Abstract
Localization of Calliphora erythrocephala chromosome 6 in a 3D nuclear space at different stages of nurse cell chromatin polytenization was analyzed by fluorescence in situ hybridization and 3D microscopy. The obtained results suggest a large-scale chromatin relocation in the C. erythrocephala nurse cell nuclei, which is accompanied by a change in the chromosome territory of chromosome 6 associated with the change in expression activity of the nucleus and formation of reticular chromatin structure. It was revealed that the relocation of chromosome 6 (nucleolus organizer chromosome) is accompanied by fragmentation of the single large nucleolus into micronucleoli, which are spread over the entire nuclear space being associated with their nucleolar organizer regions. Presumably, the chromosome 6 material during transition to a highly polytenized structure is redistributed in the nucleus so that the inactive pericentromeric regions are displaced to the nuclear periphery, while the chromosome regions carrying rDNA sequences loop out beyond the chromosome territory. Being dispersed over the entire nuclear space, rDNA sequences are likely to be amplified, thereby providing numerous small signals from the chromosome 6-specific DNA probe. Micronucleoli are formed around the actively transcribed nucleolar organizer regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Spatial organization of chromatin in the nucleus is one of the key problems in modern cell biology. The data on genomic DNA sequences of many organisms obtained so far have made it clear that these sequence data are insufficient for a complete understanding of how the genetic machinery works. Currently, the arrangement of chromosomes in the nucleus is regarded as a key player in its functioning. The chromosome arrangement in the nucleus has been studied in diploid interphase nuclei of many organisms (Lichter et al. 1990; Cremer et al. 1995; Visser et al. 2000; Rouquette et al. 2009; Cremer and Cremer 2010).
The accumulated data suggest that individual chromosomes occupy in the interphase nuclei more or less distinctly demarcated regions, the so-called territories (Zorn et al. 1979; Cremer and Cremer 2006; van Koningsbruggen et al. 2010). However, the principles underlying the functioning of the nucleus and chromosome arrangement in the nuclear space are still vague.
The study of chromosome spatial organization in the nuclei of Calliphora erythrocephala nurse cells has several advantages for clarifying the functioning of the genetic material in the nucleus. First and foremost, this is connected with polytenization of the C. erythrocephala nurse cell nuclei, during which the copy number of the chromosomes constituting the main set multiply increases without cell division. Second, the nurse cell chromatin in the course of follicle development undergoes several morphofunctional changes, which are likely to be associated with the changes in transcriptional profile. This makes it possible to study the dependence of the nuclear architecture on the changes in transcriptional profile.
The paired meroistic ovaries of the insects, such as C. erythrocephala, consist of the egg tubes, comprising germarium and vitellarium. In the germarium, mitotic divisions lead to formation of cysts interconnected via cytoplasmic bridges. After the fourth mitosis, 15 of the 16 cystocytes differentiate into nurse cells, while the remaining cell becomes an oocyte (Aizenshtadt et al. 1977). In C. erythrocephala, the nurse cell chromatin is polytenized before a young follicle separates from the germarium. The nurse cell chromatin is formed of thin fibers spread over the nuclear space (Fig. 1a). The polytenization of nurse cell chromatin is explainable by the need to supply the developing oocyte with a large amount of ribosomal RNA and proteins. The newly formed follicle moves to the vitellarium, where the oocyte grows and the egg is formed. After the oocyte has separated from the germarium, the nurse cell chromatin has a primary reticular structure, and chromatids are spread all over the nucleus (Fig. 1b). Further polytenization leads to formation of the nuclei with classical polytene chromosomes (Fig. 1c). Then the contact between chromatids partially disappears, polytene chromosomes shorten, and blob-like chromosomes are formed (Fig. 1d, e). The chromatids continue to compact considerably and completely separate from one another, now resembling mitotic chromosomes (Fig. 1f). This is followed by a gradual decompaction of chromatids, which fill the entire nuclear space; the nucleus restores its reticular structure but at a higher level of polyteny (Fig. 1g; Anan’ina et al. 2007).
