Abstract
Dwindling fresh water resources and climate change poses serious threats to rice production. Roots play crucial role in sensing water gradient and directing growth of the plant towards water through a mechanism called hydrotropism. Since very little information is available on root hydrotropism in major food crops, this study was carried out to clone and characterize an ortholog of Arabidopsis MIZU-KUSSEI1 (MIZ1) from rice. Contrasting rice genotypes for drought and salt tolerance were selected based on phenotyping for root traits. Nagina 22 and CR-262-4 were identified as most tolerant and Pusa Sugandh 5 and Pusa Basmati 1121 were identified as most susceptible varieties for both drought and salt stresses. Allele mining of MIZ1 in these varieties identified a 12 bp Indel but did not show specific allelic association with stress tolerance. Analysis of allelic variation of OsMIZ1 in 3024 rice genotypes of 3K genome lines using Rice SNP-Seek database revealed 49 InDels. Alleles with the 12 bp deletions were significantly prevalent in indica group as compared to that of japonica group. Real-time RT-PCR analysis revealed that OsMIZ1 expression levels were upregulated significantly in tolerant cv. Nagina 22 and CR-262-4 under osmotic stress, while under salt stress, it was significantly upregulated only in CR-262-4 but maintained in Nagina 22 under salt stress. However, in the roots of susceptible genotypes, OsMIZ1 expression decreased under both the stresses. These results highlight the possible involvement of OsMIZ1 in drought and salt stress tolerance in rice. Furthermore, expression studies using publically available resources showed that enhanced expression of OsMIZ1 is regulated in response to disease infections, mineral deficiency, and heavy metal stresses and is also expressed in reproductive tissues in addition to roots. These findings indicate potential involvement of MIZ1 in developmental and stress response processes in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is a major staple food crop in India and world. Environmental stresses, such as drought and salinity limit rice productivity all over the world including Asia and sub-Saharan Africa. Approximately 34 mha of rain-fed lowland and 8 mha of upland rice in Asia, with 13.6 mha area affected in India alone, suffer from drought stress of varying intensities almost every year (Wopereis et al. 1996; Singh et al. 2016a, b). Rice production is also limited by soil salinity in most semi-arid regions of the world (Rengasamy 2010). Development of rice cultivars with an inherent capacity to tolerate drought and salinity stress is one of the most promising strategies for having sustainable yield under current and future climatic conditions.
Drought and/or salt tolerance is a complex trait governed by many physiological and biochemical properties of plants. Plants use different mechanisms to cope with drought stress, namely, dehydration avoidance, dehydration tolerance, and drought recovery. Roots contribute to dehydration avoidance (Samson et al. 2002; Wang and Yamauchi 2006). Root characteristics, particularly number of seminal roots, root length, surface area, root volume, and root biomass, increase water uptake by plants and stabilize yields in saline, dry, and other problem soils and stress environments (Serraj et al. 2009). There is increasing recognition that future grain productivity, especially under low input conditions, can be achieved through optimization of root system architecture (RSA). Rice genotypes showing greater drought tolerance are associated with deep root growth (Henry et al. 2011). Several studies showed an advantage of having superior root traits for yield stability under stress conditions (Price et al. 2002; Tuberosa et al. 2002; Ober et al. 2005; Sarker et al. 2005; Gowda et al. 2011).
Plants utilize hydrotropism to bend their roots towards moistened areas of soil in the presence of moisture gradients (Takahashi et al. 2009; Moriwaki et al. 2013) for water uptake. Genetic analysis of hydrotropism in Arabidopsis led to the identification of new players viz., AHR1, NHR1, MIZU-KUSSEI1 (MIZ1), and MIZ2, involved in sensing of moisture and hydrotropic root growth towards soil region with high soil water potential. MIZ1 is expressed in root tips and hydathodes (Kobayashi et al. 2007). MIZ1 is a member of domain of unknown function proteins (DUF617) in Arabidopsis. Interestingly, MIZ1 homologs were not found in the genomes of animals or in microbes, and thus exist only in terrestrial plants. Analysis of loss of function mutants and overexpression lines of Arabidopsis demonstrated that MIZ1 gene is essential for hydrotropism in roots and is up-regulated after exposure to osmotic or salt stress (Kobayashi et al. 2007). Transgenic plants overexpressing MIZ1 exhibit significantly enhanced hydrotropic responses (Miyazawa et al. 2012). Characteristics of hydrotropism in miz1 mutant, transgenic plants overexpressing MIZ1 (MIZ1OE), and wild-type plants showed that WT plants developed root systems in regions with higher water potential, whereas the roots of miz1 mutant plants did not show a similar response (Iwata et al. 2013). This pattern of root distribution induced by hydrotropism was more pronounced in MIZ1 overexpressing transgenic plants than in WT plants. In addition, shoot biomass and the number of plants that survived under drought conditions were much greater in MIZ1 overexpressing transgenic plants indicating an important role played by hydrotropism in drought avoidance (Iwata et al. 2013). Detailed characterization of MIZ1-GFP in root cells reveled that MIZ1 is a soluble protein in the cytoplasm and is associated with the cytoplasmic face of endoplasmic reticulum (ER). ABA has shown to regulate the MIZ1 expression. MIZ1 appears to regulate endogenous level of auxin in roots to regulate hydrotropism (Moriwaki et al. 2013). Further studies revealed that ABA-dependent SnRK2.2 kinase and MIZ1 act in the elongation zone cortical cells to regulate root hydrotropism in Arabidopsis (Dietrich et al. 2017). MIZ1 protein was found to generate slow, long-distance Ca2+ signal by inhibiting ECA1, an endoplasmic reticulum Ca2+ pump (Shkolnik et al. 2018). However, information on MIZ1 in plants other than Arabidopsis is lacking. Rice genome shows at least 13 protein similar to Arabidopsis MIZ1 having domain of unknown function (DUF617). Studies on MIZ1 homologs from other species might provide a clue to understand the evolution of drought and salt stress avoidance system in land plants and the universality of the molecular mechanism for root hydrotropism. Further, this gene has a potential to improve water mining of crops and thus drought and salinity tolerance. Therefore, the identification and characterization of such genes that mediate plant responses to water scarcity from rice may provide a powerful method to engineer or select for crop plants with enhanced tolerance for a drier and warmer climate.
