Abstract
Banana is an important fruit crop and its yield is hampered by multiple abiotic stress conditions encountered during its growth. The NAC (NAM, ATAF, and CUC) transcription factors are involved in plant response to biotic and abiotic stresses. In the present study, we studied the induction of banana NAC042 transcription factor in drought and high salinity conditions and its overexpression in transgenic banana to improve drought and salinity tolerance. MusaNAC042 expression was positively associated with stress conditions like salinity and drought and it encoded a nuclear localized protein. Transgenic lines of banana cultivar Rasthali overexpressing MusaNAC042 were generated by Agrobacterium-mediated transformation of banana embryogenic cells and T-DNA insertion was confirmed by PCR and Southern blot analysis. Our results using leaf disc assay indicated that transgenic banana lines were able to tolerate drought and high salinity stress better than the control plants and retained higher level of total chlorophyll and lower level of MDA content (malondialdehyde). Transgenic lines analyzed for salinity (250 mM NaCl) and drought (Soil gravimetric water content 0.15) tolerance showed higher proline content, better Fv/Fm ratio, and lower levels of MDA content than control suggesting that MusaNAC042 may be involved in responses to higher salinity and drought stresses in banana. Expression of several abiotic stress-related genes like those coding for CBF/DREB, LEA, and WRKY factors was altered in transgenic lines indicating that MusaNAC042 is an efficient modulator of abiotic stress response in banana.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stress in agricultural perspective has been defined as any external factor that threatens the survival of the plant along with reducing its growth, overall productivity, and reproductive capacity (Rhodes and Nadolska-Orczyk 2001). High salinity and water deficiency in the soil are major hurdles for growth and yield of many plant species (Mahajan and Tuteja 2005). High salinity is an environmental stress resulting in hyperionic and hyperosmotic conditions causing reduction in growth and photosynthesis, membrane disorganization, generation of reactive oxygen species, and overall toxicity to cell metabolism (Niu et al. 1995; Greenway and Munns 1980; Yeo 1998). Drought stress results in cell shrinkage leading to cellular membrane damage, impaired function of membrane-associated enzymes, broad level peroxidation of cellular membrane, and protein denaturation by excessive production of ROS, extensive protein aggregation, and reduced photosynthesis activity (Bowler et al. 1992; Hoekstra et al. 2001). Banana and plantain are grown worldwide with a production of around 106.7 million tonnes per year (FAO 2013, http://faostat.fao.org). Banana is sensitive to water stress due to features like shallow roots and a permanent green canopy (Xu et al. 2014; van Asten et al. 2011). Banana needs nearly 2,000- to 2,500-mm rainfall evenly distributed throughout the year for optimal yield output (Vanhove et al. 2012). In India, 51.1 Mha of agricultural land is drought prone while 6.73 Mha is affected by salinity (NRCB newsletter 2010, http://www.nrcb.res.in). Moreover, report suggests that 25 % of ground water used for irrigation in India is saline, and by 2025, nearly 11.7 Mha of agricultural land may get affected by salinity (NRCB newsletter 2010, http://www.nrcb.res.in). In Tamil Nadu (India), banana production has been reported around 4.64 million tones which was declined due to drought during 2001–2004 (Surendar et al. 2013). Field studies conducted at the NRCB (National Research Centre for Banana, India) suggested a reduction of up to 42 % in bunch weight of banana by imposition of drought for one month during flowering (Ravi et al. 2013).
Plants being sessile need to withstand environmental stresses and hence to adapt and survive under stress conditions, plants have developed various mechanisms including enhanced expression of stress protective genes which include regulatory factors like transcription factors known to regulate expression of many functional genes involved in abiotic or biotic stress tolerance. One of these plant-specific transcription factor families is the NACs (NAM, ATAF, and CUC). NAC proteins have a DNA binding domain at N-terminus and a regulatory region towards C-terminus. NAC domain containing proteins are present in large number in plants with almost 110 members in Arabidopsis and 151 members in rice (Olsen et al. 2005; Nuruzzaman et al. 2010). Few of NAC proteins from different plant species have been characterized for their functions in plant development and stress responses. Rice SNAC1 is stress inducible and its overexpression resulted in elevated drought tolerance of transgenic plants (Hu et al. 2006). Soybean NAC transcription factors, GmNAC20 and GmNAC11 have been characterized as important regulators of salinity and cold stress (Hao et al. 2011). Overexpression of rice SNAC2 and SNAC3 resulted in improved drought tolerance in transgenic plants (Hu et al. 2008; Fang et al. 2015). Hordeum vulgare NAC transcription factor, HvSNAC1, is induced strongly by abiotic stress conditions and transgenic lines showed improved drought tolerance at various growth stages (Al Abdallat et al. 2014). Wheat NAC transcription factors TaGRAB1 and TaGRAB2 inhibits DNA replication of wheat dwarf geminivirus in wheat cells by interacting with Rep A protein of the virus (Xie et al. 1999). Wheat NAC transcription factor, TaNAC4, is induced by multiple stress conditions like methyl jasmonate, infection of stripe rust pathogen, ABA, ethylene, high salinity, wounding, and low-temperature indicating that TaNAC4 is involved in response to multiple stress conditions (Xia et al. 2010). TaNAC69 expression was positively associated with stress responses of wheat and transgenic plants overexpressing TaNAC69 produced higher shoot biomass and longer roots than control plants (Xue et al. 2011; Baloglu et al. 2012). A comprehensive analysis of NAC transcription factors in banana (Musa acuminata) was reported recently resulting in identification of nearly 167 potential NAC transcription factor coding genes (Cenci et al. 2014). In the recent years, there has been growing interest on the characterization of the NAC transcription factors in banana with emphasis on secondary wall development and stress responses. MusaVND1, a banana NAC domain containing transcription factor was demonstrated to convert banana embryogenic cells into tracheary elements-like cells indicating its role in xylem development (Negi et al. 2015a). Further, MusaVND2 and MusaVND3 were reported to exhibit ability to develop ectopic secondary wall development in transgenic banana (Negi et al. 2015b). Expression of banana NAC factors, MaNAC1 and MaNAC2, was elevated in the fruit by ethylene and their interaction with ethylene insensitive 3 (EIN3)-like protein which is a component of ethylene signaling suggested that MaNAC1/2 may be involved in fruit ripening (Shan et al. 2012). While MaNAC1 is also involved in cold tolerance of banana fruits and is involved in interaction with ICE1-CBF cold signaling pathway (Shan et al. 2014). MaNAC5 in cooperation with WRKY transcription factor is involved in pathogen resistance and PR-genes regulation (Shan et al. 2015). Another banana NAC transcription factor, MusaNAC68, is induced by multiple stress conditions and positively regulates drought and salinity tolerance in banana (Negi et al. 2015c). However, exact role of NAC transcription factors in imparting tolerance to abiotic and biotic stresses needs to be elucidated in banana. Among the other NAC transcription factors, Arabidopsis thaliana NAC042/JUB1 has been proposed as central longevity regulator and its overexpression lowers intracellular H2O2 levels and increase tolerance to abiotic stress conditions besides delaying senescence (Wu et al. 2012) and enhancing heat tolerance in NAC042/JUB1 overexpressing transgenic plants (Shahnejat-Bushehri et al. 2012). In another study, Arabidopsis ANAC042 was shown to be involved in regulation of camalexin (a phytoalexin) which was substantiated by lower levels of camalexin in the T-DNA insertion mutants (Saga et al. 2012).
