelial cell injury in porcine pars oesophagea and further establish this mammal as a useful model to investigate signalling pathways involved in bile-mediated cell injury in squamous epithelium.
4 Discussion
Concepts in the stress-protein response field are providing important foundations for understanding disease progression, as cell and tissue injury are linked intimately with the environment. Adenocarcinoma of the oesophagus is one of the fastest rising cancers in western countries, and growing evidence suggests that environmental stresses play a role in tissue injury: The reflux of acidic gastric fluid plays a role in tissue injury and elevation of risk for disease development. Recent evidence has indicated that bile acids including deoxycholic acid can induce significant DNA damage to cells and highlights a role for these chemicals in mediating cell injury (Jolly et al. 2004; Wild and Hardie 2003). Further data has demonstrated the existence of a unique array of stress proteins in normal human squamous epithelium and the role of one of these proteins (SEP53) in attenuating bile-mediated cell death and associated calcium release (Darragh et al. 2006). These data together provide a rationale for developing animal models in understanding the physiological control of bile-mediated injury to oesophageal squamous epithelial tissue. Oesophageal cell line models do not exist for studying SEP53 function. Given that porcine oesophageal tissue is known to be injured by gastric reflux, these data highlight the possibility that this mammal might be the most suitable model to better define physiological control of the DNA damage induced by bile acids. To begin such an analysis, we have cloned the SEP53 gene from porcine genomic DNA to characterise its gene structure, for developing sequence information in analysing SEP53 expression in porcine tissue and in analysing SEP53 expression in porcine pars oesophagea.
The gene structure the SEP53 gene is of particular importance, as earlier work has shown that some regions evolve exceptionally rapidly (Little et al. 2007). The structure of porcine SEP53 is similar to the human locus, with three exons and two introns contained within approximately 5,000 bp. The predicted open reading frame of porcine SEP53 shows strong homology to the human gene (and indeed to all mammalian SEP53 genes), particularly in the calcium-binding domain of exon II. The function of this domain is to maintain both the anti-apoptotic function of SEP53 in bile-induced damage assays and also to maintain the cytoplasmic localisation after gene transfection (Fig. 4; Darragh et al. 2006). Regarding exon III, two striking features emerge from our analysis: Firstly, the SEP53 gene contains repeat regions, and the number of repeats present can differ between even closely related species (Fig. 3a). Secondly, the SEP53 gene has a very large region that is undergoing positive selection, i.e. the repeated fixation of amino-acid-changing mutations (Little et al. 2007). This positive selection is largely confined to the large exon III, and the repeat regions appear to show the greatest degree of adaptive evolution (Little et al. 2007). These rapidly evolving regions in the C terminus of SEP53 have no homology to known proteins, so its function is unclear. The high degree of inserted repeats (two for humans up to five for porcine) suggests a structural adaptation to an antagonising pathogen. These data together suggest that non-synonymous mutations or polypeptide insertions, when they arise, can improve SEP53 function, and thus, the new variants are driven by natural selection to fixation. Most genes do not show evidence of positive selection, but immune system genes show an exceptionally high occurrence of positive selection (Ford 2002; Little and Cobbe 2005; Little et al. 2004; Obbard et al. 2006). This has led us to propose that SEP53 could play a role during pathogenic infection of epithelial tissues (Little et al. 2007). Interestingly, it may be common that genes involved in antagonistic co-evolutionary interactions are linked to increased cancer risk, a situation that arises when genes critically involved in key physiological processes are then subject to rapid evolution and thereby lose functional capacity in their original role (Crespi and Summers 2006). Regarding SEP53, the need to co-evolve with a pathogen could alter the protein to a degree that compromises stress-response functionality or the capacity to interact with other epithelial proteins, thus leading to tissue dysfunction that is not compatible with evolutionary fitness. In summary, stress protein responses are critical to maintain tissue integrity in metazoans, and the squamous epithelium in mammals has evolved a novel type of stress protein system. The most novel gene within this stress system is the SEP53 gene (Yagui-Beltran et al. 2001; Darragh et al. 2006), which is undergoing striking adaptive evolution (Little et al. 2007). Most human genes do not show discernible evidence of positive selection, but immune system genes show an exceptionally high occurrence of positive selection.. This has led us to propose that SEP53 could play a role during “pathogenic” infection of epithelial tissues (Little et al. 2007). This hypothesis demands direct testing for SEP53–pathogen interactions and requires a physiological model to examine the function of this stress protein pathway. Porcine squamous epithelium has been forwarded as a more physiological model for oesophageal squamous cell responses than the murine equivalent precisely because the pathophysiological disease of the porcine oesophagus we used to study SEP53 expression is similar to humans. The validation of the SEP53 gene in porcine squamous tissue now raises the possibility that transgenics, which overproduce or attenuate the SEP53 protein, can be developed to begin to understand how squamous epithelium responds to a range of environmental insults, including parasites, viruses, heat shock, acid, and bile-mediated DNA damage.
