Abstract
Relaxin is a prototype of the relaxin family peptide hormones and plays important biological functions by binding and activating the G protein-coupled receptor RXFP1. To study their interactions, in the present work, we applied the newly developed bioluminescent ligand–receptor binding assay to the relaxin-RXFP1 system. First, a fully active easily labeled relaxin, in which three Lys residues of human relaxin-2 were replaced by Arg, was prepared through overexpression of a single-chain precursor in Pichia pastoris and in vitro enzymatic maturation. Thereafter, the B-chain N-terminus of the easily labeled relaxin was chemically cross-linked with a C-terminal cysteine residue of an engineered NanoLuc through a disulfide linkage. Receptor-binding assays demonstrated that the NanoLuc-conjugated relaxin retained high binding affinity with the receptor RXFP1 (K d = 1.11 ± 0.08 nM, n = 3) and was able to sensitively monitor binding of a variety of ligands with RXFP1. Using the novel bioluminescent binding assay, we demonstrated that three highly conserved B-chain Arg residues of relaxin-3 had distinct contributions to binding of the receptor RXFP1. In summary, our present work provides a novel bioluminescent ligand–receptor binding assay for the relaxin-RXFP1 system to facilitate their interaction studies, such as characterization of relaxin analogues or screening novel agonists or antagonists of RXFP1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Relaxin is a prototype of the relaxin family peptide hormones, which includes relaxin, relaxin-3 (also known as INSL7), INSL3, INSL4, INSL5, and INSL6 (Bathgate et al. 2013; Cernaro et al. 2014; Nair et al. 2012). Relaxin was discovered in the 1920s and named “relaxin” because it induces relaxation of the pubic symphysis (Hisaw 1926). In the 1970s, the primary structure of relaxin was elucidated and it was found to be similar in structure to insulin, which resulted in the coining of the term “insulin superfamily” (James et al. 1977; Schwabe and McDonald 1977). In the early 1980s, relaxin genes were cloned from different species (Hudson et al. 1981, 1983), and it was confirmed that the two-chain relaxin was synthesized in vivo in a single-chain form. In humans and other primates, there are two relaxin genes, RLN1 and RLN2, but relaxin-2 is the major form found in circulation and in storage. In rodents and other mammals, only one relaxin gene is present. Although the physiological functions of relaxin have been extensively studied since its discovery, its receptor remained unknown for almost 80 years. It was only in 2002 that the orphan G protein-coupled receptor LGR7 (renamed as RXFP1 later) was found to be the long sought relaxin receptor (Hsu et al. 2002).
Relaxin is primarily expressed in the corpus lutea in females and in the prostate in males. Relaxin has classical reproductive functions, such as softening of the pelvic ligament for delivering offspring and promotion of mammary gland development in females, and maintaining sperm motility in males. Moreover, relaxin also has important non-reproductive functions, as evidenced by its anti-fibrotic and vasodilatory effects. Given the vasodilatory effect of relaxin, its role in the treatment of acute heart failure has been under investigation (Teerlink et al. 2013).
Ligand–receptor binding assay is a widely used technique to study the interaction of hormones with their receptors (Bylund and Toews 2011; Hulme and Trevethick 2010; Maguire et al. 2012). However, the conventional assays rely on radioisotope-labeled tracers, which have drawbacks such as radioactive hazards and short shelf lives. In recent studies, we developed novel bioluminescent ligand–receptor binding assays for some protein/peptide hormones (He et al. 2014; Liu et al. 2015; Song et al. 2015; Zhang et al. 2013a) based on a small nanoluciferase (NanoLuc) reporter that has the brightest bioluminescence reported to date (Hall et al. 2012). In the present study, we applied the novel bioluminescent ligand–receptor binding assay to the relaxin-RXFP1 system to facilitate their interaction studies, such as characterization of relaxin analogues and screening of novel agonists or antagonists of RXFP1.
Materials and methods
DNA manipulation
The gene encoding the easily labeled relaxin precursor was constructed from four chemically synthesized oligonucleotide primers through annealing and elongation via T4 DNA polymerase. After cleavage by the restriction enzyme KpnI, the synthesized gene was ligated into a pPinkα-HC vector (Invitrogen, Carlsbad, CA, USA), which had been pre-cleaved by the restriction enzymes StuI (blunt end) and KpnI (sticky end), resulting in the formation of the expression construct pPinkα-HC/relaxin. The nucleotide sequence of the easily labeled relaxin precursor was confirmed by DNA sequencing.
