Abstract
Mitochondria are all-important organelles of eukaryotic cells since they are involved in processes associated with cellular mortality and human diseases. Therefore, trustworthy techniques are highly required for the identification of new mitochondrial proteins. We propose Mito-GSAAC system for prediction of mitochondrial proteins. The aim of this work is to investigate an effective feature extraction strategy and to develop an ensemble approach that can better exploit the advantages of this feature extraction strategy for mitochondria classification. We investigate four kinds of protein representations for prediction of mitochondrial proteins: amino acid composition, dipeptide composition, pseudo amino acid composition, and split amino acid composition (SAAC). Individual classifiers such as support vector machine (SVM), k-nearest neighbor, multilayer perceptron, random forest, AdaBoost, and bagging are first trained. An ensemble classifier is then built using genetic programming (GP) for evolving a complex but effective decision space from the individual decision spaces of the trained classifiers. The highest prediction performance for Jackknife test is 92.62% using GP-based ensemble classifier on SAAC features, which is the highest accuracy, reported so far on the Mitochondria dataset being used. While on the Malaria Parasite Mitochondria dataset, the highest accuracy is obtained by SVM using SAAC and it is further enhanced to 93.21% using GP-based ensemble. It is observed that SAAC has better discrimination power for mitochondria prediction over the rest of the feature extraction strategies. Thus, the improved prediction performance is largely due to the better capability of SAAC for discriminating between mitochondria and non-mitochondria proteins at the N and C terminus and the effective combination capability of GP. Mito-GSAAC can be accessed at http://111.68.99.218/Mito-GSAAC. It is expected that the novel approach and the accompanied predictor will have a major impact to Molecular Cell Biology, Proteomics, Bioinformatics, System Biology, and Drug Development.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mitochondria are popularly known as the powerhouse of the cell as well as the central unit of eukaryotic cells. Mitochondria perform key roles in composite biochemical processes such as programed cell death (Gottlieb 2000) and ionic homeostasis (Jassem et al. 2002). In addition, mitochondrial dysfunctions have been revealed to be associated with apoptosis, aging, and a number of pathological conditions. We are particularly interested in predicting mitochondrial proteins since they are affiliated with over 100 known human diseases such as Alzheimer’s disease (Hutchin and Cortopassi 1995), Type II diabetes (Gerbitz et al. 1996), and Parkinson’s disease (Wooten et al. 1997).
The prediction of mitochondrial proteins has largely been performed using machine learning and statistical approaches. Some of the interesting approaches that use machine learning and statistical approaches in conjunction with sequence information as well as biological information for the prediction of mitochondrial proteins are Target P (Emanuelsson et al. 2000), Signal P 3.0 (Bendtsen et al. 2004), WoLF PSORT (Horton et al. 2006), TargetLoc (Höglund et al. 2006), MitoProt II (Claros and Vincens 1996), MITOPRED (Guda et al. 2004), MitPred (Kumar et al. 2006) and PFMpred (Verma et al. 2009). In MitPred, first support vector machine (SVM)-based methods were developed using amino acid and dipeptide (Dp) composition of proteins (Kumar et al. 2006) and then the split amino acid composition (SAAC) was used (Kumar et al. 2006). The prediction accuracy was further improved by combining blast search and SVM method. Finally, a hybrid approach that combines Hidden Markov model profiles and SVM was used for the prediction of mitochondria proteins. On the other hand, in case of PFMpred, a hybrid model combining PSSM profile and SAAC was developed for mitochondria prediction and has achieved high performance accuracy of 92% (Kumar et al. 2006). Similarly, several approaches that employ in machine learning or statistical methods using protein sequence information have also been reported, whereby typically, avoiding any need of biological information is paid in terms of a decrease in the accuracy. A number of different computational approaches based on amino acid composition (AAC) or Dp composition (Dp) have been developed, including the covariant discriminant algorithm based (Chou and Elrod 1999), discrete wavelet transform based (Jiang et al. 2006), SVM based (Hua and Sun 2001; Chou and Cai 2002; Kumar et al. 2006; Jiang et al. 2006; Tan et al. 2006) and fuzzy kNN based (Huang and Li 2004). Tan et al. (2006) have reported the highest accuracy of 85% using pure machine learning approaches by applying genetic algorithm-partial least square (GA-PLS) on Dp features in conjunction with SVM.