Material and methods
Plant materials and stress treatment
A total number of 20 genotypes of rice were screened under 150 mM NaCl and 15% PEG in hydroponics. The seeds were sterilized with 2% Bavistin (w/v for 10 min) followed by sodium hypochlorite 2% (v/v for 10 min) followed by washing five times with sterilized distilled water and finally immersed in water for another 8 h at 20 °C in dark. The surface-sterilized seeds were germinated in aseptic Petri dishes (diameter 90 mm and height 15 mm), which were paved with damp germination paper soaked in distilled water. After sprouting for 3 days at room temperature, the seedlings were subjected to stress treatments viz. 150 mM NaCl (− 0.744 MPa at 25 °C, ionization constant of 2 was used for calculation) and 15% PEG-6000 (− 0.295 Ψs at 25 °C) for a period of 7 days in a growth chamber under long-day photoperiod (16 h light/8 h dark) at 22 °C and a relative humidity of 65–75% using germination paper roll method. On 10th day, phenotypic data on number of seminal roots, seminal root length, coleoptile length, length of longest root, and relative change in the above traits under stress conditions was recorded manually. The experiment was conducted in randomized complete block design (RCBD) with three replicates per treatment within each experiment (unstressed control, osmotic, and salt stresses). Each Petri plate containing ten seedlings was considered as an experimental unit and each genotype within a replicate was represented by five healthy and homogenous seedlings. The Stress Susceptibility Index (S) was used to characterize the relative stress tolerance of the various genotypes (S ≤ 0.50 highly stress tolerant, S > 0.50 ≤ 1.00 moderately stress tolerant and S > 1.00 susceptible).
Stress Susceptibility Index
Susceptibility Index (S) was calculated using formula of Fischer and Maurer (1978).
where
- Y:
-
mean trait of a genotype in a stress environment
- Yp:
-
trait of a genotype under stress free environment
- D:
-
stress intensity = 1–X/Xp
- X:
-
mean Y of all genotypes
- Xp:
-
mean Yp of all genotypes
Based on ‘S’ index, roots from two susceptible and two tolerant genotypes were harvested and rinsed with distilled water three times before being immersed in liquid nitrogen and stored at − 80 °C for further molecular analysis.
Further, to carry out the physiological and biochemical analysis, selected genotypes were grown hydroponically in half strength Yoshida solution, and 14-day-old seedlings were given osmotic stress using 15% PEG-6000 made in half strength Yoshida solution. Treatment was carried out for 10 days. Thereafter, various physiological and biochemical analyses were done on leaves. Relative water content (RWC) was measured according to method of Barrs and Weatherley (1962); membrane stability index (MSI) was measured according to Deshmukh et al. (1991) and chlorophyll content was measured using the method of Hiscox and Israelstam (1979). Root samples were scanned in grayscale at 300 dpi using a desktop scanner (Epson Expression 11000XL) and grayscale images obtained in tiff format were analyzed using WinRHIZO software from LA2400 (v2009, Regent Instruments, Montreal, QC, Canada). Phenotypic data on total root length (RL, cm), total root surface area (RSA, cm2), root diameter (RD, mm), root volume (RV, cm3), root dry weight (RDW), and relative change in the above traits under stress conditions were recorded. RDW (mg) was recorded after drying the root samples in a hot air oven at 70 °C for 72 h.
Identification of MIZ1 from rice
The protein sequence of Arabidopsis thaliana MIZU-KUSSEI 1 (MIZ1) gene (AT2G41660) was used to BLAST search rice genome (http://rice.plantbiology.msu.edu/) to identify homologs of MIZ1 in rice. The sequence homology between the putative OsMIZ1 sequence (Loc_Os02g47980.1) and Arabidopsis MIZ1 was compared by using Clustal Omega software (http://www.ebi.ac.uk/Tools/msa/clustalo/). NCBI, conserved domain search, was done to find if DUF617 is present in the predicted homolog (Marchler-Bauer et al. 2011).
RNA isolation, cDNA preparation, and quantitative reverse transcription-PCR (qRT-PCR)
The root tissues of the treated (after subjecting to 150 mM NaCl and 15% PEG-6000 stress for a period of 7 days) as well as control plants were harvested for RNA isolation. Total RNA was isolated using Nucleopore-Genetix RNA sure plant kit and treated with DNase I as per the instructions. For cDNA preparation, 1 μg of total RNA was used to synthesize first-strand cDNA using SuperScript™ II Reverse Transcriptase (Invitrogen) following manufacturer’s instructions. Quantitative PCR analysis of MIZ1 gene was carried out using KAPA SYBR® FAST qPCR Master Mix (2×) Kit. For qRT-PCR experiment, each reaction contained 10 μl of 2× SYBR® Premix Ex Taq™ (KAPA), 0.4 μl of 2.0 μM gene-specific primers, 0.4 μl of 50× ROX References Dye, and 1.0 μl of cDNA in a final volume of 20 μl. The PCR parameters were the following: 95 °C for 30 s; 40 cycles of 95 °C for 5 s and 59 °C for 20 s; and a dissociation step at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. StepOne™ Real-Time PCR from Applied Biosystems was used for performing quantitative expression analysis. A quantitative analysis was performed using the 2−∆∆ CT method. The gene specific primers for OsMIZ1 (forward primer: 5′-CTACTGCAACGGCCGAAAGTG-3′ and reverse primer: 5′-AGGGTTCATCATGTAGAACGCCT-3′) and rice ubiquitin reference gene (forward primer: 5′-GAAGCACAAGCACAAGAAGGTG-3′ and reverse primer: 5′-CTGGTTGTAGACGTAGGTGAG-3′) were used.