Overexpression of the easily labeled relaxin precursor
The expression construct pPinkα-HC/relaxin was linearized using the restriction enzyme SpeI and transformed into PichiaPink strain 1 using the spheroplast approach. Transformants were grown on a selective PAD plate and inoculated into 50 ml of liquid BMGY medium for small-scale methanol induction according to the manual of the PichiaPink™ Expression System (Invitrogen). Expression levels of the easily labeled relaxin precursor were analyzed using tricine SDS-PAGE. The colony with the highest expression level was used for large-scale culture in salt medium (5 g/l NH4Cl, 4 g/l KH2PO4, 4 g/l K2HPO4, 0.4 g/l CaCl2, 0.4 g/l NaCl, 2 g/l MgSO4, and 2 ml/l PTM1 trace elements) in shaking flasks and induced by addition of methanol at an interval of 12 h according to our previous procedure (Guo et al. 2015).
Purification of the easily labeled relaxin precursor
Culture broth supernatant was first loaded onto a cation ion-exchange column (sulfonic ethyl Sephadex) pre-equilibrated with 20 mM glycine buffer (pH 2.0). After loading, the column was washed with 20 mM glycine buffer (pH 2.0), and the bound easily labeled relaxin precursor was eluted from the column using 20 mM glycine buffer (pH 2.0) containing 1.0 M NaCl. The eluted fraction was then subjected to high-performance liquid chromatography (HPLC), and the easily labeled relaxin precursor was eluted from a C18 reverse-phase column (Zorbax 300SB-C18, 9.4 × 250 mm; Agilent Technologies, Santa Clara, CA, USA) by an acidic acetonitrile gradient, manually collected, lyophilized, and analyzed by mass spectrometry.
Enzymatic maturation of the easily labeled relaxin precursor
The purified easily labeled relaxin precursor was dissolved in digestion buffer (100 mM Tris–HCl, 2 M urea, pH 8.0) at a final concentration of approximately 5 mg/ml. First, endoproteinase Lys-C (Sigma-Aldrich, St. Louis, MO, USA) was added (1 U enzyme versus ~15 mg peptide), and the digestion was carried out at 30 °C overnight. Second, purified papaya glutaminyl cyclase was added (mass ratio of enzyme to peptide, 1:100) and the solution was continuously incubated at 30 °C for 3 h. Third, carboxypeptidase B was added (mass ratio of enzyme to peptide, 1:100) and the solution was continuously incubated at 30 °C for 1 h. Finally, the mixture was acidified to pH3 and subjected to HPLC. The mature easily labeled relaxin was eluted from a C18 reverse-phase column (Zorbax 300SB-C18, 4.6 × 250 mm; Agilent Technologies) by an acidic acetonitrile gradient, manually collected, lyophilized, and analyzed by mass spectrometry.
Circular dichroism analysis
The mature easily labeled relaxin was dissolved in 1.0 mM aqueous HCl (pH 3.0) and quantified by absorbance at 280 nm using the extinction coefficient of 12,490 M−1 cm−1. Thereafter, its final concentration was adjusted to 20 μM, and circular dichroism measurement was carried out on a Jasco-715 polarimeter at room temperature. The spectrum was scanned from 190 to 250 nm using a cuvette with a 1.0-mm path length. The secondary structural content was estimated from the spectrum using the software J-700 for Windows Secondary Structural Estimation (Version 1.10.00).
Receptor activation assay of the mature easily labeled relaxin
The receptor activation assays were carried out according to our previous procedure (Zhang et al. 2012b) using human embryonic kidney (HEK) 293T cells transiently cotransfected with the expression construct of human RXFP1 (pcDNA6/RXFP1) and the cAMP-response element (CRE)-controlled NanoLuc reporter vector pNL1.2/CRE (Zhang et al. 2014) that was generated in our previous work by insertion of a synthetic CRE sequence into the pNL1.2 vector (Promega, Madison, WI, USA). The transfected cells were trypsinized and seeded into a 96-well plate, and the cells were continuously cultured for 24–36 h to ~90 % confluence. Thereafter, the medium was removed and the assay solution (serum-free DMEM medium plus 1 % bovine serum albumin), containing 25 μM 3-isobutyl-1-methylxanthine and various concentrations of mature easily labeled relaxin, was added (100 μl/well). After continuous culture for 5–6 h, the cells were lysed using Lysis Solution (Promega, 100 μl/well) and the cell lysate was transferred to a white opaque 96-well plate (50 μl/well). After addition of freshly diluted substrate (50 μl/well), bioluminescence was immediately measured on a SpectroMax M5 plate reader (Molecular Devices, Sunnyvale, CA, USA) using the luminescence mode. The measured bioluminescence data were expressed as mean ± SE (n = 3) and fitted with sigmoidal curves using the SigmaPlot10.0 software.