To test whether banana NAC042 can also elevate the drought and high salinity tolerance of transgenic banana and to promote the utilization of NAC genes in genetic improvement of banana, we have characterized MusaNAC042 and shown its probable involvement in drought and high salinity tolerance. We have chosen Musa cv. Rasthali for the generation of transgenic plants as it is one of the economically important banana cultivars in India and is highly susceptible to drought as well as high salinity stress conditions resulting in considerable yield reduction. Quantitative RT-PCR indicated that expression of MusaNAC042 was elevated during multiple stress conditions suggesting its potential role in their regulation. Transgenic banana plants showed better tolerance to drought and high salinity than control plants. MusaNAC042 encodes a nuclear localized protein and transgenic banana overexpressing MusaNAC042 displayed altered expression of multiple stress-related genes. The present study suggests that MusaNAC042 as a stress-related transcription factor can be useful for engineering abiotic stress tolerance in banana.
Material and methods
In silico analysis
MusaNAC042 was employed for pBLAST search at NCBI and NAC sequences (with high BLAST score) from different plant species were chosen for generating a phylogenetic tree. Neighbor joining tree (bootstrap value of 1,000 replicates) was constructed with Clustal omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and MEGA6 software (Kumar et al. 2008). For multiple sequence alignment, MusaNAC042 was aligned with A. thaliana ANAC042 (NP_181828.1), Theobroma cacao TcNAC042 (XP_007048954.1), Oryza brachyantha ObJUB1 (XP_006651871.1), and Vitis vinifera VvJUB1 (XP_002283251.1) using Clustal omega and online available box shade server (http://www.ch.embnet.org/software/BOX_form.html).
Stress treatments and expression analysis
Expression analysis of MusaNAC042 was carried out using in vitro-grown and green house-hardened plants of banana cv. Karibale Monthan plants. Similar-aged plants (plants with growth of 2 months in green house) were used in all the stress treatments. Individual banana plants were imposed with different stress treatments and RNA was isolated from leaves for expression analysis at different time points (5, 12, and 48 h). For each time point, three plants were utilized for stress imposition and top most leaves were harvested at the defined time. Tissue of three separately treated plants was mixed in equal amount for the isolation of total RNA. The QPCR was performed at least three times for each treatment. For salinity stress, 20 ml of 250 mM NaCl was irrigated to the plants. For drought treatment, plants were dried on a blotting paper in a laminar air flow hood (Negi et al. 2015c). Total RNA isolated from leaves using Concert plant RNA reagent (Invitrogen, Waltham, MA, USA) was subsequently cleaned using RNeasy spin column supplied with the RNeasy plant mini kit (Qiagen, Hilden, Germany) following the manufacturer instructions. During RNA isolation, the genomic DNA contamination was eliminated using Qiagen on column DNAase digestion (cat. no.79254) following manufacturer’s instructions. Synthesis of cDNA was carried out with 2 μg total RNA using thermoscript AMV-RT by following manufacturer’s instructions (Invitrogen: cat. no.12236-014). Quantitative RT-PCR was carried out with 1:100 diluted cDNA and SYBR Green Jump Start Taq Ready Mix (Sigma, St. Louis, MO) following the supplier instructions. Q-PCR running condition used was: 94 °C for 4 min followed by 30 cycles of 94 °C for 20 s, 55 °C for 20 s and 72 °C for 20 s followed by a melting curve analysis. Expression of Musa EF1α (housekeeping gene) was also analyzed for normalization of different Ct values. Calculations were carried out as follows: ΔCt (Target gene) = Ct (Target gene) − Ct (EF1α) and fold change was calculated by dividing 2−ΔCt value of target gene in stressed and in control condition.
Construction of MusaNAC042 overexpression vector
Complete coding sequence of MusaNAC042 was amplified from leaf cDNA of banana cv. Karibale Monthan. PCR was carried out in a 50-μl volume containing forward and reverse primer (10 pmol each), Taq DNA polymerase (1 unit), 0.5 μl dNTP (10 mM stock), 1x PCR buffer, 2 mM MgCl2 and 1 μL of cDNA (1:100 diluted). PCR running conditions were: 94 °C for 5 min for initial denaturation followed by 35 cycles with each cycle of 94 °C for 30 s, 55 °C for 45 s and 72 °C for 1 min after which final extension was done at 72 °C (for 10 min). Binary vector pCAMBIA1301 was digested with NcoI and PmlI to release the GUS coding sequence and the resultant vector backbone was gel purified (Roche, Penzberg, Bavaria, Germany, catalogue number. 11732676001). MusaNAC042 coding sequence was digested with BspHI and PmlI and ligated with above-generated vector backbone. Recombinant plasmid (pCAMBIA1301- MusaNAC042) was confirmed by restriction digestion as well as by sequencing of cloned insert. Agrobacterium tumefaciens strain EHA105(Hood et al. 1993) was electroporated with the binary vector. Primer sequences employed in this study are available in Electronic supplementary material (ESM) 1.