Morphological transformations of the C. erythrocephala nurse cell chromatin during polytenization: a separation of a young follicle from the germarium, b germarium and a young follicle, c polytene chromosomes of nurse cells, d blob-like chromosomes, e decompaction of blob-like chromosomes, f a fragment of the nurse cell nucleus with chromatids resembling mitotic chromosomes, g an oocyte and 15 nurse cells at the stage of secondary reticular nuclei. Bars represent 10 μm (Anan’ina et al. 2007)
The C. erythrocephala karyotype comprises six pairs of chromosomes. Ribbert (1979) constructed a cytological map of the C. erythrocephala nurse cell polytene chromosomes and proposed the following nomenclature: chromosomes 1–5 are large meta- and acrocentric chromosomes and chromosome 6, X chromosome, is a metacentric (the smallest chromosome of the set). The stage of separate polytene chromosomes makes it possible to solve a complex problem of chromosome identification in the nuclear space.
We have earlier demonstrated that the mutual chromosome arrangement in the C. erythrocephala nurse cell nuclei follows certain rules. The nurse cell chromosomes do not from a chromocenter but are rather dispersed over the nuclear space. Chromosome 6 is always connected with chromosome 2 via a thin cord. Chromosome 4 is located near chromosomes 3 and 5, while chromosome 5, near chromosome 1. Thus, the following associated chromosome arrangement is frequently observable: 6–2, 4–3, and 5–1 (Stegniy et al. 1999).
These results on a spatial organization of the C. erythrocephala nurse cell nuclei have been obtained using squash preparations of the nurse cells at the stage of separate polytene chromosomes; however, the nucleus is 3D and the chromosome arrangement should be examined in a 3D nuclear space. Would the definite patterns in chromosome arrangement observed in the nurse cell nuclei with polytene chromosomes be retained when the chromatin passes into a reticular state? Once each chromosome occupies a certain space in the nucleus, what takes place there at the stages other than the stage of separate polytene chromosomes?
To answer these questions, we set a challenge to analyze the localization of C. erythrocephala chromosome 6 in a 3D nuclear space at different stages of nurse cell chromatin polytenization. It is currently feasible owing to the combination of fluorescence in situ hybridization (FISH) and 3D microscopy (Cremer et al. 2000, 2008).
Chromosome 6 was chosen purposefully. Boyes and Shewell (1975) have demonstrated that the sex chromosome (chromosome 6) is a nucleolus organizer chromosome, i.e., it contains rRNA genes, the transcription of which is associated with formation of the nucleolus. Consequently, studying the changes in the location of chromosome 6 and the nucleolus in the nuclear space during polytenization, it is also possible to clarify how the spatial organization of the nucleolus organizer chromosome in the nucleus influences the formation and distribution of the nucleolus, i.e., how this influences the activity of rRNA genes during the chromatin polytenization.
In this work, we have analyzed the localization of chromosome 6 in a 3D nuclear space at different stages of polytenization and morphofunctional transformations of the C. erythrocephala nurse cell chromatin as well as the formation of the nucleolus during chromatin polytenization.
Materials and methods
The object of the study was C. erythrocephala Mg. (Diptera: Calliphoridae) from wildlife populations of the city of Tomsk (Russia). Imagoes were bred under standard conditions (Vinogradova 1984) at a temperature of 20–22°C. The nurse cells of the ovaries from the imago 2 to 5 days after emerging from the puparium were examined. Both freshly isolated C. erythrocephala ovary nurse cells and the samples fixed in Carnoy’s fluid (mixture of 100% ethanol with glacial acetic acid at a ratio of 3:1) were used.
Producing chromosome 6-specific DNA probe
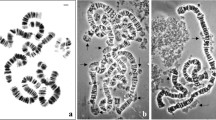
The chromosome 6 DNA was isolated by sampling the material from an air-dry preparation of C. erythrocephala nurse cell polytene chromosomes using a microhandler. A 2D FISH of the DNA probe with polytene chromosomes demonstrated an intensive signal on the arms of chromosome 6; however, the signal was absent in the centromeric densely compacted region. The absence of the signal on the other chromosomes suggests that the probe is chromosome-specific and appropriate for unambiguous identification of chromosome 6 (Fig. 2; Vasserlauf et al. 2003; Kokhanenko et al. 2010).