Cloning, sequencing, and phylogenetic analysis
In order to clone the precise ortholog of Arabidopsis MIZ1 from rice, a BLAST search was performed using AtMIZ1 as a query against rice genome sequences (Genes in MSU RGAP release 7) using RGAP (Rice Genome Project Annotation Project) BLAST search tool. This led to the identification of DUF617 domain containing protein (Loc_0247980.1) showing highest hit score (491) and maximum identity (56.74%) at e-value 7e-60. As per RGAP, the gene is located at chromosome 2 with coding sequence (5′-3′) coordinates 29364087-29365510. For cloning of rice MIZ1 gene, PCR primers were designed using gene sequence, Loc_Os02g47980.1. PCR amplification was performed using cDNA from roots tissues of Nagina 22 and CR-262-4, and genomic DNA of Pusa Basmati 1121, Pusa Sugandh 5, and Rasi as template with gene specific forward (5′-CTTGCCATGGCGAGAGCGTTC-3′) and reverse primers (5′-GTCAGACTCTGAGCAGGTAGATG-3′). The amplified PCR product of expected size was gel purified using DNA GelElute kit. The purified PCR amplicon was cloned in pTZ57R/T vector using TA cloning procedure. Positive clones were confirmed by colony PCR and restriction digestion and were then subjected to DNA sequencing. The sequences obtained from three different OsMIZ1 clones from each variety were checked using MultAlin, a multiple sequence alignment program (Corpet 1988) as well as BLASTn. Gene structure analysis was illustrated using Gene Structure Display Server (GSDS) program (Hu et al. 2015). For phylogenetic analysis, OsMIZ1 amino acid sequences and their orthologs in different crops were aligned using ClustalW program (Thompson et al. 1994) with alignment parameters as pair wise alignment, gap opening penalty 10, and gap extension penalty 0.1: for multiple alignments, gap opening penalty 10.0 and gap extension penalty 0.2. The resulting alignment was used in Molecular Evolutionary Genetics Analysis 6 (MEGA6) program (Tamura et al. 2013) for generation of unrooted phylogenetic tree using neighbor-joining method with 1000 bootstrap replicates.
Bioinformatics analysis
For expression and co-expression studies of OsMIZ1, publically available online resources (expression data) either rice genome annotation project (Kawahara et al. 2013) or Genevestigator (Hruz et al. 2008) were used. Rice SNP-Seek database (Alexandrov et al. 2015; Mansueto et al. 2016) was used to get insight into allele count and frequency in 3024 rice genome resequencing data. Transcriptional co-regulation of OsMIZ1 was analyzed by using RiceFREND database (Sato et al. 2013) with two hierarchy and the mutual rank was set as 10. The co-expressed gene network was constructed by using Cytoscape version 3.6.1 (Shannon et al. 2003).
Results
Root phenotyping under osmotic and salt stresses
The mean reduction in the seminal root length (SRL) of 20 genotypes analyzed under osmotic (15% PEG) and salt (150 mM NaCl) was 27 and 41%, respectively. Known tolerant rice cv. Nagina 22 and CR-262-4 showed least reduction in seminal root length (SRL), while susceptible cv. Pusa Basmati 1121 and Pusa Sugnadh 5 showed > 55% reduction under salt stress and > 37% reduction under PEG-induced drought stress (Table 1). On the basis of ‘S’ index for total SRL/seedling, Nagina 22 and CR-262-4 were selected as most tolerant, while Pusa Basmati 1121 and Pusa Sugandh 5 as most susceptible genotypes for both osmotic and salinity stress (Table 2).
The total root length and root surface area were enhanced under osmotic stress in tolerant genotypes Nagina 22 and CR-262-4, while total root length and root surface area were reduced under these stresses in susceptible Pusa Basmati 1121 and Pusa Sugandh 5. Highest total root length and root surface were produced by drought-tolerant CR-262-4 under PEG stress, followed by Nagina 22 (Fig. 1a, b, Suppl. Fig. S1). On the other side, Pusa Basmati 1121 and Pusa Sugandh 5 showed a decline in total root length and root surface area under both the stresses as compared to control (Fig. 1a, b, Suppl. Fig. S1). In case of root diameter, mixed trend was observed with one of the selected susceptible as well as tolerant varieties showing significant decrease in diameter under drought stress over control (Fig. 1c, Suppl. Fig. S1). However, root volume under stress was reduced by about 50% in susceptible varieties Pusa Basmati 1121 and Pusa Sugandh 5, while it increased by 1.25-fold in Nagina 22 under stress as compared to control (Fig. 1d, Suppl. Fig. S1). Root biomass of Pusa Basmati 1121 and Pusa Sugandh 5 was drastically reduced (43–46%) under stress conditions, while tolerant cultivars maintained root biomass under stress conditions (Fig. 1e, Suppl. Fig. S1).
Physiological responses under osmotic stress
The content of photosynthetic pigments (chlorophyll a, chlorophyll b, and carotenoids), relative water content, and membrane stability index were measured in tolerant and susceptible genotypes under osmotic stress. Osmotic stress reduced RWC in all genotypes with tolerant lines showing less reduction as compared with susceptible ones under osmotic stress. Similarly, MSI also followed the same trend (Fig. 2a). It declined by 20–25% in sensitive Pusa Basmati 1121 and Pusa Sugandh 5, while drought-tolerant Nagina 22 and CR-262-4 showed only 5.4–7.2% decline in MSI under osmotic stress as compared with control plants (Fig. 2b). The content of chlorophyll a, chlorophyll b, and carotenoids was although more in Pusa Basmati 1121 and Pusa Sugandh 5 compared to Nagina 22 and CR-262-4 under control conditions, the susceptible genotypes showed more decline in the content of chlorophyll a, chlorophyll b, carotenoids under stress treatment. Nagina 22 and CR-262-4 maintained the contents of chlorophyll a, chlorophyll b, and higher levels of carotenoids under osmotic stress (Fig. 2c–e).
Cloning of MIZ1 ortholog in rice
BLASTp search of the rice genome RGAP7 with AtMIZ1 protein sequence showed that among the DUF617 proteins in rice, Loc_Os02g47980 with highest hit score, identities (56%) and positives (71), and lowest expect value (7e-60). Hence, we identified Loc_Os02g47980 as closest orthologs of AtMIZ1 and named it as OsMIZ1. As per the RGAP, the open reading frame of OsMIZ1 is 873 bp and codes for a protein with 291 amino acids. The predicted molecular weight and pI values of rice MIZ ortholog are 30.819 kDa and 11.1033, respectively. Although AtMIZ1 gene does not contain intron, OsMIZ1 gene contains one intron of 124 bp long (Suppl. Fig. S2).