Construction of pCAMBIA1302-MusaNAC042 for sub-cellular localization
Full length coding sequence of MusaNAC042 was cloned in pCAMBIA1302 employing BglII and SpeI restriction sites, generating a translational fusion of MusaNAC042 and GFP. Embryogenic cells of banana cultivar Rasthali were transiently transformed using A. tumefaciens (EHA105) harboring pCAMBIA1302-MusaNAC042. GFP fluorescence was monitored after 5 days of transformation with the help of fluorescent microscope (Eclipse 80i, Nikon, Shinagawa, Tokyo, Japan). Hoechst 33258 staining was carried out to identify position of nuclei in the cells. Binary vector was sequenced to confirm the cloning of MusaNAC042 in correct coding frame. Primer sequences for generation of recombinant vector are provided in ESM 1.
Generation of transgenic banana plants
Agrobacterium-mediated transformation of banana cv Rasthali embryogenic cells was carried out as described previously (Ganapathi et al. 2001). Briefly, 0.5 ml PCV (packed cell volume) of embryogenic cells was co-cultivated with A. tumefaciens strain EHA105 for 30 min and later aspirated onto glass fiber filter which was incubated on semi-solid M2-medium in dark for 3 days (Cote et al. 1996). Following this, Agrobacterium was eliminated by culturing the cells in light on M2-medium added with antibiotic Cefotaxime (400 mg/l). Selection of transformed cells and growth of embryos was carried out on embryo development medium (BEM) with cefotaxime (400 mg/l) and selection agent hygromycin (5 mg/l). Conversion of somatic embryos into shoot was carried on MS-medium supplemented with BA (0.5 mg/l).
Confirmation of transgenic lines
Genomic DNA of putative transgenic lines was isolated using Plant Genomic DNA Kit (Sigma, USA; G2N350) and subjected to PCR amplification of hpt-II (hygromycin phosphotransferase) using PCR conditions: 94 °C for 5 min (initial denaturation), 35 cycles of 94 °C for 30 s, 55 °C for 45 s, and 72 °C for 45 s and final extension at 72 °C (10 min). Southern blotting was carried out to determine copy number of T-DNA transferred in different transgenic lines. Genomic DNA isolated as described above was digested (20 μg) with NcoI for 16 h at 37 °C. Digested genomic DNA was resolved on 0.8 % agarose TAE (pH 8.0 tris acetate buffer consisting of tris-acetate (0.04 M) and EDTA (0.001 M)) gel. Transfer of digested DNA onto Hybond-N nylon membrane (Amersham, Catalogue number RPN.203 N) was carried out with 10x SSC (0.45 M NaCl and 0.045 M tri-sodium citrate with final pH 7.0) buffer by capillary transfer. DNA was fixed onto membrane by baking at 120 °C for 30 min. DIG-labeled probe against hpt-II coding sequence was prepared using DIG labeling kit (Roche, cat. no. 11585614910) as per manufacturer’s instruction. In brief, 1 μg hpt-II (heat denatured) was mixed with 4 μl of DIG high prime (supplied with the kit) in a reaction of 20 μl and incubated at 37 °C for 3 h. Hybridization of probe with membrane-bound DNA was carried overnight at 42 °C using DIG Easy Hyb granules. Excess and non-specific binding of probe was washed by stringency washes at room temperature (with 2× SSC with 0.1 % SDS) and at 65 °C (with 0.5× with 0.1 % SDS). Binding of probe was visualized by anti-DIG antibody (1:5,000) labeled with alkaline phosphatase. Chemiluminescence signal was detected as per manufacturer’s protocol (Roche). Transcript level of MusaNAC042 in confirmed transgenic lines was analyzed by quantitative RT-PCR as described above. Amplification of banana EF1α was used for normalization of the Ct values.
Leaf disc assay
Leaf discs (1.0 cm diameter) excised from healthy leaves of control and transgenic lines were incubated on different concentrations of NaCl (0, 100, 200, and 250 mM) for imposing salinity stress. Treatment was carried out for 6 days under continuous white light at 27 ± 1 °C after which visual injury to discs was recorded. Total chlorophyll estimation was carried out as reported earlier (Arnon 1949). Briefly, leaf disc homogenized in 80 % acetone was centrifuged at 5,000 rpm (10 min) and the absorbance was recorded at 652 nm. Total chlorophyll content was expressed as microgram per milligram of fresh weight. Thiobarbituric-acid method was used for analyzing the MDA content as described earlier (Negi et al. 2015c). Briefly, leaf tissue was homogenized and incubated at 95 °C for 15 min with 0.5 % TBA in 20 % TCA. Reaction was terminated by cooling on ice and subsequently supernatant was collected by centrifugation. Absorbance was recorded at 600 and 532 nm and the reading of 600 nm was subtracted with reading at 532 nm. MDA content was estimated with the help of extinction-coefficient of 155 mM−1cm−1 as reported earlier (Heath and Packer 1968). Experiment was carried out in triplicate. MDA and chlorophyll content analysis was done in triplicate after mixing the tissue of at least three separate treatments.
Drought and high salinity tolerance assay
Two-month-old control and transgenic lines maintained in the green house were used for stress tolerance analysis. Plants were exposed to salinity stress by irrigating plants with 20 ml of 250 mM NaCl on every alternate day up to 15 days. For drought imposition, water was withheld for 14 days and stress symptoms were recorded. Recovery potential of transgenic and control plants was monitored after 1 month of regular re-watering with tap water. MDA, FV/FM ratio, and proline content were calculated for stressed plants during treatment. Photosynthetic efficiency was monitored in terms of Fv/Fm ratio using plant efficiency analyzer (Hansatech Instruments, Norfolk, UK; model: Handy-Pea). MDA content was estimated as described above. Proline content was estimated using ninhydrin reaction procedure. Stress assay were carried out with three biological replications. Tissues of the three replications were mixed in equal amount and three technical replications were performed for biochemical estimation of MDA and proline. Soil gravimetric water content was estimated as described earlier (Gaxiola et al. 2001) with three biological replications. Relative water content in the leaves of control and transgenic lines was performed as described earlier (Gaxiola et al. 2001). For relative water content, youngest fully expanded leaves were used and three biological replications were tested.