3D fluorescence in situ hybridization (3D FISH) of chromosome 6-specific DNA probe with C. erythrocephala nurse cell chromatin
The protocol (Bantignies et al. 2007) was used as a basic. Some modifications were introduced to adjust it to 3D FISH using C. erythrocephala ovary nurse cells.
All procedures with the tissue were performed in 1.5-ml centrifuge tubes using either a rotation stirrer with a medium rotation amplitude or a thermostirrer with different rotation amplitudes. At all the stages, the volume of solution in tubes should be twofold larger than the sample volume. When necessary, a gentle centrifugation (1 min at 500 rpm) was performed previously to each change of solution.
Tissue fixation
The ovaries were isolated in 1× PBS (standard) and fixed with 4% paraformaldehyde in PBT (0.1% Tween 20 in 1× PBS) for 20 min at a room temperature.
Tissue prehybridization
The tissue was incubated in the RNase A solution (100–200 μg/ml) in PBT for 2 h in a rotation stirrer at a room temperature, transferred into PBS-Tr (0.3% Triton X-100 in 1× PBS), and incubated for 1 h at a room temperature. The sample was transferred to hybridization mixture (50% formamide, 10% dextran sulfate, and 1% Tween 20 in 2× SSC; pH 7.0); before this, the sample was successively treated with the hybridization mixture–PBS–Tr at ratios of 1: 5, 1: 1, and 5: 1 (20 min with each).
Hybridization
DNA probe was produced from microdissected DNA of chromosome 6 according to a standard protocol (Rubtsov et al. 1999). The sample in a centrifuge tube was supplemented with the DNA probe (dissolved in hybridization mixture) and jointly denatured at 80°C in a shaker (450 rpm) for 15 min. The tissue hybridization was performed in a shaker (450 rpm) for 14–17 h.
Posthybridization washing and preparation of slides
The sample was washed in 7 washing buffers (20 min in each), namely, twice in buffer 1 (50% formamide Sigma, 2× SSC, and 0.3% CHAPS) and once in buffer 2 (40% formamide Sigma, 2× SSC, and 0.3% CHAPS) and 3 (30% formamide Sigma and 70% PBT) and 4 (20% formamide Sigma and 80% PBT); all at 37°C in a shaker at 800 rpm; and once in buffer 5 (10% formamide Sigma and 90% PBT), 6 (100% PBT), and 7 (100% PBS-Tr) in a rotation stirrer at a room temperature. The centrifuge tube was supplemented with a drop of DAPI-Vectashield and incubated for 1 h in a rotation stirrer at a room temperature. The sample was placed onto a glass slide and covered with cover glass in a drop of DAPI-Vectashield.
Producing 3D C. erythrocephala nurse cell specimens stained with silver nitrate
All procedures were performed in 1.5-ml centrifuge tubes. Freshly isolated nurse cell sample was fixed with 4% paraformaldehyde for 30 min at a room temperature, supplemented with 3 μl of Triton X-100, and incubated for 30 min at a room temperature at 450 rpm. The sample was washed twice with water for 15 min each time at 600 rpm at a room temperature, supplemented with 50% silver nitrate solution, and incubated overnight at 37°C at 600 rpm. Then the sample was again washed twice with water for 15 min each time at 600 rpm at a room temperature and placed onto a glass slide in a drop of water. Follicles were separated with a microscopic needle. To avoid deformation of the 3D sample by a cover glass, the sample was placed into a chamber formed of a cover glass glued to a glass slide with a biadhesive tape and filled with glycerin.
Studying the ultrastructure of C. erythrocephala nurse cell nuclei
Ultrathin sections of the C. erythrocephala nurse cells were made as described by Wickly et al. (1975). The ultrastructure was examined by transmission electron microscopy (Karupu 1984).
Microscopic analysis
-
(1)
The 3D FISH data were analyzed and processed using an LSM 510 META (Carl Zeiss) and an Axio Imager Z1 microscope equipped with an ApoTome slider module (Carl Zeiss). An AxioCam MRm (Carl Zeiss) camera was used for microphotography. The images were analyzed using the AxioVision rel. 4.7 software.