Chemical conjugation of the easily labeled relaxin with NanoLuc
For N-succinimidyl-3-(2-pyridyldithiol)propionate (SPDP) modification, the mature easily labeled relaxin was dissolved in dimethyl sulfoxide at a final concentration of ~1.5 mM. The modification reagent SPDP was dissolved in anhydrous N,N-dimethyl formamide at a final concentration of 80 mM. To initiate modification, one volume of the relaxin stock solution was sequentially mixed with 0.73 volumes of 0.2 M aqueous phosphate buffer (pH 7.5), 0.17 volumes of 6 M guanidine chloride, and 0.1 volumes of the SPDP stock solution. The modification reaction was carried out at 30 °C for 1–2 h. Thereafter, the reaction mixture was diluted 10-fold with water, acidified to pH3 with trifluoroacetic acid, and subjected to HPLC. The peaks eluted from a C18 reverse-phase column (Zorbax 300SB-C18, 4.6 × 250 mm; Agilent Technologies) by an acidic acetonitrile gradient were manually collected, lyophilized, and analyzed by mass spectrometry.
For conjugation with the engineered NanoLuc (Luc-Cys) carrying a unique exposed Cys at the C-terminus (Liu et al. 2015), the SPDP-modified relaxin was dissolved in 1.0 mM aqueous HCl solution (pH 3.0) at a final concentration of ~0.3 mM. Thereafter, one volume of the modified relaxin stock solution was sequentially mixed with one volume of reaction buffer (300 mM Tris–HCl, 3.0 mM EDTA, and 6.0 M urea, pH 7.5) and one volume of the purified NanoLuc solution (eluted from a DEAE column with a concentration of ~0.1 mM). The conjugation reaction was carried out at 30 °C for 30 min. Thereafter, the reaction mixture was 10-fold diluted with 20 mM phosphate buffer (pH 7.5) and loaded onto a DEAE ion-exchange column (TSKgel DEAE-5PW, 7.5 × 75 mm, from Sigma Aldrich). The peaks eluted by a sodium chloride gradient were manually collected and analyzed by non-reducing SDS-PAGE.
Receptor-binding assays
HEK293T cells were transiently transfected with the expression construct of human RXFP1 (pcDNA6/RXFP1). The next day, the transfected cells were trypsinized and seeded into a 96-well plate. After the cells grew to confluence (24–36 h), the medium was removed and binding solution (serum-free DMEM medium plus 1 % bovine serum albumin) was added (100 μl/well). In the saturation binding assays, the binding solution contained various concentrations of the NanoLuc-conjugated relaxin (R2-Luc); in the competition binding assays, the binding solution contained 0.5 nM of R2-Luc and various concentrations of the competitor. Nonspecific binding was determined by competition with 1.0 μM of the mature easily labeled relaxin. After incubation at 21 °C for 1–2 h, the assay solution was removed and the cells were washed twice with ice-cold phosphate-buffered saline (200 μl/well for each wash). Subsequently, the cells were lysed using Lysis Solution (100 μl/well, from Promega), and the cell lysate was transferred to a white opaque 96-well plate (50 μl/well). After addition of the freshly diluted substrate (50 μl/well), bioluminescence was immediately measured on a SpectroMax M5 plate reader (Molecular Devices) using the luminescence mode. The measured bioluminescence data were expressed as mean ± SE (n = 3) and fitted with one-site binding curves using the SigmaPlot10.0 software.
Results and discussion
Design of an easily labeled relaxin for site-specific conjugation with NanoLuc
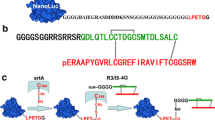
To apply the novel bioluminescent ligand–receptor binding assay to the relaxin-RXFP1 system, we designed an easily labeled relaxin, in which three Lys residues of human relaxin-2 were replaced by Arg, for convenient site-specific conjugation with the NanoLuc reporter (Fig. 1). The easily labeled relaxin has a unique primary amine moiety at the B-chain N-terminus due to the formation of a primary amine-less pyroglutamate residue at its A-chain N-terminus and lack of internal primary amine moieties. Thus, it could be conveniently conjugated with an engineered NanoLuc reporter carrying a unique exposed C-terminal Cys residue (Liu et al. 2015) using the bifunctional cross-linker SPDP, which carries a primary amine-specific N-hydroxysuccinimidyl (NHS) ester moiety and a sulfhydryl-specific pyridyldithiol moiety, according to our previous procedure (Zhang et al. 2013a). To prepare the easily labeled relaxin using the Pichia expression system, which can efficiently overexpress a chimeric R3/I5 precursor as found in our previous work (Guo et al. 2015), we designed a single-chain precursor in which the B-chain C-terminus and the A-chain N-terminus of the easily labeled relaxin were joined together by a short peptide linker (Fig. 1a). After secretory overexpression in Pichia pastoris and purification, the precursor was converted to mature easily labeled relaxin through sequential treatment with endoproteinase Lys-C, papaya glutaminyl cyclase and carboxypeptidase B according to our previous procedures (Zhang et al. 2012a, b, 2013b).