In order to clone the coding sequences of putative OsMIZ1, RT-PCR amplification of OsMIZ1 was attempted from cDNA of identified drought tolerant (Nagina 22 and CR-262-4), susceptible (Pusa Basmati 1121 and Pusa Sugandh 5) cultivars as well as Rasi which showed medium susceptibility index both for drought and salt stress. Successful RT-PCR amplification of expected size was observed from cultivars Nagina 22 and CR-262-4. No amplification was observed from cDNA in case of Pusa Basmati 1121, Pusa Sugandh 5 and Rasi, which possibly could be due to low level of MIZ1 expression in these cultivars. Therefore, for these three cultivars, OsMIZ1 was PCR amplified by using genomic DNA as template. Amplified PCR product from both cDNA as well as genomic DNA was cloned in cloning vector, and plasmids were subjected for DNA sequencing. OsMIZ1 sequences from these five rice genotypes were submitted to NCBI GenBank with accession numbers KT285509-KT285512 and KT306830. Sequence comparison of OsMIZ1 coding sequences from these cultivars revealed 12-bp deletion in coding sequence of Rasi, Nagina 22, CR-262-4, and Pusa Basmati 1121 as compared to that of Cultivar Nipponbare and Pusa Sugandh 5. This deletion resulted in deletion of four alanine residues in MIZ1 protein; however, the further coding frame remained the same (Fig. 3). There was no other sequence variation among these five varieties and between Nipponbare. In order to get a deeper insight into allelic variation of OsMIZ1 in larger number of genotypes/accessions, we used Rice SNP-Seek database (Alexandrov et al. 2015; Mansueto et al. 2016) and extended the search to 3024 rice genome resequencing data. Interestingly, among the 3024 accessions, the (Loc_Os0247980) locus does not have any SNPs, however, harbors 49 InDels. The 12 bp InDel observed in Rasi and Nagina 22 was conspicuous in 3K genome data. In total, 3024 varieties, from the total major allele count of 5970, the count of alleles with first 9 bp of 12-bp deletion on chromosome 2 from location 29365043 to 29365051 was 1976 with 33.10% allele frequency. In tandem, the allele count with the next 3-bp deletion (29365052 to 29365054) was 1974 with allele frequency of 33.07% (Fig. 4; Suppl. Fig. S3). Further, alleles with the 12-bp deletions were significantly prevalent in indica group of rice with allele frequency ranging from 52.48 to 62.32% in different sub-populations of indica rice; while in the sub-populations of japonica group of rice, the allele frequency ranged from 0 to 7.77%.
Multiple alignment of translated amino acid sequence of OsMIZ1 from five drought and salt tolerant/susceptible varieties with Nipponbare (Loc_Os02g47980). Protein sequences with accession number, ALM55558.1, ALM55559.1, ALM55560.1, ALM55561.1, and 5AL010968.1, were used for multiple sequence alignment. DUF617 domain in MIZ1 protein is underlined. Unfilled box indicates 12-bp deletion at nucleotide level resulting in deletion of four Adenine residues
Allele frequency of OsMIZ1 (Loc_Os02g47980) in 3024 accessions available at SNP-seek database. Allele frequency of OsMIZ1 in different rice subpopulations have been indicated by distinctly colored lines. On X-axis, numbers represent positions of Indels. Number on Y-axis indicates allele frequency in percentage. Table in the bottom shows frequency of OsMIZ1 alleles with 12-bp deletion in different indica and japonica sub-populations
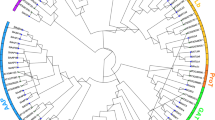
Phylogenetic analysis of rice MIZ1 with other plants of Poaceae and Arabidopsis showed that rice MIZ1 is close to its counterpart in sorghum, maize, and foxtail and most distant from wheat among Poaceae family, whereas it is farthest from Arabidopsis MIZ1 among the selected plant species (Fig. 5).
Phylogenetic relationship between rice MIZ1 and orthologs of MIZ1 from different plants. Full-length amino acid sequences from rice and other plants from Poaceae and Arabidopsis were aligned using ClustalW. The neighbor-joining tree was prepared with 1000 bootstraps alignments using Molecular Evolutionary Genetics Analysis 6 (MEGA6) and bootstrap consensus tree is shown
MIZ1 shows genotype-specific drought and salt stress-induced expression in rice
To find out possible role of MIZ1 in drought and salt stress, expression of OsMIZ1 was studied in response to drought and salt stress in tolerant and susceptible varieties using qRT-PCR. OsMIZ1 showed strong drought-induced expression in root tissue of identified drought-tolerant varieties Nagina 22 and CR-262-4 (Fig. 6a). Level of induction of OsMIZ1 expression was higher in Nagina 22 than CR-262-4, whereas no significant expression was observed in both the susceptible varieties even after drought stress. In case of salt stress-induced expression, the identified salt-tolerant varieties behaved distinctly. Higher expression of OsMIZ1 in response to salt stress was observed only in CR-262-4, while, in Nagina 22, the expression was induced by only about 1.5 times. However, in susceptible genotypes, OsMIZ1 expression levels were drastically reduced under salt stress (Fig. 6a). These results show that drought and salt stress-induced expression of OsMIZ1 are genotype specific, and genotypes which maintained better root traits have relatively higher expression levels. Expression profiles of OsMIZ1 studied using Genevestigator data also showed drought and salt stress induced as well as genotype-specific expression of OsMIZ1 (Suppl. Figs. S4 and S5).