References
Anderson SI, Lopez-Corrales NL, Gorick B, Archibald AL (2000) A large-fragment porcine genomic library resource in a BAC vector. Mamm Genome 11:811–814
Ayles HL, Friendship RM, Ball RO (1996) Effect of dietary particle size on gastric ulcers, assessed by endoscopic examination, and relationship between ulcer severity and growth performance of individually fed pigs. Swine Health 4:211–216
Blake MJ, Gershon D, Fargnoli J, Holbrook NJ (1990) Discordant expression of heat shock protein mRNAs in tissues of heat-stressed rats. J Biol Chem 265:15275–15279
Blake MJ, Fargnoli J, Gershon D, Holbrook NJ (1991) Concomitant decline in heat-induced hyperthermia and HSP70 mRNA expression in aged rats. Am J Physiol 260:R663–667
Christie KN, Thomson C, Hopwood D (1995) A comparison of membrane enzymes of human and pig oesophagus; the pig oesophagus is a good model for studies of the gullet in man. Histochem J 27:231–239
Ciocca DR, Calderwood SK (2005) Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones 10:86–103
Crespi BJ, Summers K (2006) Positive selection in the evolution of cancer. Biol Rev 81:407–424
Darragh J, Hunter M, Pohler E, Dillon JF, Ross P, Kernohan N, Hupp TR (2006) The calcium binding domain of SEP53 is required for survival in response to DCA-mediated stress. FEBS J 273:1930–1947
Fitzgerald RC, Farthing MJ (2003) The pathogenesis of Barrett’s esophagus. Gastrointest Endosc Clin N Am 13:233–255
Ford MJ (2002) Applications of selective neutrality tests to molecular ecology. Mol Ecol 11:1245–1262
Friendship RM (1999) Gastric ulcers. In: Straw BE et al (ed) Diseases of swine. Iowa State Univesity Press, Ames Iowa, pp 685–694
Friendship RM (2003) Gastric ulcers; an under-recognized cause of mortality and morbidity. Adv Pork Prod 14:159–163
Hopwood D, Moitra S, Vojtesek B, Johnston DA, Dillon JF, Hupp TR (1997) Biochemical analysis of the stress protein response in human oesophageal epithelium. Gut 41:156–163
Humphray SJ, Scott C, Clark R, Marron B, Plumb R, Bender C, Camm N, Davis J, Jenks A, Noon A et al (2007) A high utility integrated map of the pig genome. Genome Biology 8:139, DOI 10.1186/gb-2007-8-7-r139
Hupp TR (2000) Development of physiological models to study stress protein responses. Methods Mol Biol 99:465–483
Hurst LD (2002) The Ka/Ks ratio:diagosing the form of sequence evolution. Trends Biochem Sci 18:486–487
Imai FL, Uzawa K, Nimura Y, Moriya T, Imai MA, Shiiba M, Bukawa H, Yokoe H, Tanzawa H (2005) Chromosome 1 open reading frame 10 (C1orf10) gene is frequently down-regulated and inhibits cell proliferation in oral squamous cell carcinoma. Int J Biochem Cell Biol 37:1641–1655
Jolly AJ, Wild CP, Hardie LJ (2004) Acid and bile salts induce DNA damage in human oesophageal cell lines. Mutagenesis 19:319–324
Lakhotia SC, Singh BN (1996) Synthesis of a ubiquitously present new HSP60 family protein is enhanced by heat shock only in the Malpighian tubules of Drosophila. Experientia 52:751–756