Quantification of stress-related gene expression
Multiple members of CBF/DREB, WRKY, and LEA family were identified from NCBI and banana genome sequence database (http://banana-genome.cirad.fr/). Total RNA was isolated from leaves of control and transgenic lines as described above. The cDNA synthesis was performed as described earlier. Quantitative RT-PCR with 1:50 diluted cDNA as template was carried out and the data obtained was used for calculations of fold change in expression as described above. The quantitative RT-PCR analysis was carried out in triplicate. Primer sequences employed in this study are available in ESM 1. Sequences of primers for quantitative RT-PCR are provided in ESM 1.
Results
Phylogenetic analysis of MusaNAC042
MusaNAC042 was identified employing pBLAST in banana genome sequence database (http://banana-genome.cirad.fr/) using Arabidopsis NAC042 (ANAC042: At2g43000.1 in Arabidopsis Genome Initiative) as search query. Sequence with the highest pBLAST score was designated as MusaNAC042. Coding sequence of MusaNAC042 contained 861 bases and encoded a 287-amino-acids-long protein with predicted molecular weight of 32.7 kDa and a theoretical pI of 6.4. MusaNAC042 shared high sequence similarity with other NAC042/JUNGBRUNNEN1 proteins from different plant species. MusaNAC042 exhibited identities of 52 % with GrJUB1, 50 % with CsJUB1, 54 % with JcJUB1, 52 % with VrJUB1, 52 % with NsJUB1, 55 % TcNAC042, 56 % with NnJUB1, 55 % with VvJUB1, 62 % with PdJUB1, 60 % with BdJUB1, 68 % with ObJUB1, 54 % with SiJUB1, 51 % with ZmJUB1, 51 % with ANAC042, 50 % with BrJUB1, 34 % with AtNST1, 23 % with AtVND6 and 31 % with AtSND1 (Fig. 1). N-terminal end of MusaNAC042 possessed a conserved NAC domain while the C-terminal end was relatively divergent (Fig. 2).
Phylogenetic analysis of banana MusaNAC042 with selected NAC proteins from different plant species. Amino acid sequences of NAC proteins in the tree are: Gossypium raimondii GrJUB1 (XP_012441089.1), Citrus sinensis CsJUB1 (XP_006475534.1), Jatropha curcas JcJUB1 (XP_012087398.1), Vigna radiata VrJUB1 (XP_014500826.1), Nicotiana sylvestris NsJUB1 (XP_009778771.1), Theobroma cacao TcNAC042 (XP_007048954.1), Nelumbo nucifera NnJUB1 (XP_010272267.1), V. vinifera VvJUB1 (XP_002283251.1), Phoenix dactylifera PdJUB1 (XP_008806688.1), Brachypodium distachyon BdJUB1 (XP_003559373.1), O. brachyantha ObJUB1 (XP_006651871.1), Setaria italica SiJUB1 (XP_004981565.1), Zea mays ZmJUB1 (XP_008644461.1), A. thaliana ANAC042 (NP_181828.1), Brassica rapa BrJUB1 (XP_009142897.1), A. thaliana AtNST1 (NP_182200.2), A. thaliana AtSND1 (AEE31527.1), and A. thaliana AtVND6 (AED96207.1). Neighbor joining tree was built with bootstrap value of 1,000. MusaNAC042 is boxed in blue
Expression analysis of MusaNAC042
MusaNAC042 was induced in the leaves of banana cultivar Karibale Monthan under high salinity and drought. During drought (drying of the plant on blotting paper), expression of MusaNAC042 was maximum at 5 h and then it was decreased at 48 h but remain higher than control level (Fig. 3a). Similarly, upon application of 250 mM NaCl (Fig. 3b) expression of MusaNAC042 peaked at 12 h and then reduce towards 48 h but remain higher than the expression value in control.
Sub-cellular localization of MusaNAC042
MusaNAC042 was fused with GFP in pCAMBIA1302 and transiently overexpressed in banana cultivar Rasthali embryogenic cells displayed uniform distribution of green fluorescence (Fig. 4a) indicating GFP distribution throughout the cell. Cells transformed with pCAMBIA1302-MusaNAC042 displayed preferential localization of green fluorescence in nucleus suggesting nuclear localization of MusaNAC042 (Fig. 4c). Position of nucleus in cells was determined after staining with Hoechst 33258 (Fig. 4b, d).
Subcellular localization of MusaNAC042 in banana embryogenic cells. a GFP fluorescence observed in pCAMBIA1302 transformed cells. b Nuclear position in the cells detected by Hoechst 33258 staining. c Green fluorescence observed in cells transformed with pCAMBIA1302-MusaNAC042. d Hoechst 33258 staining of cell transformed with pCAMBIA1302- MusaNAC042. The scale bar measures 50 μm
Regeneration of transgenic banana lines
Complete coding sequence of MusaNAC042 was cloned downstream of CaMV35S promoter in NcoI and PmlI sites of pCAMBIA1301 for achieving constitutive expression of MusaNAC042 in transgenic lines (Fig. 5a). Embryogenic cells of banana cultivar Rasthali were transformed with pCAMBIA1301-MusaNAC042 by Agrobacterium-mediated method and further growth was monitored (Fig. 5b). Growth of transformed embryogenic cells on hygromycin selection medium resulted in white and globular embryos (Fig. 5c). Further culturing of embryos on shoot development medium resulted in the conversion into putatively transformed shoots (Fig. 5d). Individual shoots were clonally propagated on shoot multiplication medium (Fig. 5e) and rooting was carried on MS-medium supplemented with 1 mg/l NAA (Fig. 5f) which were subsequently hardened in the green house (Fig. 5g).