Recently, Hu and Fan (2009) have proposed the physiochemical encoding method that maps protein sequences into feature vector composed of the locations and length of the amino acid groups (AAGs) with similar physiochemical properties. Their method yields an improvement of about 20% than that of the method based on simple ACC. An extended version of the pseudo-amino acid composition has been also been employed for the submitochondria subcellular localization and a good prediction performance has been achieved (Du and Li 2006). Similarly, Nanni and Lumini (2008a) have achieved high prediction performance using an interesting approach based on genetic programming (GP) for creating Chou’s pseudo amino acid-based features for sub-mitochondria localization. To improve the mitochondria prediction ability, this study employed GP for generating an effective decision space from that of the individual classifiers’ spaces. It has been observed that the utilization of ensemble classifiers is increasing for predicting protein subcellular localization. There are protein sequences that have multiple protein subcellular localizations for which interesting ensemble classifiers have been developed (Shen and Chou 2007; Chou and Shen 2007). Rotation forest has been proposed, which is based on investigating the diversity-accuracy landscape for ensemble classifier (Rodríguez et al. 2006). Similarly, RotBoost, which is a relatively new ensemble technique, has been proposed by combining rotation forest and Adaptive Boosting (AdaBoost) and has yielded lower prediction error than either of rotation forest and Adaboost (Zhang and Zhang 2008). Recently, (Nanni et al. 2010) have proposed an effective ensemble approach for protein subcellular localization using a high performance set of PseAAC and sequence-based descriptor.
For approximately incorporating the sequence-order effects, the idea of the pseudo amino acid composition (PseAAC) has been proposed (Chou 2001, 2005a, b). PseAAC has then been used in conjunction with various machine learning approaches to enhance the prediction quality (Chou and Cai 2006; Chou and Shen 2006a; Guo et al. 2006; Xiao et al. 2005, 2006a). However, the percent composition of the whole sequence does not give proper weight to the computational bias, which is known to be present in mitochondrial protein termini. Therefore, the concept of SAAC was introduced where the protein sequence is divided into three parts: N terminus, C terminus and a region between these two termini (Chou and Shen 2006a, b). SAAC has thus proved better accuracy for mitochondria predictions as it is giving greater weight to the proteins that have a signal at either N or C terminus (Kumar et al. 2006).
In the present study, our aim is to develop a novel high performance prediction system that can employ both selection of an effective feature extraction strategy and construction of an ensemble approach for mitochondria classification using sequence information only. For this purpose, we have used two recent datasets and analyzed different feature extraction strategies such as AAC, Dp, PseAAC, and SAAC. A number of different classifiers are then trained on these extracted features, i.e. SVM, k-nearest neighbor (kNN), random forest (RF), multilayer perceptron (MLP), AdaBoost, and bagging. A GP-based ensemble classifier is subsequently developed for mitochondrial prediction, which is able to develop an efficient decision space from the decision spaces of the individual classifiers.
Materials and methods
Datasets
The first dataset used in this paper is the same as in (Jiang et al. 2006), which consist of 499 mitochondrial proteins called positive examples. We denote this dataset by Mitochondria dataset (Mito_D). This dataset was obtained from Swiss-Prot release 46.6 by the keyword mitochondrial. A total of 2,833 entries were obtained. All the sequences with ambiguous words, such as POTENTIAL, BY SIMILARITY, or PROBABLE and fragments were then excluded. Moreover, 681 proteins (so called negative examples) belonging to locations other than mitochondrial site were selected by taking one out of every 250 entries in Swiss-Prot. Mitochondrial protein sequences or fragments were then deleted from the negative examples.
To validate the performance of our proposed method, we have used another dataset taken from (Verma et al. 2009). We denote this dataset by Malaria Parasite Mitochondria dataset (MP_Mito_D). This dataset consists of total 175 instances out of which 40 are mitochondrial proteins called positive examples and 135 examples belong to other locations, i.e. cytoplasm, extracellular, apicoplast, and are called negative examples. The homologies between sequences are checked. To remove the homologous sequences from the benchmark dataset, a 25% cut-off threshold is imposed and only those protein sequences are considered that have less than 25% sequence identity to any other protein sequences in a same subset (Chou and Shen 2006b, 2007, 2008).
Performance measures
We have assessed the performance of our method using the following performance measures.
-
(a)
Sensitivity or coverage of positive examples It is the percentage of mitochondrial proteins, which are correctly predicted as mitochondria.
$$ {\text{Sensitivity}} = {\frac{\text{TP}}{\text{TP} + \text{FN}}} \times 100 $$(1)where, TP and TN are correctly predicted mitochondrial and non-mitochondrial proteins, respectively, whereas FP and FN are wrongly predicted mitochondrial and non- mitochondrial proteins, respectively.