Little TJ, Cobbe N (2005) The evolution of immune-related genes from disease carrying mosquitoes: diversity in a peptidoglycan- and a thioester-recognizing protein. Insect Mol Biol 14:599–605
Little TJ, Colbourne JK, Crease TJ (2004) Molecular evolution of daphnia immunity genes: polymorphism in a gram-negative binding protein gene and an alpha-2-macroglobulin gene. J Mol Evol 59:498–506
Little TJ, Nelson L, Hupp TR (2007) Adaptive evolution of a stress response protein. PLos One 2:e1003
Metzler B, Hu Y, Dietrich H, Xu Q (2000) Increased expression and activation of stress-activated protein kinases/c-Jun NH(2)-terminal protein kinases in atherosclerotic lesions coincide with p53. Am J Pathol 156:1875–1886
Mosser DD, Morimoto RI (2004) Molecular chaperones and the stress of oncogenesis. Oncogene 23:2907–2918
Obbard DJ, Jiggins FM, Halligan DL, Little TJ (2006) Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol 16:580–585
Olliver JR, Hardie LJ, Dexter S, Chalmers D, Wild CP (2003) DNA damage levels are raised in Barrett’s oesophageal mucosa relative to the squamous epithelium of the oesophagus. Biomarkers 8:509–521
Reid BJ, Blount PL, Rabinovitch PS (2003) Biomarkers in Barrett’s esophagus. Gastrointest Endosc Clin N Am 13:369–397
Roosendaal R, Vos JH, Roumen T, van Vugt R, Cattoli G, Bart A, Klaasen HL, Kuipers EJ, Vandenbroucke-Grauls CM, Kusters JG (2000) Slaughter pigs are commonly infected by closely related but distinct gastric ulcerative lesion-inducing gastrospirilla. J Clin Microbiol 38:2661–2664
Schlenke TA, Begun DJ (2003) Natural selection drives Drosophila immune system evolution. Genetics 164:1471–1480
Schook LB, Beever J, Rogers J, Humphray S, Archibald A, Chardon P, Milan D, Rohrer G, Eversole K (2005) Swine Genome Sequencing Consortium (SGSC): a strategic roadmap for sequencing the pig genome. Compar Funct Genom 6:251–255
Vogelstein B, Kinzler KW (2004) Cancer genes and the pathways they control. Nat Med 10:789–799
Wild CP, Hardie LJ (2003) Reflux, Barrett’s oesophagus and adenocarcinoma: burning questions. Nat Rev Cancer 3:676–684
Xu Z, Wang MR, Xu X, Cai Y, Han YL, Wu KM, Wang J, Chen BS, Wang XQ, Wu M (2000) Novel human esophagus-specific gene c1orf10: cDNA cloning, gene structure, and frequent loss of expression in esophageal cancer. Genomics 69:322–330
Yagui-Beltran A, Craig AL, Lawrie L, Thompson D, Pospisilova S, Johnston D, Kernohan N, Hopwood D, Dillon JF, Hupp TR (2001) The human oesophageal squamous epithelium exhibits a novel type of heat shock protein response. Eur J Biochem 268:5343–5355
Acknowledgements
This work was supported in part by grants from the Cancer Research UK, the BBSRC, and the Association for International Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nelson, L., Anderson, S., Archibald, A.L. et al. An animal model to evaluate the function and regulation of the adaptively evolving stress protein SEP53 in oesophageal bile damage responses. Cell Stress and Chaperones 13, 375–385 (2008). https://doi.org/10.1007/s12192-008-0037-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12192-008-0037-1