Regeneration of transgenic banana lines. a T-DNA region of pCAMBIA1301-MusaNAC042 designed to overexpress MusaNAC042. b Development of embryogenic cells of banana cultivar Rasthali transformed with pCAMBIA1301-MusaNAC042 on glass fiber filter. c Close up of developing embryos on selection medium. d Shoot emergence on selection medium. e Multiple shoots of different putatively transgenic banana lines. f Rooting of different putative transgenic lines. g Hardened plants of different transgenic lines in green house
Molecular confirmation of T-DNA integration in transgenic lines
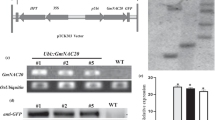
Genomic DNA of putative transgenic lines was analyzed for PCR amplification of hpt-II (Hygromycin phosphotransferase). PCR amplification of a 788-bp band from coding region of hpt-II indicated successful integration of T-DNA in genome of transgenic lines (Fig. 6a) while no amplification was observed in the wild-type (control, non transformed) plants. Southern blot analysis carried out with digoxin labeled hpt-II probe revealed two to five copies of T-DNA transferred in different transgenic lines (Fig. 6b). Further, overexpression of MusaNAC042 due to T-DNA copies analyzed by quantitative RT-PCR indicated that lines L2, L4, L7, and L11 accumulated 8.18-, 6.12-, 12.88-, and 9.23-fold of MusaNAC042 transcript respectively over control value (Fig. 6c).
Molecular confirmation of transgenic lines. a PCR amplification of hpt-II from genomic DNA of different lines. b Analysis of number of T-DNA insertions in different lines by Southern blot analysis. The blot was probed by digoxin labeled probe against hpt-II present within the T-DNA region. c MusaNAC042 transcript analysis in different transgenic lines by quantitative RT-PCR. The Ct-value of EF1α was utilized for normalization of MusaNAC042 Ct-values. Values indicated are mean ± SD of three technical replicates. (WT: wild type; L2, L4, L7, and L11: transgenic lines)
Leaf disc method for analysis of salinity tolerance
Tolerance of transgenic banana lines towards salinity stress was analyzed by leaf disc method. For this, leaf disc punched using cork borer were incubated in different concentrations of NaCl. Visual injury to leaf disc due to chlorophyll degradation was lower in transgenic lines than control suggesting better tolerance to salinity in the transgenic lines. While bleaching in the control discs was profound even in 100 mM NaCl, leaf discs of transgenic lines showed remarkably less bleaching and senescence even in 200 mM NaCl (Fig. 7a) after 6 days of treatment. Higher tolerance of transgenic lines in leaf disc method was confirmed by estimation of total chlorophyll and MDA content. Transgenic leaf discs retained significantly more chlorophyll than control at different concentrations of NaCl (Fig. 7b). Extent of oxidative damage in term of MDA content was significantly lower in transgenic discs than the control plant leaf disc suggesting enhanced salinity tolerance of transgenic lines (Fig. 7c).
Salinity tolerance test by leaf disc method. a Chlorophyll loss from leaf disc of control and transgenic lines floated on different concentration of NaCl (0, 100, 200, 250 mM). b Total chlorophyll content (in microgram per milligram FW) in leaf disc after salinity treatment. c MDA (malondialdehyde) content (in nanomoles of MDA per gram of FW) in leaf disc after NaCl treatment. Values indicated are mean ± SD. (VC: vector control; L2, L4, L7, and L11: transgenic lines). Statistical significant events at 5 % are represented with an asterisk (*)
High salinity and drought-tolerance analysis of transgenic banana lines
Superior stress tolerance of transgenic banana lines observed in leaf discs assay was further confirmed by stress-tolerance analysis of hardened plants (Figs. 8a, 9a). Salinity (250 mM NaCl) and drought stress (Soil gravimetric water content was 0.21 and 0.15 on 7th and 14th day, respectively) was imposed on plants and symptoms appeared in the form of leaf yellowing and wilting. However, the stress symptoms were less prominent in transgenic lines than control suggesting that overexpression of MusaNAC042 could reduce the stress-induced damage (Figs. 8b, 9b). Recovery of transgenic lines was much better than control in case of both salinity and drought stress. Compared to recovery in NaCl-stressed plants, control plants failed to recover and ceased to grow while majority of transgenic lines recovered and resumed normal growth (Fig. 8c). Transgenic lines stressed by drought treatment recovered better than control indicating superior drought tolerance ability of transgenic lines overexpressing MusaNAC042 (Fig. 9c). Biochemical responses of control and transgenic lines were recorded during and after stress treatment in terms of photosynthetic efficiency (Fv/Fm) and content of proline and malondialdehyde (MDA). Transgenic lines displayed better photosynthetic efficiency than control as measured by significantly higher Fv/Fm ratio during imposition of salinity (Fig. 8d) and drought (Fig. 9d). Stress-induced oxidative damage was significantly lower in transgenic lines as shown by lower MDA (marker for membrane damage) content in transgenic lines under salinity (Fig. 8e) and drought stress (Fig. 9e). Level of osmoprotectant proline was higher in transgenic lines than control which further suggested that performance of transgenic lines was better under salinity (Fig. 8f) and drought stress (Fig. 9f). Relative water content of all the transgenic lines was remarkably higher than control plants at the same level of drought stress indicating that transgenic lines could survive better than control under such stress conditions (Fig. 9g).