-
(b)
Specificity or coverage of negative examples It is the percentage of non-mitochondrial proteins, which are correctly predicted as non-mitochondria.
$$ {\text{Specificity}} = {\frac{\text{TN}}{\text{FP} + \text{TN}}} \times 100 $$(2) -
(c)
Accuracy It is the percentage of correctly predicted proteins (mitochondrial and non-mitochondrial proteins).
$$ {\text{Accuracy}} = {\frac{\text{TP} + \text{TN}}{\text{TP} + \text{FP} + \text{FN} + \text{TN}}} \times 100 $$(3) -
(d)
Mathew’s correlation coefficient It is considered as one of the most robust performance parameter. MCC equal to one is regarded as perfect prediction while zero for completely random prediction.
$$ {\text{MCC}} = {\frac{\text{TP} \times \text{TN} - \text{FP} \times \text{FN}}{{\sqrt {[\text{TP} + \text{FP}][\text{TP} + \text{FN}][\text{TN} + \text{FP}][\text{TN} + \text{FN}]} }}} $$(4) -
(e)
Q-Statistics To measure the diversity of the classifiers Q-statistic is considered as a promising performance parameter (Nanni and Lumini 2008b). The Q-statistic of any two base classifiers C and D is defined as:
$$ Q_{i,j} = {\frac{ad - bc}{ad + bc}} $$(5)where, a and d represent the number of correct and incorrect prediction of both classifiers. However, b is the correct prediction of classifier first and incorrect prediction of classifier second. In contrast, c is the correct prediction of classifier second but incorrect prediction of classifier first. The value of Q varies between −1 and 1.
Feature extraction strategies
Amino acid composition and dipeptide composition
The aim of calculating composition of proteins is to transform the variable length of protein sequence into a fixed-length feature vector (Hayat and Khan 2010). This is a most crucial step during classification of proteins using machine-learning techniques because they require fixed length patterns. The information of proteins can be encapsulated to a vector of 20 dimensions using AAC of the protein. In addition to AAC, Dp has also been used for classification that gives a fixed pattern length of 400. The advantage of Dp over AAC is that it encapsulates information about the fraction of amino acids as well as their local order. The AAC as well as Dp-based features have been generated as described by (Garg et al. 2005). Both compositions have been used as features to classify mitochondrial and non-mitochondrial proteins.
Split amino acid composition
In SAAC method (Chou and Shen 2006a, b), the protein sequence is divided in parts and composition of each part is calculated separately. Recently, (Verma et al. 2009), have developed SAAC-based method to predict mitochondrial proteins of malaria parasite and have achieved reasonable accuracy. In our SAAC model, each protein is divided into three parts: (i) 25 amino acids of N-termini, (ii) 25 amino acids of C-termini, and (iii) region between these two terminuses. There are some small sequences in the datasets whose length is less than 50 amino acids in the protein sequence. Therefore, to accommodate these sequences, we have divided these sequences into three parts with 10 amino acids of N and C terminus and the region between these two terminuses.
Pseudo amino acid composition
Amino acid composition model has been widely used in conjunction with quite a few statistical methods for predicting protein attributes. However, in case of AAC, all the sequence-order information is lost. To compensate this problem, the concept of the PseAAC has been proposed, which incorporates the sequence-order effects (Chou 2000, 2001). Simple AAC contains the composition of the 20 amino acids while the PseAA composition contains a set of greater than 20 discrete factors. The first 20 of these represent the components of its basic AAC and the additional factors carry some sequence-order information. For example
where λ is the numbers of tiers used in PseAA. The optimal value of tiers and the selection of the best physiochemical properties can influence the classification performance. In our case, we have selected λ = 21 and analyzed the performance using different combination of physiochemical properties. We have considered λ = 21 because it is yielding best results. The first 20 elements, i.e. P 1, P 2,…, P 20 just represent the occurrence of frequencies of the 20 amino acids. Whereas, P 21 is the 1st correlation order factor, P 22 is the second correlation order factor, and so on. These elements are determined based on the physiochemical properties. In this study, we have used three physiochemical properties, i.e. hydrophobicity, electronic, and bulk properties. There are various models for representing these properties. We have used FH, EIIP, and CPV models, respectively.
Classification approaches
Support vector machine
Support vector machine is a machine learning approach and is based on statistical learning theory (Vapnik 1998). A brief and clear description on how to use SVM for classification (Chou and Cai 2002; Cai et al. 2003). It has also been reported that SVM in conjunction with feature selection provides quite interesting results (Huang et al. 2008). In this study, we have implemented SVM using the LIBSVM 2.88-1 package, which allows us to choose a number of parameters and kernels (e.g. linear, polynomial, radial basis function, and sigmoid). In this particular work, the mitochondrial proteins were defined as one class (labeled as +1) and the non-mitochondrial proteins were defined as another class (labeled as −1). SVM was implemented in MATLAB 7.7 and a third degree polynomial was chosen as the kernel function. Quadratic programming method was employed to solve the optimization problem. All the parameters were kept constant except C (regulatory parameter) and s (the kernel width parameter). In the training process, C and s were optimized by parameter optimization (Guo et al. 2006; Khan et al. 2008a).
k-nearest neighbor
The k-nearest neighbor algorithm is a method which classifies objects based on k-nearest training examples in the feature space. The Euclidean distance of test sample to all other samples in the feature space is calculated and k samples are selected based on minimum Euclidian distance. The value of k is usually taken as odd. The Euclidian distance is calculated using the Eq. 5.