a Schematic presentation of the amino acid sequence of the easily labeled relaxin precursor overexpressed in Pichia pastoris. The B-chain is shown in red, and the A-chain in green. Cys residues are shown as filled circles, and disulfide bonds as rods. The positions of Lys replaced by Arg are shown in blue. The cleavage sites of endoproteinase Lys-C are indicated by arrows. The A-chain N-terminal Gln residue that was converted to pyroglutamate by papaya glutaminyl cyclase after Lys-C cleavage is indicated by an arrowhead. The B-chain C-terminal Lys residue removed by carboxypeptidase B after Lys-C cleavage is indicated by an asterisk. b Schematic presentation of the NanoLuc-conjugated easily labeled relaxin (color figure online)
Preparation of the easily labeled relaxin
After the synthesized gene of the easily labeled relaxin precursor was cloned into the Pichia expression vector pPinkα-HC and transformed into PinkPichia strain 1, the transformants were subjected to a small-scale expression test and tricine SDS-PAGE analysis confirmed secretory expression of the easily labeled relaxin precursor. Thereafter, the transformant with the highest expression level was used for large-scale culture in salt medium in shaking flasks. As analyzed by tricine SDS-PAGE (Fig. 2a, inner panel), band density of the easily labeled relaxin precursor (indicated by an asterisk) gradually increased after methanol induction and reached a plateau around 72–84 h. The precursor yield estimated from the band density was 50–60 mg/l in the shaking flasks. If cultured in a fermenter, the yield would be significantly increased, typically over fivefold, because high cell density could be reached in fermentation.
Purification and enzymatic maturation of the easily labeled relaxin precursor. a HPLC purification of the easily labeled relaxin precursor. The eluent from the cation ion-exchange chromatography was loaded on a C18 reverse-phase column and eluted by an acidic acetonitrile gradient. Inner panel tricine SDS-PAGE analysis. The culture supernatant (20 μl) after methanol induction was loaded onto a 16.5 % tricine SDS-gel. After electrophoresis, the gel was stained with Commasie Brilliant Blue R250. The band of the easily labeled relaxin precursor is indicated by an asterisk. b HPLC purification of the mature easily labeled relaxin. After the purified precursor was sequentially treated with endoproteinase Lys-C, papaya glutaminyl cyclase and carboxypeptidase B, the mixture was loaded onto a C18 reverse-phase column and eluted by an acidic acetonitrile gradient. Inner panel mass spectrometry analysis of the major fraction
For purification, the easily labeled relaxin precursor in the culture supernatant was first absorbed by a cation ion-exchange column. The eluted precursor fraction from the ion-exchange column was then subjected to reverse-phase HPLC, and a major peak (indicated by an asterisk) was eluted from a C18 reverse-phase column (Fig. 2a). Mass spectrometry analysis confirmed that the major peak represented the easily labeled relaxin precursor fraction (measured value 7519.0; theoretical value 7519.5). From 1 l of the Pichia culture broth, approximately 30 mg of the purified precursor could be obtained. Thus, the easily labeled relaxin precursor could be efficiently prepared through overexpression in Pichia pastoris.
Thereafter, the purified easily labeled relaxin precursor was sequentially treated by endoproteinase Lys-C (cleavage of the peptide bond at the C-terminal of Lys residues), papaya glutaminyl cyclase (conversion of the A-chain N-terminal Gln to pyroglutamate after Lys-C cleavage), and carboxypeptidase B (removal of the additional B-chain C-terminal Lys residue after Lys-C cleavage). After the digestion mixture was subjected to HPLC, a major peak (indicated by an asterisk) was eluted from a C18 reverse-phase column (Fig. 2b). As analyzed by mass spectrometry (Fig. 2b, inner panel), the major peak had the expected molecular mass (measured value 6117.0; theoretical value 6118.1), which suggests that mature easily labeled relaxin was obtained after in vitro enzymatic maturation. From 10 mg of the purified precursor, typically 5–6 mg of mature peptide was obtained, suggesting that the enzymatic maturation approach was efficient.
Characterization of the mature easily labeled relaxin
To test whether the mature easily labeled relaxin was correctly folded, we analyzed its secondary structure by circular dichroism (Fig. 3a). The mature peptide showed a typical helix-dominated conformation similar to that of the recombinant relaxin-3. Its α-helix content estimated from the spectrum was 48 %, which is consistent with the crystal structure of human relaxin-2 (Eigenbrot et al. 1991). Thus, we deduced that the Pichia-overexpressed easily labeled relaxin had a native conformation with correct disulfide linkages.