High salinity tolerance analysis of transgenic banana lines. a Two-month-old control and transgenic lines used for imposing salinity stress. b Control and transgenic lines subjected to 250 mM NaCl after 15 days of stress initiation displaying the salinity induced stress symptoms. c Control and transgenic lines after watering with tap water for one month. d Photosynthetic efficiency (Fv/Fm) of control and transgenic lines at initiation and end of stress treatment. e MDA content (nmols MDA/g FW) of control and transgenic lines at different time period. f Proline content of control and transgenic lines at different time period. Values indicated are mean ± SD. (VC: vector control; L2, L4, L7, and L11: transgenic lines). Statistical significant events at 5 % are represented with an asterisk (*)
Drought tolerance analysis of transgenic banana lines. a Two-month-old control and transgenic lines used for imposing drought. b Control and transgenic lines subjected to drought displaying the drought symptoms. c Control and transgenic lines after watering with tap water for one month. d Photosynthetic efficiency (Fv/Fm) of control and transgenic lines subjected to drought at indicated time points. e MDA content (in nanomole of MDA per gram of FW) of control and transgenic lines at different time period. f Proline content of control and transgenic lines during drought. g Percentage relative water content of transgenic lines and control plants before and after the drought stress treatment. Values indicated are mean ± SD. (VC: vector control; L2, L4, L7, and L11: transgenic lines). Statistical significant events at 5 % are represented with an asterisk (*)
Expression of stress-related genes in transgenic lines
Probable involvement of MusaNAC042 in stress response pathways in banana was monitored by analyzing expression of members of CBF/DREB, LEA, and WRKY family by quantitative RT-PCR. A total of 55 members of CBF/DREB, 30 members of LEA, and 50 members of WRKY gene family were analyzed by quantitative RT-PCR. At least expression of nine CBF/DREBs, five WRKY, and three LEA coding genes was found to be altered in transgenic lines suggesting that MusaNAC042 might be directly or indirectly affecting their expression. The quantum of expression change among these nine CBF/DREBs was as follows: FL659594 (3.3-fold), ES433029 (5.1-fold), FF561178 (3.2-fold), GSMUA_Achr11T15810_001 (14.2-fold), GSMUA_Achr6T28760_001 (2.3-fold), GSMUA_Achr4T26080_001 (7.1-fold), GSMUA_Achr11T03210_001 (9.9-fold), GSUA_Achr8T09110_001 (3.4-fold), and GSMUA_Achr1T15350_001 (3.1-fold). While the expression alteration for 5 WRKY was: GSMUA_Achr5G04870_001 (2.9-fold), GSMUA_Achr10G23420_001 (2.5-fold), GSMUA_Achr6G13630_001 (3.9-fold), GSMUA_Achr3G05670_001 (2-fold), and GSMUA_AchrUn_randomG17570_001 (3.1-fold). Fold change in transcript level of three LEA was: GSMUA_Achr11T05130_001 (4.6-fold), GSMUA_Achr10T21680_001 (7.4-fold), and GSMUA_Achr5T16860_001 (2-fold) (Fig. 10).
Transactivation activity of MusaNAC042. Quantitative RT-PCR of stress-related genes in transgenic banana overexpressing MusaNAC042. Change in expression of at least 9 CBF/DREB, 5 WRKY, and 3 LEA coding genes was observed in transgenic banana plants. Corresponding banana genome database locus identifier or NCBI accession number is indicated on top of graph. Values indicated are mean ± SD). Statistical significant events at 5 % are represented with an asterisk (*)
Discussion
In this report, we have characterized NAC042 transcription factor from banana and investigated its involvement in salinity and drought stress responses. Only few reports (Wu et al. 2012; Shahnejat-Bushehri et al. 2012; Saga et al. 2012) are available on functions of NAC042/JUB1 suggesting necessity of investigations into functions of this important NAC protein. Expression analysis and cloning of MusaNAC042 was carried out in Musa cv. Karibale Monthan (ABB genome) while transformation for generation of transgenic banana lines was carried out with Musa cv. Rasthali (AAB genome). Banana cultivars with more copies of B-genome (cultivars with ABB genome) have been documented to be more robust towards tolerance of stress conditions compared to banana cultivars with single B-genome or having only A-genome, i.e., cultivars with AAB or AAA genome (Placide et al. 2012; Robinson and Sauco 2010). Recently, the higher stress tolerance ability of banana cultivars with B-genome than cultivars with A-genome has been experimentally tested using molecular techniques (Vanhove et al. 2012). MusaNAC042 is induced by abiotic stress conditions like drought and high salinity. Abiotic stress conditions lead to generation of harmful reactive oxygen species (ROS) which also acts as signaling molecules for triggering stress responses (Choudhury et al. 2013). Arabidopsis JUB1 is induced by many abiotic stress conditions like high salinity, stress induced by methyl viologen, hydrogen peroxide (H2O2), ozone, Alternaria alternata fungal toxin, and other conditions leading to generation of H2O2, a type of ROS (Wu et al. 2012). Induction of MusaNAC042 by high salinity indicates that Arabidopsis NAC042/JUB1 and MusaNAC042 may be sharing similar function of conferring abiotic stress tolerance.
The data obtained from Q-PCR experiments are generally represented after 2−ΔΔCt, 2−ΔCt, or 2−Ct transformation (Schmittgen and Livak 2008; Livak and Schmittgen 2001). The comparative Ct method (2−ΔΔCt) gives the “fold change” in expression of a particular gene while the 2−ΔCt transform the QPCR readings into “normalized expression” relative to expression of an internal control (Schmittgen and Livak 2008). We have calculated fold change in the expression of genes analyzed in this study by dividing the normalized expression (2−ΔCt) under treatment and control conditions. Expression of Musa EF1α was monitored for normalization of Ct values. One of the earlier reports has evaluated different reference genes in banana suggesting that banana EF1α is one the most optimal reference genes among others for quantitative RT-PCR experiments as it shows minimum variation in different tissues including leaves of greenhouse plants (Podevin et al. 2012). The suitability of EF1α as a reference gene has been well-documented in many other plant systems supporting the utilization of EF1α as a reference gene in banana (Ray and Johnson 2014; Silveira et al. 2009; Galeano et al. 2014; Reid et al. 2006).