Expression of OsMIZ1 (Loc_Os02g47980) in rice. aOsMIZ1 expression under drought and salt stress in rice varieties using quantitative RT-PCR. Expression levels were normalized against expression of ubiquitin gene as an internal control and are shown relative to untreated control plants. Values are presented as the mean and the error bars indicate standard deviation of three independent experiments. b Expression of OsMIZ1 in different tissue types. The expression level of OsMIZ1 was plotted based on information in rice expression matrix from next-generation sequencing transcriptome data available at in the Rice Genome Annotation Project database. *Statistically significant difference at P0.05
In order to get more insight in gene expression pattern of OsMIZ1, gene expression data of the rice genome annotation project was searched for tissue-specific gene expression, OsMIZ1 expression can be observed in different tissues including leaves, inflorescence, anthers, pistil, and embryo (Fig. 6b). Differential expression of OsMIZ1 was also found in mature roots, leaves, and pollens of Nipponbare in different transcriptome libraries (data not shown). These expression patterns were also cross-validated using Genevestigator database, where higher OsMIZ1 expression was observed mostly in root and leaf tissues followed by inflorescence of different developmental stages (Suppl. Fig. S6). To have an idea of cis-regulatory elements (CRE) in promoter sequence of MIZ1, a 2-kb promoter sequence upstream of transcription start site (TSS) was used to find out CRE from PlantCARE database (Lescot et al. 2002). The promoter analysis showed abundance of abiotic stress response elements such as ABRE, CGTCA-motif, DRE, MYB, MYC, STRE, and TGACG-motifs etc. in MIZ1 promoter (Suppl. Table S1).
Other facets of OsMIZ1 and possible involvement plant development and stress response
We also came across interesting features of OsMIZ1 expression using Genevestigator. In an independent report, a transcriptomics approach was employed to identify/map the aroma QTLs in basmati rice employing bi-parental mapping population derived from cross between Pusa Basmati 1121 and Pusa 1342 (non-aromatic variety) (Pachauri et al. 2014). Expression profile of OsMIZ1 in these gene expression studies revealed that OsMIZ1 showed higher expression in non-aromatic rice variety, Pusa 1342 than basmati variety, Pusa Basmati 1121 in all the four replicates. Similarly, the expression of OsMIZ1 was higher in non-aromatic RILs than the aromatic RILs, albeit with little lower magnitude (Fig. 7).
Expression pattern of OsMIZ1 (Loc_Os02g47980) in aromatic, Pusa 1121 and non aromatic rice variety, Pusa 1342 and in bulk pool of aromatic and non-aromatic RILS derived from their cross. The expression data is retrieved from Genevestigator micro array data in Experiment ID, OS-00109. The scale represents percentage of expression potential with the maximum value is displayed as dark colored and minimum value is displayed as no color. The heat map was plotted using Genevestigator
In an experiment, expressions of genes in two susceptible varieties of rice (Nipponbare and IR24) were studied in response to different strains and/or mutants of Xanthomonas oryzae pv. oryzae (Xoo), a causal organism of bacterial blight disease in rice. The expression of OsMIZ1 in these experiments was checked making use of Genevestigator data (Suppl. Fig. S7). Interestingly, OsMIZ1 expression was particularly higher in all the replicates of both the varieties at 24 h post-inoculation of Xoo strains PXO99AME1 (double mutant of effectors, a pthXo6 and avrXa27) and PXO99AME2 (effector, pthXo1 mutant) in comparison to wild types (PXO99A, T7174, and PXO86) as well as other strains of Xoo, PXO99ME5 (reduced virulence strain with uncharacterized mutation in a TAL effector) and PXO99AME7 (nonfunctional type III secretion system, non-pathogenic).
Higher expression of OsMIZ1 was also found upon heavy metal treatment especially arsenic (As-V) and lead (Pb) at 100 mM concentrations post 24 h (Suppl. Fig. S8). Similarly elevated expression of OsMIZ1 was observed in response to heat treatment (42 °C) (Suppl. Fig. S9).
Since there is little information about possible role of DUF617 domain containing proteins and their interacting proteins in plants especially about OsMIZ1, we used the transcriptome data from iron and phosphorus interaction in rice seedling experiment (Zheng et al. 2009) to check rice gene co-expression (GSE17245-Turquoise Module) from rice genome annotation project database. Thirteen genes among a total of 3573 genes showed strong co-expression (0.95 and 1) with OsMIZ1 (Loc_Os02g47980) as their expression levels elevated in rice roots grown in nutrient media deficient in phosphorus (+Fe-P) and peaked when nutrient media was deficient in both iron and phosphorus (−Fe-P) (Suppl. Fig. S10). Additionally, co-expression analysis showed that gene such as ‘Isp4 protein like’ which is upregulated in Fe deficiency in rice roots co-expresses with MIZ1 (Suppl. Fig. S11). These results indicate possible role of OsMIZ1 in mineral uptake mechanism. Co-expression analysis revealed that OsMIZ1 co-expresses with ferritin/ribonucleotide reductase-like family protein, which may be involved in iron homeostasis and oxidative stress responses (Suppl. Fig. S10, S11).
Discussion
In terrestrial environments, plant’s survival largely depends on ability of its roots to draw water and essential nutrients from soil. Plants have evolved a mechanism to direct their root growth in response to water gradient through a mechanism called hydrotropism (Takahashi et al. 2009; Miyazawa et al. 2011). Root hydrotropism is thought to have role in drought avoidance as well as efficient uptake of water and nutrients from soil (Eapen et al. 2005; Takahashi et al. 2009). In Arabidopsis, it was shown that the MIZU-KUSSEI (MIZ) 1 and 2 have crucial role in hydrotropism (Kobayashi et al. 2007; Miyazawa et al. 2012). Arabidopsis MIZ1 (AtMIZ1) which harbors plant specific, domain of unknown function (DUF617) is considered to be the positive regulator of hydrotropism. The AtMIZ1 was also shown to potentiate the ability of drought avoidance, increase the root cell viability under hydro-stimulated conditions in Arabidopsis (Miyazawa et al. 2012). In rice, the closest ortholog of AtMIZ1 is a gene encoding a DUF domain containing protein at locus Loc_Os02g47980. In the present study, we cloned the gene from five rice varieties and named it as OsMIZ1 based on its orthology to AtMIZ1. Through physiological studies and root screening under drought and salt stress, we identified drought and salt tolerant as well as susceptible varieties. The allele mining of coding region of OsMIZ1 in identified drought/salt tolerant and susceptible rice varieties revealed no allelic variation specific to drought/salt tolerant and susceptible varieties (Fig. 3). However, a 12 bp insertion was observed in one of the drought and salt sensitive rice variety, Pusa Sugandh 5 and Nipponbare, which is also sensitive to salt stress (Ferdose et al. 2009). To know more about OsMIZ1 alleles in known drought/salt tolerant and susceptible rice varieties, we extended our search using SNP-Seek database (Mansueto et al. 2016, 2017). Interestingly, Pokkali and Nona Bokra which are known salt/drought-tolerant cultivars (Akbar et al. 1986; Puram et al. 2017) harbor OsMIZ1 allele having 12-bp deletion, whereas the sensitive varieties such as IR-6 and Pusa Basmati 1 (Khan et al. 2013; Singh et al. 2018) were found to have OsMIZ1 alleles with 12 bp insertion. However, these indications of association of OsMIZ1 alleles with 12 bp deletion to salt/drought tolerance would require more elaborate enquiry. Nevertheless, it is important to note two observations in this regard; one, the high-frequency OsMIZ1 alleles with 12 bp deletion in several sub-populations of indica rice (Fig. 4, 52.48–62.32%) than that of japonica rice sub-populations (0–7.7%). Other being, the higher salinity tolerance observed in indica rice than japonica rice (Lee et al. 2003).