Characterization of the mature easily labeled relaxin. a Circular dichroism spectra. b Receptor activation assay. The expression construct of human RXFP1 and a CRE-controlled NanoLuc reporter were cotransfected into HEK293T cells, and the NanoLuc activity was measured after these cells were treated with the mature easily labeled relaxin or relaxin-3. The measured bioluminescence data were expressed as mean ± SE (n = 3) and fitted to sigmoidal curves using the software SigmaPlot 10.0. Data are representative of two independent experiments that gave essentially the same results
Activity of the mature easily labeled relaxin was measured by receptor activation assay using a CRE-controlled NanoLuc reporter (Fig. 3b). The easily labeled relaxin activated the receptor RXFP1 in a typical sigmoidal manner, with a calculated pEC50 of 10.52 ± 0.11 (n = 3), consistent with the previously reported pEC50 value (10.60 ± 0.04, n = 4) of the chemically synthesized relaxin-2 (Bathgate et al. 2006). The measured activity of the easily labeled relaxin was higher than that of the recombinant relaxin-3 (pEC50 = 9.70 ± 0.07, n = 3), consistent with the previous study which showed that relaxin-2 has higher activity than relaxin-3 towards RXFP1 (Bathgate et al. 2006; Sudo et al. 2003). Thus, the easily-labeled relaxin retained full activity and was suitable for conjugation with the NanoLuc reporter.
Conjugation of the easily labeled relaxin with NanoLuc
To prepare a bioluminescent relaxin tracer for ligand–receptor binding assays, we chemically conjugated the fully active easily labeled relaxin with an engineered NanoLuc (Luc-Cys) that carries a long hydrophilic arm and a unique exposed Cys at the C-terminus (Liu et al. 2015). First, an active disulfide bond (pyridyldithiol moiety) was introduced into the B-chain N-terminus of the easily labeled relaxin through reaction with a bifunctional reagent SPDP. After the modification mixture was subjected to HPLC, several peaks were eluted from a C18 reverse-phase column (Fig. 4a). After lyophilization, mass spectrometry analysis (Fig. 4a, inner panel) revealed that the asterisk-indicated peak represented the modified relaxin carrying a single pyridyldithiol moiety (measured value 6314.0; theoretical value 6315.4). Thereafter, the modified relaxin was conjugated with Luc-Cys through disulfide exchange of the unique exposed Cys of Luc-Cys with the introduced active disulfide bond of the easily labeled relaxin. After the conjugation mixture was loaded onto an ion-exchange column, an asymmetric peak was eluted using a sodium chloride gradient (Fig. 4b). We manually collected the peak into three fractions (labeled as 1–3) and analyzed them using non-reducing SDS-PAGE (Fig. 4b, inner panel). The fraction 1 had a major band that represented the unreacted monomeric Luc-Cys (indicated by the letter m). The fraction 2 had a major band that represented the expected conjugate (indicated by an asterisk) and two minor bands that represented monomeric and dimeric Luc-Cys (indicated by the letters m and d, respectively). The fraction 3 had a single band of the expected conjugate (R2-Luc) with an apparent molecular weight of approximately 30 kDa (theoretical value 29.2 kDa) and was used for later experiments. The bioluminescence of R2-Luc was 1.5 × 105 counts/fmol when measured on a SpectroMax M5 plate reader using a white opaque 96-well plate, confirming that the NanoLuc reporter was fully active after conjugation with relaxin.
Conjugation of NanoLuc with the easily labeled relaxin. a HPLC purification of the SPDP-modified easily labeled relaxin. After the mature easily labeled relaxin was modified by SPDP, the reaction mixture was loaded onto a C18 reverse-phase column and eluted by an acidic acetonitrile gradient. The peak of SPDP-modified relaxin is indicated by an asterisk. Inner panel mass spectrometry analysis of the SPDP-modified relaxin. b HPLC purification of the NanoLuc-conjugated relaxin. After reaction of the SPDP-modified relaxin with Luc-Cys, the conjugation mixture was loaded onto a DEAE ion-exchange column and eluted by a gradient of sodium chloride. The eluted peak was manually collected into three fractions labeled as 1, 2 or 3. Inner panel non-reducing SDS-PAGE analysis of the eluted fractions. An aliquot of each fraction was loaded onto a 12 % non-reducing SDS-gel. After electrophoresis, the gel was silver stained. The R2-Luc band is indicated by an asterisk, and the monomeric or dimmeric Luc-Cys bands are indicated by the letters m and d, respectively
Binding affinity of the NanoLuc-conjugated relaxin with the receptor RXFP1