The sample under question X is then assigned to the category, which is found in majority among the k samples. The kNN is considered as a simple classifier, based on instance-based learning and has been commonly employed in protein prediction problems (Khan et al. 2008a).
Random forest
Random forest is designed to produce accurate predictions that do not overfit the data (Breiman 2001). RF employs the statistical technique bootstrap in which samples are drawn to construct multiple trees. Each tree is grown using some form of randomization. The leaf nodes of each tree are labeled by estimates of the posterior distribution. Each internal node contains a test that best divides the space of data to be classified. A protein sample is classified by sending it down every tree and aggregating the reached leaf distributions. Out-of-bag samples can be employed to compute an unbiased error rate and variable importance, eliminating the need for a test set or cross-validation. Because a large number of trees are grown, there is limited generalization error (that is, the true error of the population as opposed to the training error only), which means that overfitting is unlikely.
By growing each tree to maximum size without pruning and selecting only the best split among a random subset at each node, RF tries to maintain some prediction strength while inducing diversity among trees (Breiman 2001). Random predictor selection decreases the correlation among un-pruned trees and keeps the bias low. By taking an ensemble of un-pruned trees, variance is also reduced. Another advantage of RF is that the predicted output depends only on one user-selected parameter which is the number of predictors to be chosen randomly at each node. In this work, the parameters of RF are set with number-of-trees equal to 15 and iterations equal to 25.
AdaBoost
AdaBoost is one of the most popular and successful implementations of boosting. Its name is an acronym created from its description, i.e. Adaptive Boosting. We have used Adaboost.M1 (Freund and Schapire 1996) provided in Weka 3.6.2, where REPTree has been used as a weak learner with number of iterations equal to 25 and the rest of the parameters are set with the default values.
Bagging
One of the simple ensemble classifier is a method that generates multiple versions of a predictor and employs these to develop an aggregated predictor. The aggregation averages over the versions when predicting a numerical outcome and employs a plurality vote when predicting a class (Breiman 1996). Weka 3.6.2 is used for simulation of bagging approach with REPTree as a weak learner. The numbers of iterations were set to 25 and the remaining parameters are set with default values.
Multilayer perceptron
The MLP consists of a system of simple interconnected neurons, which yields a model representing a nonlinear mapping between an input and an output vector (Khan et al. 2008b). The nodes are connected by weights and output signals, which function as the sum of the inputs to the node modified by a simple nonlinear transfer, or activation function. The superposition of many simple nonlinear transfer functions enables the MLP to approximate non-linear functions. An MLP has one or more hidden layers between input and output layers. In this work, we have used the Matlab-based neural networks toolbox for MLP implementation. An MLP having one input, two hidden, and one out layers has been used. The training algorithm was set to trainl-based backpropagation approach. The rest of the parameters were used as default.
Predicting mitochondria proteins
Developing individual mitochondria classifier
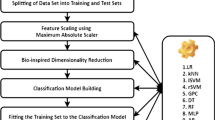
Several mitochondria prediction methods employing individual classifiers have been proposed in the literature so far. In our proposed method, we have trained different individual classifiers on several feature extraction strategies. First, mitochondria protein sequences are converted into features using the feature extraction strategies. These features are finally provided to the different individual classifiers for training and prediction performance for each individual classifier is determined as shown in Fig. 1. We have trained six different individual classifiers such as SVM, kNN, MLP, RF, AdaBoost, and bagging on four different feature extraction strategies: AAC, Dp, PseAAC and SAAC.
Developing GP based ensemble classifier (Mito-GSAAC)
GP introduced by Koza (1992) is an evolutionary algorithm designed for automatically constructing and evolving computer programs. It differs from genetic algorithm in the ability to evolve variable length solutions. GP has emerged as a powerful tool not only for evolving a classifier but also for optimum combination of classifiers (Khan et al. 2005). GP works by evolving a population of randomly created initial population using a fitness measure. It selects the fittest one to take part in the evolution and thus efficiently searches for the desired solution.
Several interesting combination strategies in classification of protein sequences have been employed recently. Out of these, combination strategies majority voting-based strategies have been widely used (Khan et al. 2010). GP, on the other hand, has shown promising results when used for combination of binary classifiers due to its inherent learning capability and its tree structure (Khan et al. 2005). Therefore, this work capitalizes the learning capabilities of GP, whereby it is used as a stacking-based ensemble approach and thus is different from the majority voting technique. We have employed GP to develop a complex but efficient decision space when provided with decision spaces of the individual trained classifiers as shown in Fig. 2.