-
(2)
Nurse cell specimens stained with silver nitrate were examined using an Axio Imager Z1 (Carl Zeiss) microscope in transmission light.
-
(3)
A JEM-100 CXII JEOL electron microscope was used for electron microscopy examination of ultrathin sections of nurse cell.
Results
Changes in the spatial localization of chromosome 6 in the C. erythrocephala nurse cell nuclei at different stages of polytenization
FISH was used to determine the position of chromosome 6-specific DNA probe in the nuclear space of C. erythrocephala at different stages of polytenization. At the early stages of oogenesis, 16 cell cysts are formed in the C. erythrocephala germarium; then these cells differentiate into 1 oocyte and 15 nurse cells. The nurse cell chromatin is arranged in thin fibers; light microscopy does not detect any distinct structure of these fibers. Separate chromosomes are not yet formed (Fig. 3a1). Analysis of nurse cell optical sections and their graphical reconstruction have demonstrated that chromosome 6 has a compact structure and occupies the central position in the nucleus (Fig. 3a1, a2).
Central position of chromosome 6 in the C. erythrocephala nurse cell nuclear space at different stages of chromatin polytenization: a stage of germarium cystocytes, b stage of the nuclei with primary reticular chromatin structure, c the nucleus with separate polytene chromosomes, d stage with compact blob-like chromosomes; a 1–d 1 serial optical sections of the whole nucleus demonstrating a central position of chromosome 6 in the nucleus; a 2, b 2, d 2 graphical reconstruction of optical sections of a half nucleus in the nuclear space. c 3 Graphical reconstruction of optical sections of the whole nucleus. Chromatin is colored red (DAPI) and DNA probe, yellow. Bars represent 5 μm
Endoreduplication of the nurse cell chromatin starts in the formed follicle. In the nuclei with primary reticular chromatin structure, chromosome 6 still occupies a central position in the nucleus; however, the morphology of chromosome territory changes. Two isolated labeled areas are detectable in the center; they are immediately adjacent, which suggests an active hybridization of the DNA probe to the chromosome 6 arms and the absence of the label in densely compacted pericentromeric heterochromatin regions of chromosome 6 (Fig. 3b1, b2). At this stage, the region unstained with DAPI is detectable around chromosome 6 (Fig. 3b1). Since chromosome 6 is a nucleolus organizer chromosome, it is likely that formation of the nucleolus associated with chromosome 6 starts at this stage, displacing the material of the remaining chromosomes to the nuclear periphery.
The continuing endoreduplication leads to formation of polytene chromosomes; their further compaction gives blob-like chromosomes. At these stages, both chromosome 6 and the region unstained with DAPI (putatively the region occupied by the nucleolus associated with this chromosome) are still located in the central part of the nucleus (Fig. 3c1, c2, d1, d2).
Considerable changes in the intranuclear localization of chromosome 6 commence when the blob-like chromosomes begin to separate into chromatids and the chromatids begin their decompaction (Fig. 4a1). This is accompanied by a gradual relocation of chromosome 6 to the nuclear periphery (Fig. 4a2).
The changes in spatial organization of chromosome 6 associated with decompaction of chromatids and restoration of reticular structure of the nucleus during polytenization: a 1 serial optical sections of the nucleus demonstrating the displacement of chromosome 6 material to the nuclear periphery at the stage of decompaction of blob-like chromosomes, a 2 graphical reconstruction of the whole nucleus at the stage of decompaction of blob-like chromosomes, b 1 serial optical sections of the whole nucleus at the stage of secondary reticular structure, b2 distribution of chromosome 6 material over the space of highly polytenized reticular nucleus, and b3 graphical reconstruction of the nucleus with reticular chromatin structure. Chromatin is colored red (DAPI) and DNA probe, yellow. Bars represent 5 μm
After completion of the chromatid decompaction, chromatin uniformly fills the entire nuclear space, and the nucleus restores its reticular structure but at a higher level of polyteny. At this stage, a dense compact region of chromosome 6 is detectable in the nuclear periphery (Fig. 4b1, b3). In addition, numerous small signals of chromosome 6-specific DNA probe located at a distance from the compact region are observed in the nuclear space (Fig. 4b1, b2). The area unstained with DAPI becomes undetectable in the central part of the nucleus. Instead, numerous small areas free of chromatin, frequently containing small signals of the DNA probe, are observed in the nuclear space (Fig. 4b1).