To test whether R2-Luc retained high binding affinity with the receptor RXFP1, we carried out a saturation binding assay using HEK293T cells transiently overexpressing human RXFP1 as the receptor source. As shown in Fig. 5a, R2-Luc bound the living HEK293T cells overexpressing RXFP1 in a typical saturation manner, with a calculated dissociation constant (K d) of 1.11 ± 0.08 nM (n = 3). The measured K d of R2-Luc was similar to the previously reported K d values of the europium-labeled relaxin (0.85 ± 0.08 nM for B-chain labeling and 0.50 ± 0.04 nM for A-chain labeling) (Shabanpoor et al. 2012) and the radioisotope-labeled relaxin (0.209 ± 0.025 nM for 33P-labeling and 0.18 nM for 125I-labeling) (Sudo et al. 2003; Kuei et al. 2007), thus the conjugated NanoLuc had no serious detriments to the receptor-binding affinity of relaxin. In contrast, no specific binding was detected on the non-transfected HEK293T cells (Fig. 5c), which suggests that the HEK293T cells did not express the endogenous relaxin receptor. NanoLuc itself could not bind the HEK293T cells overexpressing RXFP1 (Fig. 5c), which suggests that the detected specific R2-Luc binding with these cells was attributed to the easily labeled relaxin. Thus, R2-Luc retained high binding affinity with the receptor RXFP1, although a large NanoLuc moiety was attached. The measured maximal binding capacity (B max) was 137000 ± 3000 counts/well (n = 3), equal to the average receptor density of ~11,000 receptors/cell (the specific activity of NanoLuc was 1.5 × 105 counts/fmol and ~5×104 cells/well). Moreover, nonspecific binding of R2-Luc was quite low due to the highly hydrophilic nature of the NanoLuc reporter. The Scatchard plot of the specific binding data was a linear curve (Fig. 5b), which suggests the one-site binding of R2-Luc with the receptor RXFP1.
Binding of the NanoLuc-conjugated relaxin towards the receptor RXFP1. a Saturation binding with the receptor RXFP1. HEK293T cells transiently overexpressing human RXFP1 were used as the receptor source. Nonspecific binding data were obtained by competition with 1.0 μM of mature easily labeled relaxin. The measured bioluminescence data were expressed as mean ± SE (n = 3). Total binding data were fitted to Y = B maxX/(K d + X) + NsX, specific binding data to Y = B maxX/(K d + X), and nonspecific binding data to a linear curve, using the SigmaPlot10.0 software. b Scatchard plot of the specific binding data. c Binding of R2-Luc with non-transfected HEK293T cells and binding of Luc-Cys with HEK293T cells overexpressing RXFP1. The measured bioluminescence data were expressed as mean ± SE (n = 3). All data in this figure are representative of two independent experiments that gave essentially the same results
Novel bioluminescent ligand–receptor binding assay for relaxin-RXFP1 system
As R2-Luc retained high binding affinity with the receptor RXFP1, we used it as a novel bioluminescent tracer in competition assays to determine the binding potencies of various RXFP1 ligands (Fig. 6a). Competition binding of the mature easily-labeled relaxin showed a typical sigmoidal curve, with a calculated pIC50 of 9.36 ± 0.02 (n = 3), which was higher than the measured pIC50 (8.82 ± 0.03, n = 3) of relaxin-3. This is consistent with the finding that relaxin-2 has higher activity than relaxin-3 towards RXFP1 (Bathgate et al. 2006; Sudo et al. 2003). Thus, the novel bioluminescent relaxin tracer could sensitively monitor the binding potencies of different ligands towards the receptor RXFP1.
Bioluminescent ligand–receptor binding assays for relaxin-RXFP1 system. a Competition binding using the NanoLuc-conjugated relaxin as a tracer. The measured relative bioluminescence data were expressed as mean ± SE (n = 3) and fitted to sigmoidal curves using the SigmaPlot10.0 software. Data are representative of two independent experiments that gave essentially the same results. b Amino acid sequence alignment of relaxin-3 B-chains from different species. The three highly conserved Arg residues are shown in gray
Relaxin-3 (also known as INSL7) can also bind and activate RXFP1 in vitro. It has three highly conserved Arg residues in the B-chain—B12Arg, B16Arg, and B26Arg (Fig. 6b). In our previous work, we replaced these positively charged Arg residues with negatively charged Glu and measured their binding potencies towards RXFP3 and RXFP4 (Wang et al. 2014; Zhang et al. 2014). In the present work, we determined their binding potencies towards RXFP1 by using the novel bioluminescent binding assay (Fig. 6a and the calculated pIC50 values were summarized in Table 1). When the B-chain C-terminal B26Arg was replaced by Glu, the mutant retained similar binding potency towards the receptor RXFP1 compared with the wild-type relaxin-3. Thus, B26Arg was not required for binding with RXFP1 although it is critical for binding with RXFP3 and RXFP4 (Table 1). When the B-chain central B12Arg or B16Arg was replaced by Glu, both mutants almost lost their binding potencies with RXFP1 (pIC50 < 6), which suggests that both Arg residues were critical for relaxin-3 to bind RXFP1. These two B-chain central Arg residues are also involved in binding with RXFP3 and RXFP4, but their roles are less important than those of B26Arg towards RXFP3 and RXFP4 (Table 1). Thus, the three highly conserved B-chain Arg residues play distinct roles in binding with RXFP1, RXFP3, and RXFP4.