Results and discussion
Mitochondria prediction using different amino acid-based features
This work aims at predicting eukaryotic mitochondrial proteins using SAAC and ensemble classifier without using any biological information. However, in order to create the ensemble classifier, first we have tested a number of classifiers using different feature extraction strategies like AAC, Dp, PseAAC, and SAAC. The performance of the various classification algorithms is evaluated through various performance measures such as accuracy, sensitivity, specificity, MCC and Q-Statistics. We have found that all the classifiers, SVM, kNN, MLP, RF, AdaBoost and bagging have yielded better performance on SAAC as compared to other feature extraction strategies (Table 1). This means that SAAC offers greater discrimination power in comparison with the rest of feature extraction strategies and is largely due to the composition difference at the N and C terminus in mitochondria and non-mitochondria. We have achieved a classification accuracy of 90.34% on SAAC using RF as a classifier with number of iterations equal to 25 and number of trees equal to 15. Finally, as detailed in Sect. 4.3, we have developed a GP-based ensemble classifier using the predictions of individual classifiers, SVM, kNN, RF and AdaBoost on SAAC features.
Using the first feature extraction strategy AAC, the classification accuracy of AdaBoost is better compared to the other classification algorithms. In case of Dp, SVM obtained the highest accuracy among the various classifiers. Similarly using PseAA, the classification accuracy of SVM is the highest among the various classifiers. In case of SAAC, the accuracies of the individual classifiers are: AdaBoost 88.64%, bagging 88.64%, kNN 84.58%, MLP 85.54%, RF 90.34%, and SVM 88.05%. Thus, the classification performance of the various classification algorithms that we used has improved in case of SAAC.
We have also used a hybrid feature-extraction strategy to analyze the performance of the various classification algorithms, whereby the different features are just concatenated. In case of this hybrid feature-extraction strategy, the accuracy of AdaBoost, Bagging, kNN, and MLP decreases, while slight improvement in the accuracy of RF and SVM has been observed. Therefore, it is observed that petite improvement in accuracy is obtained for some of the classifiers at the cost of quite high dimensionality of the feature vector space.
Is SAAC really better for mitochondria classification?
It is reported in literature that mitochondria have a large difference in composition from non-mitochondria mostly in the N- and C-terminus (Kumar et al. 2006). N-terminus or amine terminus is the initial portion of some amino acids in the protein sequence. Similarly, C-terminus or carboxyl-terminus is the end portion of amino acids in the sequence. Therefore, splitting the protein sequence into three parts i.e. N-terminus, C-terminus and the portion of sequence between these two termini, and then calculating the AAC for all three parts would provide a better discrimination for mitochondria versus non-mitochondria as can be observed from Table 1. Hence, it shows that overall protein sequence composition avoids the high signals in some parts of the protein sequence, which can better discriminate the mitochondria from non-mitochondria.
GP ensemble and SAAC-based mitochondria classification
We observed that for mitochondria prediction, SAAC performs better than the various feature extraction strategies that we have used. Therefore, for better exploitation of the SAAC features for mitochondria prediction, we have developed an ensemble classification approach using GP. Previously, GP has been used for the optimum combination of classifiers for gender classification problem and it has been observed that it provides better prediction performance (Khan et al. 2005; Khan and Mirza 2007). Thus, by employing GP-based ensemble approach, a high accuracy of 92.62% is obtained for the mitochondria prediction. We denote our proposed GP ensemble as Mito-GSAAC.
We have used conventional functions in the GP tree: a set of four binary arithmetic operators (+, −, * and a protected division), if less than (IFLT), if greater than (IFGT), and absolute. We have combined the predictions of kNN, SVM, RF, and AdaBoost using GP to develop an optimal decision space. The dataset is divided into two portions, i.e., training and testing. Two-third of training data is given to GP for training and then it is validated on the remaining one-third data (Table 2). First, an initial population of 100 polynomials is generated. Fitness for each new individual is calculated using area under receiver operating curve (AUROC).
ROC and MCC as fitness criteria in GP
ROC and MCC have been used as fitness criteria in GP simulation for developing ensemble classifier. ROC is a graph plotted between true positive rate (TPR) and false positive rate (FPR) for different threshold values. TPR represents the number of correct positive cases divided by the total number of positive cases whereas, FPR represents the number of negative cases predicted as positive cases, divided by the total number of negative cases (Khan et al. 2005). The area under the ROC curve i.e. AUROC is then computed and is considered as the fitness in GP simulation. A GP individual with the highest value of AUROC is chosen the best individual in the population.
MCC is also considered as a rigorous performance measure in classification applications. Therefore, we have also used MCC as fitness criteria in some of the GP simulations. It is observed that both the fitness criterion i.e. AUROC and MCC yield almost the same performance on the testing data (Table 3). The accuracy versus complexity graph and the best individual GP tree for the MCC-based fitness criteria are demonstrated in Fig. 3.