Functional analysis of MusaNAC042 was carried out by generating transgenic banana lines overexpressing MusaNAC042. Four transgenic lines were generated by transformation of embryogenic cells of banana cultivar Rasthali and confirmed by PCR and Southern blot. Genomic DNA was restricted with NcoI for Southern blot analysis as NcoI digest the T-DNA of pCAMBIA1301-MusaNAC042 only once, resulting in a direct interrelationship between number of bands observed during autoradiography and copies of T-DNA transferred to genome of transgenic lines. Moreover, difference in band size in autoradiograph suggested that each transgenic line has originated by an independent transformation event. Data regarding the overexpression of MusaNAC042 could not be correlated with copies of T-DNA in transgenic lines indicating that different copies of T-DNA might have been incorporated into regions of genomic DNA with differences in transcriptional activities. Some of NAC domain proteins are characterized as positive regulators of senescence. Notable among these are ORS1 (Balazadeh et al. 2011), AtNAP (Guo and Gan 2006), and ANAC092 (ORE1) (Balazadeh et al. 2010). However, many of the NAC domain factors are known to repress the senescence process. Arabidopsis NAC genes ANAC019, ANAC055, and ANAC072 overexpression resulted in superior drought tolerance (Fujita et al. 2004; Tran et al. 2004). Arabidopsis JUB1 delayed senescence in overexpression lines while jub1-1 mutant plants showed precocious senescence indicating that JUB1/ANAC042 can be an important candidate gene for stress responses (Wu et al. 2012). In our studies, transgenic banana plants overexpressing MusaNAC042 were significantly more tolerant to drought and high salinity than control, which is further supported by the induction pattern of MusaNAC042 in response to high salinity and drought. Overexpression of some of the transcription factors like rice NAC6 and DREB1, resulted in growth retardation (Ito et al. 2006; Nakashima et al. 2007) which can lead to reduction of productivity. However, such retardation was not observed in case of MusaNAC042 indicating that MusaNAC042 can be useful for increasing salinity and drought tolerance of banana. Abiotic stress conditions generally lead to proline accumulation (Surender Reddy et al. 2015) which is known to act as molecular chaperon preventing protein denaturation along with maintaining osmotic balance (Székely et al. 2007). Similarly in our study, transgenic banana lines overexpressing MusaNAC042 contained higher proline level after salinity and drought stress indicating that elevated proline can be one of the possibilities for enhanced stress tolerance of transgenic lines. Transgenic lines retain higher relative water content than control plants at the same level of drought stress suggesting better drought tolerance in transgenic lines. Relative water content has been commonly employed to measure the water content in plants and is a useful parameter to indicate the turgidity of plants under water scarcity conditions (Smart and Bingham 1974). Possible molecular mechanisms by which MusaNAC042 overexpression confers elevated salinity and drought tolerance was studied by probing expression of multiple stress-related genes. Elevated expression levels were detected for at least nine CBF/DREB (C-repeat-binding/dehydration-responsive element-binding), five WRKY, and three LEA (Late Embryogenesis Abundant) coding genes. CBF/DREB transcription factors are important class of transcription factors involved in stress responses and regulate expression of multiple stress-responsive genes (Akhtar et al. 2012). WRKY transcription factors are one of the major regulators of disease responses, abiotic stress responses, development, and senescence in plants (Banerjee and Roychoudhury 2015). LEA proteins accumulate during seed maturation for acquisition of desiccation tolerance in embryo and during dehydration in vegetative organs suggesting their protective functions during abiotic stress conditions mainly drought stress (Rorat 2006).
In conclusion, it is revealed that MusaNAC042 is a stress-responsive NAC transcription factor as it is rapidly induced under high salinity and drought stress. MusaNAC042 is a nuclear localized protein was demonstrated by transiently overexpressing MusaNAC042::GFP in banana embryogenic cells. Further, the study dealt with morphological and physiological features of MusaNAC042 overexpression in banana plants. Role of MusaNAC042 in increasing salinity and drought tolerance was demonstrated in transgenic banana plants and results were supported with elevated proline and reduced MDA content in transgenic lines. Expression pattern of abiotic stress responsive genes suggests potential molecular mechanism of MusaNAC042 function as well as its transactivation activity. Present study will expand our knowledge about the role of NAC transcription factors in understanding the mechanisms involved in abiotic stress responses in banana.
Abbreviations
- GUS:
-
Beta-glucuronidase
- cDNA:
-
Complementary DNA
- hpt:
-
Hygromycin phosphotransferase
- SSC:
-
Saline sodium citrate
- TAE:
-
Tris-acetate buffer
- ABA:
-
Abscisic Acid
- RT-PCR:
-
Reverse transcription PCR
- NAA:
-
α-Naphthaleneacetic acid
- PCR:
-
Polymerase chain reaction
- MDA:
-
Malondialdehyde
- GFP:
-
Green fluorescent protein
References
Akhtar M, Jaiswal A, Taj G, Jaiswal JP, Qureshi MI, Singh NK (2012) DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J Genet 91:385–395
Al Abdallat AM, Ayad JY, Abu Elenein JM, Al Ajlouni Z, Harwood WA (2014) Overexpression of the transcription factor HvSNAC1 improves drought tolerance in barley (Hordeum vulgare L.). Mol Breed 33:401–414
Arnon DI (1949) Copper enzymes in isolated chloroplasts: poly-phenol oxidase in Beta vulgaris. Plant Physiol 24:1–15
Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor M-I, Kohler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62:250–264
Balazadeh S, Kwasniewski M, Caldana C, Mehrnia M, Zanor MI, Xue GP, Mueller-Roeber B (2011) ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol Plant 4:346–360
Baloglu MC, Oz MT, Oktem HA, Yucel M (2012) Expression analysis of TaNAC69-1 and TtNAMB-2, wheat NAC family transcription factor genes under abiotic stress conditions in durum wheat (Triticum turgidum). Plant Mol Biol Rep 30:1246–1252
Banerjee A, Roychoudhury A (2015) WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci World J 2015:807560
Bowler C, Montagu MV, Inze D (1992) Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol 43:83–116
Cenci A, Guignon V, Roux N, Rouard M (2014) Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Mol Biol 85:63–80
Choudhury S, Panda P, Sahoo L, Panda SK (2013) Reactive oxygen species signaling in plants under abiotic stress. Plant Signal Behav 8:e23681
Cote FX, Domergue R, Monmarson S, Schwendiman J, Teisson C, Escalant JV (1996) Embryogenic cell suspensions from the male flower of Musa AAA cv. Grand Nain Physiol Plant 97:285–290
Fang Y, Liao K, Du H, Xu Y, Song H, Li X, Xiong L (2015) A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J Exp Bot. doi:10.1093/jxb/erv386