Application of the bioluminescent ligand–receptor binding assay to other protein/peptide hormones
Protein/peptide hormones are the largest group of endogenous signaling molecules, including growth factors, cytokines and neuropeptides. By binding specific cell membrane receptors, proteins/peptide hormones play important biological functions and some of them have been developed as drugs for the treatment of diseases. Our present bioluminescent ligand–receptor binding assay could be applied to other protein/peptide hormones in the future studies due to its high sensitivity, safety, and convenience. There are two general approaches to prepare the NanoLuc-based bioluminescent tracers: chemical conjugation and genetic fusion. In the chemical conjugation approach, a recombinant or chemically synthesized protein/peptide hormone was covalently cross-linked with an engineered NanoLuc through a suitable cross-linker. Using this approach, we have established bioluminescent ligand–receptor binding assays for INSL3 (Zhang et al. 2013a), ghrelin (Liu et al. 2015), and relaxin (present work). In the genetic fusion approach, a protein/peptide hormone was genetically fused at the N-terminal or C-terminal of NanoLuc and the fusion protein was overexpressed in suitable host cells. Using this approach, we have established bioluminescent ligand–receptor binding assays for leukemia inhibitor factor (He et al. 2014) and erythropoietin (Song et al. 2015). In summary, the novel bioluminescent ligand–receptor binding assay has wide applications in interaction studies of protein/peptide hormones with their receptors in future.
References
Bathgate RA, Lin F, Hanson NF, Otvos L Jr, Guidolin A, Giannakis C, Bastiras S, Layfield SL, Ferraro T, Ma S, Zhao C, Gundlach AL, Samuel CS, Tregear GW, Wade JD (2006) Relaxin-3: improved synthesis strategy and demonstration of its high-affinity interaction with the relaxin receptor LGR7 both in vitro and in vivo. Biochemistry 45:1043–1053
Bathgate RA, Halls ML, van der Westhuizen ET, Callander GE, Kocan M, Summers RJ (2013) Relaxin family peptides and their receptors. Physiol Rev 93:405–480
Bylund DB, Toews ML (2011) Radioligand binding methods for membrane preparations and intact cells. Methods Mol Biol 746:135–164
Cernaro V, Lacquaniti A, Lupica R, Buemi A, Trimboli D, Giorgianni G, Bolignano D, Buemi M (2014) Relaxin: new pathophysiological aspects and pharmacological perspectives for an old protein. Med Res Rev 34:77–105
Eigenbrot C, Randal M, Quan C, Burnier J, O’Connell L, Rinderknetct E, Kossiakoff AA (1991) X-ray structure of human relaxin at 1.5 Å: comparison to insulin and implications for receptor binding determinants. J Mol Biol 221:15–21
Guo YQ, Wu QP, Shao XX, Shen T, Liu YL, Zeng GX, Guo ZY (2015) Secretory overexpression and isotopic labeling of the chimeric relaxin family peptide R3/I5 in Pichia pastoris. Amino Acids 47:1117–1125
Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV (2012) Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol 7:1848–1857
He SX, Song G, Shi JP, Guo YQ, Guo ZY (2014) Nanoluciferase as a novel quantitative protein fusion tag: application for overexpression and bioluminescent receptor-binding assays of human leukemia inhibitory factor. Biochimie 106:140–148
Hisaw F (1926) Experimental relaxation of the pubic ligament of the guinea pig. Proc Soc Exper Biol Med 23:661–663
Hsu SY, Nakabayashi K, Nishi S, Kumagai J, Kudo M, Sherwood OD, Hsueh AJ (2002) Activation of orphan receptors by the hormone relaxin. Science 295:671–674
Hudson P, Haley J, Cronk M, Shine J, Niall H (1981) Molecular cloning and characterization of cDNA sequences coding for rat relaxin. Nature 291:127–131
Hudson P, Haley J, John M, Cronk M, Crawford R, Haralambidis J, Tregear G, Shine J, Niall H (1983) Structure of a genomic clone encoding biologically active human relaxin. Nature 301:628–631
Hulme EC, Trevethick MA (2010) Ligand binding assays at equilibrium: validation and interpretation. Br J Pharmacol 161:1219–1237
James R, Niall H, Kwok S, Bryand-Greenwood G (1977) Primary structure of porcine relaxin: homology with insulin and related growth factors. Nature 267:544–546