Comparison with existing state of the art approaches
Performance comparisons on the Mito_D dataset
We have compared our proposed Mito-GSAAC with the existing prediction methods using the Mito_D dataset and jackknife test. All prediction performances are listed in Table 4. The results show that Mito-GSAAC can identify mitochondrial proteins from other proteins with a relatively high accuracy of 92.62% and MCC of 0.85. MITOPRED, which uses biological information, also shows high performance with an accuracy of 95.68%. However, we have achieved highest accuracy without using any biological information like that of one which has achieved accuracy of 85%. In practice, not all the biological information can easily be attained. Once such information is absent, our method will be influenced little. Like MITOPRED, MitoProt also has some limitations; it can only predict the sequences starting by a methionine and the mature proteins. The discrete wavelet transforms method (Jiang et al. 2006) based on the sequence-scale similarity measurement does not rely on subcellular locations information and can directly predict protein sequences with different length. Although the performance in terms of the specificity is relatively higher, the accuracy is poor. It is usually due to the specific properties of mitochondrial protein that make it difficult to discriminate it from other proteins by just one method, or simply because the number of proteins in the mitochondrion is immoderate (Cameron et al. 2005). With the increase of the exact experimental mitochondrial proteins, the performance may also be improved significantly.
Performance analysis of the proposed Mito-GSAAC on the MP_Mito_D dataset
We also evaluate the performance of the proposed approach Mito-GSAAC on the MP_Mito_D dataset using two important statistical tests; jackknife and fivefold cross validation test.
In case of jackknife test, RF using AAC yielded the highest accuracy and MCC values of 85.14% and 0.53, respectively (Table 5). While, in case of Dp composition, SVM achieved better accuracy than the rest of the classifiers. On the other hand, in case of SAAC, both SVM and kNN yielded better accuracies as compared to the rest of the classifiers. Using the hybrid features (when all the different features are just concatenated) and jackknife test, the accuracy of SVM is 89.21%, which is slightly high as compared to that obtained using SAAC. On the other hand, the performance of the rest of the classifiers decreased when used in conjunction with the hybrid features, mainly due to the curse of dimensionality.
In case of fivefold cross validation, the performance of RF, SVM, and kNN using individual feature extraction strategy is shown in Table 6. When the Mitochondria prediction performance using different feature-extraction strategies is analyzed, enhanced performance is observed using SAAC in case of all the different classification algorithms.
In case of the hybrid feature-extraction strategy, the classification accuracy obtained by AdaBoost, bagging, MLP, RF, SVM, and kNN are 85.22, 85.10, 83.14, 87.01, 92.23, and 90.71%, respectively. It has been observed again that using the hybrid feature-extraction strategy, SVM has achieved slight improvement in accuracy as compared to that using SAAC. The performance improvement for SVM is only 0.23%; however, the dimensionality of the feature space is increased greatly. On the other hand, the performance of the rest of the classifiers decreased when trained on the hybrid features.
It has thus been observed that a mitochondria protein can be efficiently discriminated based on the differences in the amino acid at their N- and C-terminus by the SAAC feature extraction strategy. Therefore, the predicted results of AdaBoost, RF, SVM and kNN using the SAAC features are then combined through GP. The results of GP-ensemble are analyzed using both the tests; jackknife, and fivefold cross-validation as shown in Table 7.
In case of jackknife test, Mito-GSAAC has obtained an accuracy of 90.05%. In the GP simulation, the population size and no. of generations were kept equal to 200 and 100, respectively. The accuracy of Mito-GSAAC is 1.98% higher than the highest individual classifier’s result using all the feature extraction strategies.
Similarly, the predicted results for fivefold cross-validation test are also combined through GP. Mito-GSAAC obtained an accuracy of 93.21%. The accuracy of Mito-GSAAC using fivefold cross-validation test is 1.71% higher than the highest individual classifier’s result using all the feature extraction strategies.
Comparison with existing approaches on the MP_Mito_D dataset
We have also compared our proposed approach Mito-GSAAC with already published methods using the MP_Mito_D dataset (Table 8). In case of the MP_Mito_D dataset, Verma et al. (2009) have employed Dp and PSSM composition using SVM and have reported 92.57% accuracy and 0.78 MCC. Further, they have applied the combination of SAAC and PSSM using SVM as a classifier and have reported an accuracy of 92.00% and 0.81 MCC. However, the values of the classification accuracy and MCC of our proposed approach Mito-GSAAC are 93.21%, respectively, which are better compared to those obtained by Verma et al. 2009. Thus, the predicted results show that the performance of our proposed approach is not only better from RF, SVM, and kNN but also higher than the existing approaches.