The drought stress-inducible expression of OsMIZ1 in tolerant rice varieties (Fig. 5) is in incongruence with findings in Arabidopsis (Miyazawa et al. 2012) about possible involvement of MIZ1 orthologs in drought stress mechanism. Further, drought-inducible expression of OsMIZ1 was found to be genotype specific; therefore, it would be interesting to study promoter of tolerant and susceptible varieties or presence/absence of specific drought inducible cis-acting elements, if any. In case of salt stress, OsMIZ1 expression significantly increased in one of the tolerant varieties, CR-262-4 while in Nagina 22, it showed a moderate increase. Such distinct pattern of expression of several genes involved in stress responses has been observed in tolerant varieties of rice (Wei et al. 2017) indicating genotypic differences in adaptation to stress environment. Indeed, salinity and drought tolerance have been found to be complex processes comprising several changes such as reduced growth, increased gene expressions, higher ABA levels, increased level of compatible solutes, protective proteins, antioxidants and suppression of energy-consuming pathways (Bartels and Sunkar 2007).
Other than drought and salt stress, MIZ1 expression in several RNAseq and microarray experiments in rice could give further directions to ponder over its possible involvement in distinct processes of growth, development, and biotic-abiotic stress responses. Genevestigator data revealed the higher expression of OsMIZ1 in non-aromatic rice variety as well as well as RILs with non-aroma trait (Fig. 7); several tissue types including reproductive organs under normal as well as stress conditions (Fig. 6b, Suppl. Fig. S5, S6); in response to Xoo strains mutant for effector proteins (Suppl. Fig. 7) and heavy metals especially arsenic (As-V) and lead (Pb) (Suppl. Fig. S8). These encouraging findings although not painstaking are preliminary glimpses which solicit further investigation on role of MIZ1 not only in drought and or salt stress but also in growth, development, biotic, and abiotic stress tolerance mechanisms. These reflections further underpin the importance of ‘domain of unknown functions’ proteins in elucidating hitherto unknown candidates in complex biological processes. It is rightly quoted “DUFs remain a treasure trove of novel biology waiting to be plundered” (Bateman et al. 2010).
References
Akbar M, Gunawardena IE, Ponnamperuma FN (1986) Breeding for soil stresses: In progress in rainfed lowland rice. Institute Rice Research Institute, Manila, pp 263–272
Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes RR, Ulat VJ, Chebotarov D, Zhang G, Li Z, Mauleon R, Hamilton RS, McNally KL (2015) SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res 43:D1023–D1027
Barrs HD, Weatherley PE (1962) A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust J Biol Sci 15:413–428
Bartels D, Sunkar R (2007) Drought and Salt Tolerance in Plants. Crit Rev Plant Sci 24(1):23–58
Bateman A, Coggill P, Finn RD (2010) DUFs: families in search of function. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1148–1152
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
Deshmukh PS, Sairam RK, Shukla DS (1991) Measurement of ion leakage as a screening technique for drought resistance in wheat genotypes. Indian J Plant Physiol 34:89–91
Dietrich D, Pang L, Kobayashi A, Fozard JA, Boudolf V, Bhosale R, Antoni R, Nguyen T, Hiratsuka S, Fujii N, Miyazawa Y, Bae TW, Wells DM, Owen MR, Band LR, Dyson RJ, Jensen OE, King JR, Tracy SR, Sturrock CJ, Mooney SJ, Roberts JA, Bhalerao RP, Dinneny JR, Rodriguez PL, Nagatani A, Hosokawa Y, Baskin TI, Pridmore TP, De Veylder L, Takahashi H, Bennett MJ (2017) Root hydrotropism is controlled via a cortex-specific growth mechanism. Nat Plants 3:17057
Eapen D, Barroso ML, Ponce G, Campos ME, Cassab GI (2005) Hydrotropism: root growth responses to water. Trends Plant Sci 10:44–50
Ferdose J, Kawasaki M, Taniguchi M, Miyake H (2009) Differential Sensitivity of Rice Cultivars to Salinity and Its Relation to Ion Accumulation and Root Tip Structure. Plant Production Science 12:453–461
Fischer RA, Maurer R (1978) Drought resistance in spring wheat cultivars. I Grain yield response. Aust J Agic Res 29:897–907
Gowda VRP, Henry A, Yamauchi A, Shashidhar HE, Serraj R (2011) Root biology and genetic improvement for drought avoidance in rice. Field Crop Res 122:1–13
Henry A, Gowda VRP, Torres RO, McNally KL, Serraj R (2011) Variation in root system architecture and drought response in rice (Oryza sativa): Phenotyping of the OryzaSNP panel in rainfed lowland fields. Field Crop Res 120:205–214
Hiscox JD, Israelstam GF (1979) A method for extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57:1332–1334
Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008:420747
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Iwata S, Miyazawa Y, Fujii N, Takahashi H (2013) MIZ1 regulated hydrotropism functions in the growth and survival of Arabidopsis thaliana under natural conditions. Ann Bot 13:1–12
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4
Khan SK, Iqbal J, Saeed M (2013) Comparative study of grain yield and biochemical traits of Different rice varieties grown under saline and normal conditions. J Anim Plant Sci 23:575–588
Kobayashi A, Takahashi A, Kakimoto Y, Miyazawa Y, Fujii N, Higashitani A, Takahashi H (2007) A gene essential for hydrotropism in roots. Proc Natl Acad Sci U S A 104:4724–4729
Lee KS, Choi WY, Ko JC et al (2003) Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta 216:1043