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39:863–876
Galeano E, Vasconcelos TS, Ramiro DA, De Martin VF, Carrer H (2014) Identification and validation of quantitative real-time reverse transcription PCR reference genes for gene expression analysis in teak (Tectona grandis L.f.). BMC Res Notes 7:464
Ganapathi TR, Higgs NS, Balint Kurti PJ, Arntzen CJ, May GD, Van Eck JM (2001) Agrobacterium mediated transformation of embryogenic cell suspensions of the banana cultivar Rasthali (AAB). Plant Cell Rep 20:157–162
Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, Alper SL, Fink GR (2001) Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci U S A 98:11444–11449
Greenway H, Munns R (1980) Mechanisms of salt tolerance in nonhalophytes. Annu Rev Plant Physiol 31:149–190
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 68:302–331
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198
Hoekstra FA, Golovina EA, Buitink J (2001) Mechanisms of plant desiccation tolerance. Trends Plant Sci 6:431–438
Hood EE, Gelvin SB, Melchers LS, Hoekama A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2:208–218
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A 103:12987–12992
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta T)) method. Methods 25:402–408
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Negi S, Tak H, Ganapathi TR (2015a) Cloning and functional characterization of MusaVND1 using transgenic banana plants. Transgenic Res 24:571–585
Negi S, Tak H, Ganapathi TR (2015b) Functional characterization of secondary wall deposition regulating transcription factors MusaVND2 and MusaVND3 in transgenic banana plants. Protoplasma. doi:10.1007/s00709-015-0822-5
Negi S, Tak H, Ganapathi TR (2015c) Expression analysis of MusaNAC68 transcription factor and its functional analysis by overexpression in transgenic banana plants. Plant Cell Tiss Organ Cult Doi. doi:10.1007/s11240-015-0929-6
Niu X, Bressan RA, Hasegawa PM, Pardo JM (1995) Ion homeostasis in NaCl stress environments. Plant Physiol 109:735–742
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Placide R, Christian CS, Rony S (2012) Development of in vitro technique to screen for drought tolerant banana varieties by sorbitol induced osmotic stress. Afr J Plant Sci 6:416–425
Podevin N, Krauss A, Henry I, Swennen R, Remy S (2012) Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol Breed 30:1237–1252
Ravi I, Uma S, Vaganan MM, Mustaffa MM (2013) Phenotyping bananas for drought resistance. Front Physiol. 4:9
Ray DL, Johnson JC (2014) Validation of reference genes for gene expression analysis in olive (Olea europaea) mesocarp tissue by quantitative real-time RT-PCR. BMC Res Notes 7:304
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27
Rhodes D, Nadolska-Orczyk A (2001) Plant stress physiology. In Encyclopaedia of Life Sciences (eLS). doi:10.1038/npg.els.0001297
Robinson JC, Sauco VG (2010) Bananas and plantains, 2nd edn. CABI, Wallingford
Rorat T (2006) Plant dehydrins—tissue location, structure and function. Cell Mol Biol Lett 11:536–556
Saga H, Ogawa T, Kai K, Suzuki H, Ogata Y, Sakurai N, Shibata D, Ohta D (2012) Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol Plant Microbe Interact 25:684–696
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108
Shahnejat-Bushehri S, Mueller-Roeber B, Balazadeh S (2012) Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal Behav 7:1518–1521
Shan W, Kuang JF, Chen L, Xie H, Peng HH, Xiao YY, Li XP, Chen WX, He QG, Chen JY, Lu WJ (2012) Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J Exp Bot 63:5171–5187
Shan W, Kuang JF, Lu WJ, Chen JY (2014) Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ 37:2116–2127
Shan W, Chen JY, Kuang JF, Lu WJ (2015) Banana fruit NAC transcription factor MaNAC5 cooperates with MaWRKYs to enhance the expression of pathogenesis-related genes against Colletotrichum musae. Mol Plant Pathol. doi:10.1111/mpp.12281
Silveira ED, Alves-Ferreira M, Guimarães LA, da Silva FR, Carneiro VT (2009) Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol 9:84
Smart RE, Bingham GE (1974) Rapid estimates of relative water content. Plant Physiol 53:258–260
Surendar KK, Devi DD, Ravi I, Krishnakumar S, Kumar SR, Velayudham K (2013) Water stress in banana—a review. Bull Env Pharmacol Life Sci 2:01–18
Surender Reddy P, Jogeswar G, Rasineni GK, Maheswari M, Reddy AR, Varshney RK, Kavi Kishor PB (2015) Proline over-accumulation alleviates salt stress and protects photosynthetic and antioxidant enzyme activities in transgenic sorghum [Sorghum bicolor (L.) Moench]. Plant Physiol Biochem 94:104–113
Székely G, Abrahám E, Cséplo A, Rigó G, Zsigmond L, Csiszár J, Ayaydin F, Strizhov N, Jásik J, Schmelzer E, Koncz C, Szabados L (2007) Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 53:11–28
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress inducible NAC transcription factors that bind to a drought responsive cis-element in the early responsive to dehydration stress. Plant Cell 16:2481–2498
van Asten PJA, Fermont AM, Taulya G (2011) Drought is a major yield loss factor for rainfed East African highland banana. Agric Water Manage 98:541–552
Vanhove AC, Vermaelen W, Panis B, Swennen R, Carpentier SC (2012) Screening the banana biodiversity for drought tolerance: can an in vitro growth model and proteomics be used as a tool to discover tolerant varieties and understand homeostasis. Front Plant Sci 3:176
Wu A, Allu AD, Garapati P, Siddiqui H, Dortay H, Zanor MI, Asensi-Fabado MA, Munné-Bosch S, Antonio C, Tohge T, Fernie AR, Kaufmann K, Xue GP, Mueller-Roeber B, Balazadeh S (2012) JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24:482–506
Xia N, Zhang G, Liu X, Deng L, Cai G et al (2010) Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol Biol Rep 37:3703–3712
Xie Q, Sanz-Burgos AP, Guo H, García JA, Gutiérrez C (1999) GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol Biol 39:647–656
Xu Y, Hu W, Liu J, Zhang J, Jia C, Miao H, Xu B, Jin Z (2014) A banana aquaporin gene, MaPIP1;1, is involved in tolerance to drought and salt stresses. BMC Plant Biol 14:59
Xue G, Way H, Richardson T, Drenth J, Joyce P et al (2011) Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 4:697–712
Yeo AR (1998) Molecular biology of salt tolerance in the context of whole plant physiology. J Exp Bot 49:915–929
Acknowledgment
Authors thank Head, Nuclear Agriculture and Biotechnology Division, BARC, for his motivation and support. The study was funded by Department of Atomic Energy, India. Authors are thankful to Dr. P. Suprasanna for critically going through the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Handling Editor: Néstor Carrillo
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 182 kb)
Rights and permissions
About this article
Cite this article
Tak, H., Negi, S. & Ganapathi, T.R. Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 254, 803–816 (2017). https://doi.org/10.1007/s00709-016-0991-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-016-0991-x