Conclusions
In this paper, we have shown that a GP-based ensemble classifier can be developed for better exploitation of the advantages of the individual classifier trained on SAAC-based features. We have investigated four kinds of protein representations for mitochondria prediction namely, AAC, Dp, PseAAC and SAAC and used two datasets for performance analysis. First different individual classifiers such as SVM, kNN, MLP, RF, AdaBoost and bagging are trained and their prediction performances are determined. In case of Jackknife test, RF among all individual classifiers has given the highest accuracy of 90.34% in conjunction with SAAC-based features on the Mito_D dataset while SVM obtained the highest accuracy of 92.0% using SAAC and fivefold cross validation test on the MP_Mito_D dataset. Consequently, SAAC has performed better than the rest of feature extraction strategies. This better performance might be due to the high signals in the parts of mitochondria protein sequence. The proposed Mito-GSAAC has achieved a high performance accuracy of 92.62% on the Mito_D dataset and 93.21% on the MP_Mito_D dataset. Until now, most of existing studies employed only a single individual classifier to predict mitochondrial proteins. However, in the current work, we have first employed different individual classifiers and then, finally, we have developed a GP-based ensemble classifier for mitochondria prediction. This work thus proposes an effective mitochondria prediction method, Mito-GSAAC that uses raw sequence data only and thus can be helpful in the research related to cell biology and drug discovery.
References
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Breiman L (1996) Bagging predictors. Mach Learn 24:123–140
Breiman L (2001) Random forests. Mach Learn 45:5–32
Cai YD, Zhou GP, Chou KC (2003) Support vector machines for predicting membrane protein types by using functional domain composition. Biophys J 84:3257–3263
Cameron JM, Hurd T, Robinson BH (2005) Computational identification of human mitochondrial proteins based on homology to yeast mitochondrially targeted proteins. Bioinformatics 21:1825–1830
Chou KC (2000) Prediction of protein subcellular locations by incorporating quasi-sequence-order effect. Biochem Biophys Res Commun 278:477–483
Chou KC (2001) Prediction of protein cellular attributes using pseudo amino acid composition. Proteins Struct Funct Genet 43:246–255, erratum 44:60
Chou KC (2005a) Review: progress in protein structural class prediction and its impact to bioinformatics and proteomics. Curr Protein Pept Sci 6:423–436
Chou KC (2005b) Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 21:10–19
Chou KC, Cai YD (2002) Using functional domain composition and support vector machines for prediction of protein subcellular location. J Biol Chem 277:45765–45769
Chou KC, Cai YD (2006) Predicting protein–protein interactions from sequences in a hybridization space. J Proteome Res 5:316–322
Chou KC, Elrod DW (1999) Protein subcellular location prediction. Protein Eng 12:107–118
Chou KC, Shen HB (2006a) Predicting protein subcellular location by fusing multiple classifiers. J Cell Biochem 99:517–527
Chou KC, Shen HB (2006b) Hum-PLoc: a novel ensemble classifier for predicting human protein subcellular localization. Biochem Biophys Res Commun 347:150–157
Chou KC, Shen HB (2007) Euk-mPLoc: a fusion classifier for large-scale eukaryotic protein subcellular location prediction by incorporating multiple sites. J Proteome Res 6:1728–1734
Chou KC, Shen HB (2008) Cell-PLoc: a package of web-servers for predicting subcellular localization of proteins in various organisms. Nat Protoc 3:153–162
Claros MG, Vincens P (1996) Computational method to predict mitochondrial proteins and their targeting sequences. Eur J Biochem 241:779–786
Du P, Li Y (2006) Prediction of protein submitochondria locations by hybridizing pseudo-amino acid composition with various physicochemical features of segmented sequence. BMC Bioinform 7:518
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016
Freund Y, Schapire RE (1996) Experiments with a new boosting algorithm. In: Proceedings of the Thirteenth International Conference on Machine Learning. Morgan Kaufmann, Massachusetts, pp 48–156
Gerbitz KD, Gempel K, Brdiczka D (1996) Mitochondria and diabetes: genetic, biochemical, and clinical implications of the cellular energy circuit. Diabetes 45:113–126
Gottlieb RA (2000) Programmed cell death. Drug News Perspect 13:471–476
Guda C, Fahy E, Subramaniam S (2004) MITOPRED: a genome-scale method for prediction of nucleus-encoded mitochondrial proteins. Bioinformatics 20:1785–1794
Guo YZ, Li M, Lu M, Wen Z, Wang K, Li G, Wu J (2006) Classifying GPCRs and NRs based on protein power spectrum from fast fourier transform. Amino Acids 30:397–402
Hayat M, Khan A (2010) Predicting membrane protein types by fusing composite protein sequence features into pseudo amino acid composition. J Theor Biol 271(1):10–17