Kuei C, Sutton S, Bonaventure P, Pudiak C, Shelton J, Zhu J, Nepomuceno D, Wu J, Chen J, Kamme F, Seierstad M, Hack MD, Bathgate RA, Hossain MA, Wade JD, Atack J, Lovenberg TW, Liu C (2007) R3(BDelta23-27)R/I5 chimeric peptide, a selective antagonist for GPCR135 and GPCR142 over relaxin receptor LGR7: in vitro and in vivo characterization. J Biol Chem 282:25425–25435
Liu Y, Shao XX, Zhang L, Song G, Liu YL, Xu ZG, Guo ZY (2015) Novel bioluminescent receptor-binding assays for peptide hormones: using ghrelin as a model. Amino Acids 47:2237–2243
Maguire JJ, Kuc RE, Davenport AP (2012) Radioligand binding assays and their analysis. Methods Mol Biol 897:31–77
Nair VB, Samuel CS, Separovic F, Hossain MA, Wade JD (2012) Human relaxin-2: historical perspectives and role in cancer biology. Amino Acids 43:1131–1140
Schwabe C, McDonald JK (1977) Relaxin: a disulfide homolog of insulin. Science 197:914–915
Shabanpoor F, Bathgate RA, Belgi A, Chan LJ, Nair VB, Wade JD, Hossain MA (2012) Site-specific conjugation of a lanthanide chelator and its effects on the chemical synthesis and receptor binding affinity of human relaxin-2 hormone. Biochem Biophys Res Commun 420:253–256
Song G, Wu QP, Xu T, Liu YL, Xu ZG, Zhang SF, Guo ZY (2015) Quick preparation of nanoluciferase-based tracers for novel bioluminescent receptor-binding assays of protein hormones: using erythropoietin as a model. J Photochem Photobiol, B 153:311–316
Sudo S, Kumagai J, Nishi S, Layfield S, Ferraro T, Bathgate RA, Hsueh AJ (2003) H3 relaxin is a specific ligand for LGR7 and activates the receptor by interacting with both the ectodomain and the exoloop 2. J Biol Chem 278:7855–7862
Teerlink JR, Cotter G, Davison BA, Felker GM, Filippatos G, Greenberg BH, Ponikowski P, Unemori E, Voors AA, Adams KF Jr, Dorobantu MI, Grinfeld LR, Jondeau G, Marmor A, Masip J, Pang PS, Werdan K, Teichman SL, Trapani A, Bush CA, Saini R, Schumacher C, Severin TM, Metra M (2013) Serelaxin, recombinant human relaxin-2, for treatment of acute heart failure (RELAX-AHF): a randomised, placebo-controlled trial. Lancet 381:29–39
Wang XY, Guo YQ, Zhang WJ, Shao XX, Liu YL, Xu ZG, Guo ZY (2014) The electrostatic interactions of relaxin-3 with receptor RXFP4 and the influence of its B-chain C-terminal conformation. FEBS J 281:2927–2936
Zhang WJ, Gao XJ, Liu YL, Shao XX, Guo ZY (2012a) Design, recombinant preparation and europium-labeling of a fully active easily-labeled INSL3 analogue for receptor-binding assays. Process Biochem 47:1856–1860
Zhang WJ, Luo X, Song G, Wang XY, Shao XX, Guo ZY (2012b) Design, recombinant expression and convenient A-chain N-terminal europium-labelling of a fully active human relaxin-3 analogue. FEBS J 279:1505–1512
Zhang L, Song G, Xu T, Wu QP, Shao XX, Liu YL, Xu ZG, Guo ZY (2013a) A novel ultrasensitive bioluminescent receptor-binding assay of INSL3 through chemical conjugation with nanoluciferase. Biochimie 95:2454–2459
Zhang WJ, Jiang Q, Wang XY, Geng S, Shao XX, Guo ZY (2013b) A convenient method for europium-labeling of a recombinant chimeric relaxin family peptide R3/I5 for receptor-binding assays. J Pept Sci 19:350–354
Zhang WJ, Wang XY, Guo YQ, Luo X, Gao XJ, Shao XX, Liu YL, Xu ZG, Guo ZY (2014) The highly conserved negatively charged Glu141 and Asp145 of the G-protein-coupled receptor RXFP3 interact with the highly conserved positively charged arginine residues of relaxin-3. Amino Acids 46:1393–1402
Acknowledgments
We thank Promega Corporation for providing the plasmids encoding NanoLuc. This work was supported by the National Natural Science Foundation of China (31470767, 31270824) and the Fundamental Research Funds for the Central Universities (2000219098).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: J. D. Wade.
Q.-P. Wu and L. Zhang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wu, QP., Zhang, L., Shao, XX. et al. Application of the novel bioluminescent ligand–receptor binding assay to relaxin-RXFP1 system for interaction studies. Amino Acids 48, 1099–1107 (2016). https://doi.org/10.1007/s00726-015-2146-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-015-2146-3