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Mansueto L, Fuentes RR, Chebotarov D, Borja FN, Detras J, Miguel AJ, Palis K, Poliakov A, Dubchak I, Solovyev V, Sackville HR, McNally K, Alexandrov N, Mauleon R (2016) SNP-Seek II: A resource for allele mining and analysis of big genomic data in Oryza sativa. Curr Plant Biol 7-8:16–25
Mansueto L, Fuentes RR, Borja FN, Detras J, Abriol-Santos JM, Chebotarov D, Sanciangco M, Palis K, Copetti D, Poliakov A, Dubchak I, Solovyev V, Wing RA, Hamilton RS, Mauleon R, McNally KL, Alexandrov N (2017) Rice SNP-seek database update: new SNPs, indels, and queries. Nucleic Acids Res 45(D1):D1075–D1081
Marchler-Bauer A et al (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39(D):225–229
Miyazawa Y, Yamazaki T, Moriwaki T, Takahashi H (2011) Root tropism: its mechanism and possible functions in drought avoidance. Adv Bot Res 57:350–375
Miyazawa Y, Moriwaki T, Uchida M, Kobayashi A, Fujii N, Takahashi H (2012) Overexpression of MIZU -KUSSEI 1 enhances root hydrotropic response by retaining cell viability under hydrostimulated condition in Arabidopsis thaliana. Plant Cell Physiol 53:1926–1933
Moriwaki T, Miyazawa Y, Kobayashi A, Takahashi H (2013) Molecular mechanism of hydrotropism in seedling roots of Arabidopsis thaliana. Am J Bot 100:25–34
Ober E, Le Bloa M, Rajabi A, Smith C (2005) Genotypic differences in rooting patterns and soil water extraction related to drought tolerance in sugar beet. Comp Biochem Physiol A Mol Integr Physiol 141:S302
Pachauri V, Mishra V, Mishra P, Singh AK, Singh R, Singh NK (2014) Identification of Candidate Genes for Rice Grain Aroma by Combining QTL Mapping and Transcriptome Profiling Approaches. Cereal Res Commun 42:376–388
Price AH, Steele KA, Gorham J, Bridges JM, Moore BJ, Evans JL, Richardson P, Jones RGW (2002) Upland rice grown in soil-filled chambers and exposed to contrasting water deficit regimes. I. Root distribution, water use and plant water status. Field Crop Res 76:11–24
Puram VRR, Ontoy J, Linscombe S, Subudhi PK (2017) Genetic Dissection of Seedling Stage Salinity Tolerance in Rice Using Introgression Lines of a Salt Tolerant Landrace Nona Bokra. J Hered 108:658–670
Rengasamy P (2010) Soil processes affecting crop production in salt affected soils. Funct Plant Biol 37:255–263
Samson BK, Hasan H, Wade LJ (2002) Penetration of hardpans by rice lines in the rainfed lowlands. Field Crop Res 76:175–188
Sarker A, Erskine W, Singh M (2005) Variation in shoot and root characteristics and their association with drought tolerance in lentil landraces. Genet Resour Crop Evol 52:87–95
Sato Y, Namiki N, Takehisa H, Kamatsuki K, Minami H, Ikawa H, Ohyanagi H, Sugimoto K, Itoh J, Antonio BA, Nagamura Y (2013) RiceFREND: a platform for retrieving coexpressed gene networks in rice. Nucleic Acids Res 41:D1214–D1221
Serraj R, Kumar A, McNally KL, Slamet-Loedin I, Bruskiewich R, Mauleon R, Cairns J, Hijmans RJ (2009) Improvement of drought resistance in rice. Adv Agron 103:41–98
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Shkolnik D, Nuriel R, Bonza MC, Costa A, Fromm H (2018) MIZ1 regulates ECA1 to generate a slow, long-distance phloem-transmitted Ca2+ signal essential for root water tracking in Arabidopsis. Proc Natl Acad Sci U S A 115:8031–8036
Singh K, Neelam K, Kaur A, Kaur K (2016a) Rice. In: Singh M, Kumar S (eds) Broadening the Genetic Base of Grain Cereals. Springer, Berlin, pp 27–65
Singh R, Singh Y, Xalaxo S, Verulkar S, Yadav N, Singh S, Singh N, Prasad KSN, Kondayya K, Ramana Rao PV, Girija Rani M, Anuradha T, Suraynarayana Y, Sharma PC, Krishnamurthy SL, Sharma SK, Dwivedi JL, Singh AK, Singh PK, Nilanjay SNK, Kumar R, Chetia SK, Ahmad T, Rai M, Perraju P, Pande A, Singh DN, Mandal NP, Reddy JN, Singh ON, Katara JL, Marandi B, Swain P, Sarkar RK, Singh DP, Mohapatra T, Padmawathi G, Ram T, Kathiresan RM, Paramsivam K, Nadarajan S, Thirumeni S, Nagarajan M, Singh AK, Vikram P, Kumar A, Septiningshih E, Singh US, Ismail AM, Mackill D, Singh NK (2016b) From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci 242:278–287
Singh VK, Singh BD, Kumar A, Maurya S, Krishnan SG, Vinod KK, Singh MP, Ellur RK, Bhowmick PK, Singh AK (2018) Marker-Assisted Introgression of Saltol QTL Enhances Seedling Stage Salt Tolerance in the Rice Variety “Pusa Basmati 1”. Int J Genomics 8319879
Takahashi H, Miyazawa Y, Fujii N (2009) Hormonal interactions during root tropic growth: hydrotropism versus gravitropism. Plant Mol Biol 69:489–502
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Higgins GD, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tuberosa R, Sanguineti MC, Landi P, Giuliani MM, Salvi S, Conti S (2002) Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol Biol 48:697–712
Wang H, Yamauchi A (2006) Growth and function of roots under abiotic stress in soil. In: Huang BR (ed) Plant-Environment Interactions, 3rd edn. CRC Press, New York, pp 271–320
Wei H, Chen C, Ma X, Zhang Y, Han J, Mei H, Yu S (2017) Comparative Analysis of Expression Profiles of Panicle Development among Tolerant and Sensitive Rice in Response to Drought Stress. Front Plant Sci 8:437
Wopereis MCS, Kropff MJ, Maligaya AR, Tuong TP (1996) Drought-stress responses of two lowland rice cultivars to soil water status. Field Crop Res 46:21–39
Zheng L, Huang F, Narsai R, Wu J, Giraud E, He F, Cheng L, Wang F, Wu P, Whelan J, Shou H (2009) Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol 151:262–274
Acknowledgments
Authors acknowledge the support of Director, ICAR-National Bureau of Plant Genetic Resources (NBPGR), New Delhi, for carrying out this work.