Höglund A, Dönnes P, Blum T, Adolph HW, Kohlbacher O (2006) MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22:1158–1165
Horton P, Park KJ, Obayashi T, Nakai K (2006) Protein subcellular localization prediction with WoLF PSORT. In: Proceedings of the fourth Annual Asia Pacific Bioinformatics Conference APBC06, Taipei, Taiwan, pp 39–48
Hu J, Fan Z (2009) Improving protein localization prediction using amino acid group based physiochemical encoding. BICoB 2009. LNBI 5462:248–258
Hua SJ, Sun ZR (2001) Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17:721–728
Huang Y, Li Y (2004) Prediction of protein subcellular locations using fuzzy k-NN method. Bioinformatics 20:21–28
Huang WL, Tung CW, Ho SW (2008) ProLoc-GO: utilizing informative gene ontology terms for sequences-based prediction of protein subcellular localization. BMC Bioinform 9:80
Hutchin T, Cortopassi GA (1995) A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proc Natl Acad Sci USA 92:6892–6895
Jassem W, Fuggle SV, Rela M, Koo DD, Heaton ND (2002) The role of mitochondria in ischemia/reperfusion injury. Transplantation 73:493–499
Jiang L, Li ML, Wen ZN, Wang KL, Diao YB, Guo YZ, Liu LX (2006) Prediction of mitochondrial proteins using discrete wavelet transform. Protein J 25:241–249
Khan A, Mirza AM (2007) Genetic perceptual shaping: utilizing cover image and conceivable attack information using genetic programming. Inform Fus 8(4):354–365
Khan A, Majid A, Mirza AM (2005) Combination and optimization of classifiers in gender classification using genetic programming. Int J Knowl Based Intell Eng Syst 9:11
Khan A, Khan MF, Choi TS (2008a) Proximity based GPCRs prediction in transform domain. Biochem Biophys Res Commun 371(3):411–415
Khan A, Tahir SF, Majid A, Choi TS (2008b) Machine learning based adaptive watermark decoding in view of an anticipated attack. Pattern Recogn 41:2594–2610
Khan A, Majid A, Choi TS (2010) Predicting protein subcellular location: exploiting amino acid based sequence of feature spaces and fusion of diverse classifiers. Amino Acids 38:347–350
Koza JR (1992) Genetic programming: on the programming of computers by means of natural selection. MIT Press, Cambridge
Kumar M, Verma R, Raghava GPS (2006) Prediction of mitochondrial proteins using support vector machine and hidden Markov model. J Biol Chem 281:5357–5363
Nanni L, Lumini A (2008a) Using ensemble of classifiers in bioinformatics. In: Peters H, Vogel M Machine Learning Research Progress. Nova publisher, New York
Nanni L, Lumini A (2008b) Genetic programming for creating Chou’s pseudo amino acid based features for submitochondria localization. Amino Acids 34(4):653–660
Nanni L, Brahnam S, Lumini A (2010) High performance set of PseAAC and sequence based descriptors for protein classification. J Theor Biol 266(1):1–10
Rodríguez JJ, Ludmila IK, Carlos JA (2006) Rotation forest: a new classifier ensemble method. IEEE Trans Pattern Anal Mach Intell 28(10):1619–1630
Shen HB, Chou KC (2007) Hum-mPLoc: an ensemble classifier for large-scale human protein subcellular location prediction by incorporating samples with multiple sites. Biochem Biophys Res Commun 355:1006–1011
Tan F, Feng X, Fang Z, Li M, Guo Y, Jiang L (2006) Prediction of mitochondrial proteins based on genetic algorithm—partial least squares and support vector machine. Amino Acids (published online Oct 15 2006. doi:10.1007/s00726-006-0465-0)
Vapnik VN (1998) Statistical learning theory. Wiley, New York
Verma R, Varshney Grish C, Raghava GPS (2009) Prediction of mitochondrial proteins of malaria parasite using split amino acid composition and PSSM profile 39(1):101–110
Wooten GF, Currie LJ, Bennett JP, Harrison MB, Trugman JM, Parker WD Jr (1997) Maternal inheritance in Parkinson’s disease. Ann Neurol 41:265–268
Xiao X, Shao S, Ding Y, Huang Z, Chen X, Chou KC (2005) An application of gene comparative image for predicting the effect on replication ratio by HBV virus gene missense mutation. J Theor Biol 235:555–565
Xiao X, Shao SH, Chou KC (2006a) A probability cellular automaton model for hepatitis B viral infections. Biochem Biophys Res Commun 342:605–610
Xiao X, Shao S, Ding Y, Huang Z, Chou KC (2006b) Using cellular automata images and pseudo amino acid composition to predict protein sub-cellular location. Amino Acids 30:49–54
Zhang CX, Zhang JS (2008) RotBoost: a technique for combining Rotation Forest and AdaBoost. Pattern Recogn Lett. doi:10.1016/j.patrec.2008.03.006
Acknowledgments
This work was supported by the Department of Computer and Information Sciences (DCIS) at Pakistan Institute of Engineering and Applied Science (PIEAS), Pakistan.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Afridi, T.H., Khan, A. & Lee, Y.S. Mito-GSAAC: mitochondria prediction using genetic ensemble classifier and split amino acid composition. Amino Acids 42, 1443–1454 (2012). https://doi.org/10.1007/s00726-011-0888-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-011-0888-0