Funding
National Agriculture Science Fund (NASF), ICAR, New Delhi, Grant No. Phen 2015/2011-12 financially supported phenotyping and cloning and expression analysis.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments-VK, PP, VC; performed the experiments-VK, SKY, PP; bioinformatics work and data analysis-DPW, VK; manuscript drafting-VK, DPW, AK, VC.
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Néstor Carrillo
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Table S1
(DOCX 14 kb)
Supplementary Fig. S1
Root phenotypes of drought tolerant and susceptible varieties in response to drought stress (PEG 15%) (JPG 255 kb)
Supplementary Fig. S2
Gene structure of rice MIZ1. Coding sequence and genomic sequence of MIZ1 was used to develop graphics for visualization using Gene structure display server program. Filled boxes in maroon color indicate 5‘ and 3‘ UTR, filled boxes in green indicate exons and lines joining two exons indicate intron (JPG 17 kb)
Supplementary Fig. S3
Allele count of locus Loc_Os02g47980 (OsMIZ1) in 3024 accessions from SNP-seek database. Allele count in different rice subpopulations have been indicated by distinct lines. On X-axis numbers represent positions of Indels. Number on Y-axis indicates allele numbers (JPG 80 kb)
Supplementary Fig. S4
Expression of OsMIZ1 (Loc_Os02g47980) in rice genotypes FL478 and IR29 in response to salt stress. The expression data is retrieved from Genevestigator micro array data in Experiment ID, OS-00023. The scale represent percentage of expression potential with the maximum value is displayed as dark colored and minimum value is displayed as no color (JPG 34 kb)
Supplementary Fig. S5
Expression of OsMIZ1 (Loc_Os02g47980) in rice varieties IRAT109 and ZS97 for drought stress treatment in flag leaves. The expression data is retrieved from Genevestigator micro array data in Experiment ID, OS-00068. The scale represent percentage of expression potential with the maximum value is displayed as dark colored and minimum value is displayed as no color (JPG 69 kb)
Supplementary Fig. S6
Expression of OsMIZ1 (Loc_Os02g47980) during reproductive development in rice variety IR64. The expression data is retrieved from Genevestigator micro array data in Experiment ID, OS-00007. The scale represent percentage of expression potential with the maximum value is displayed as dark colored and minimum value is displayed as no color (JPG 112 kb)
Supplementary Fig. S7
Expression of OsMIZ1 (Loc_Os02g47980) in response to different strains/mutants of Xanthomonas oryzae in two susceptible rice cultivars, nipponbare and IR24. PXO99A, T7174, and PXO86 are wild type strains of Xanthomonas oryzae pv. oryzae (Xoo). PXO99AME1 is double mutant of effectors, a pthXo6 and avrXa27. PXO99AME2 is mutant of an effector, pthXo1. PXO99ME5 is a reduced virulence strain with uncharacterized mutation in a TAL effector; and strain PXO99AME7 has nonfunctional type III secretion system, non-pathogenic. The expression data is retrieved from Genevestigator micro array data in Experiment ID, OS-00061. The scale represent percentage of expression potential with the maximum value is displayed as dark colored and minimum value is displayed as no color (JPG 276 kb)
Supplementary Fig. S8
OsMIZ1 (Loc_Os02g47980) expression in roots of rice variety IR64 upon treatment with different heavy metals. The expression data is retrieved from Genevestigator micro array data in Experiment ID, OS-00040. The scale represent percentage of expression potential with the maximum value is displayed as dark colored and minimum value is displayed as no color (JPG 58 kb)
Supplementary Fig. S9
OsMIZ1 (Loc_Os02g47980) expression in rice cultivar Zhonghua11 in response to heat (42°C) treatment. The expression data is retrieved from Genevestigator micro array data in Experiment ID, OS-00024. The scale represent percentage of expression potential with the maximum value is displayed as dark colored and minimum value is displayed as no color (JPG 27 kb)
Supplementary Fig. S10
Co-expression of genes with OsMIZ1 (Loc_Os02g47980) in response nutrient media deficient in either Phosphorus (P) / Iron (Fe) or both. Lines of different colors shows co-expression patterns of genes. OsMIZ1 expression is indicated with line in red color. The locus ID of co-expressing genes along with functional annotation and their respective line color in plot on left side is mentioned in table on the right side. X axis shows different combination of nutrient media with/without Fe/P. Y axis indicates expression values in terms of Z-score. Cut off value for gene co-expression was between 0.95 and 1. Gene co-expression plot was drawn using experiment ‘Iron and phosphorus interaction in rice seedlings’ in module GSE17245-turquoise at Rice Genome annotation project database (JPG 123 kb)
Supplementary Fig. S11
Co-expression analysis of OsMIZ1 (Loc_Os02g47980) (JPG 226 kb)
Rights and permissions
About this article
Cite this article
Kaur, V., Yadav, S.K., Wankhede, D.P. et al. Cloning and characterization of a gene encoding MIZ1, a domain of unknown function protein and its role in salt and drought stress in rice. Protoplasma 257, 475–487 (2020). https://doi.org/10.1007/s00709-019-01452-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-019-01452-5