Multiple nucleoli in the highly polytene nurse cell nuclei with reticular chromatin structure
Since chromosome 6 is a nucleolus organizer chromosome, we have analyzed the location of the nucleoli in a 3D space of the nurse cell nuclei. For this purpose, we used silver nitrate staining of 3D C. erythrocephala nurse cell specimens. Silver nitrate gives dark color to the protein component of the nucleoli encompassing the nucleolar organizer, whereas the chromatin remains considerably lighter. It has been demonstrated that a large number of micronucleoli are spread over the nuclear space in the nuclei with secondary reticular structure at the final stage of chromatin polytenization, when chromatids uniformly fill the entire nuclear space. Optical sections of the whole nucleus illustratively demonstrate this distribution (Fig. 5). Silver nitrate staining detects the nucleolar proteins associated with transcriptionally active part of the DNA of nucleolar organizer regions. They are absent in the regions of untranscribed spacers (Rubtsov 2006).
The small signals of the DNA probe detected by 3D FISH (Fig. 4b), spread over the nuclei with reticular structure, are likely to represent expressed nucleolar organizer regions of chromosome 6, while the small micronucleoli in the nuclear space are their protein component.
Electron microscopic examination of the C. erythrocephala nurse cell nuclei has demonstrated the presence of numerous micronucleoli in the nurse cell nuclei with secondary reticular chromatin structure. The micronucleoli in an ultrathin section are seen as the regions with a higher electron density as compared with the chromatin. These micronucleoli are spread over the entire nuclear space (Fig. 6a). At the final stage of polytenization, the micronucleoli of nurse cell nuclei contain one to four fibrillar centers. Chromatin cords in the nucleolar organizer regions of chromosome 6, contacting the protein components of the nucleolus, become visible at a magnification of ×10,000 (Fig. 6b).
Electron microscopy examination of the structure of C. erythrocephala nurse cell nuclei: a distribution of the micronucleoli in the nucleus with secondary reticular chromatin structure and b structure of the micronucleoli in highly polytenized nurse cell reticular nuclei. Ch chromatin, MN micronucleoli, FC fibrillar center, DFC dense fibrillar chromatin, GC granular component; bars represent 10 μm in a, 1 μm in b
At the early stages of chromatin polytenization, namely, at the stage with primary reticular chromatin structure, a single large nucleolus associated with chromosome 6 is formed in the center of the nucleus (Fig. 3b). Such location and structure of the nucleolus are retained at the stages when chromosome 6 occupies a central position, i.e., at the stages of polytene chromosomes and compact blob-like chromosomes. At the stage of decompaction of blob-like chromosomes and restoration of reticular state of the nucleus, the relocation of chromosome 6 is accompanied by fragmentation of the single large nucleolus into micronucleoli, which are spread over the entire nuclear space being associated with their nucleolar organizer regions.
Discussion
The experimental data accumulated so far demonstrate that the chromosomes enriched for genes and their actively transcribed regions are located closer to the central part of the nucleus in the tissue cells with spherical nuclei. The regions of chromosome territories that are poor in genes as well as the chromosome centromeric regions tend to be localized to the periphery of interphase nuclei near the nuclear membrane. Such chromosome arrangement is established at the early stage of the cell cycle and is retained during its entire course (Croft et al. 1999; Cremer and Cremer 2001; Parada and Misteli 2002; Neusser et al. 2007; Koehler et al. 2009). In addition, the chromatin with DNA replicating in the early S phase of the cell cycle is predominantly localized to the central part of the nuclei, while the late replicating chromatin is associated with the nuclear periphery (Habermann et al. 2001; Alexandrova et al. 2003; Mayr et al. 2003; Postberg et al. 2005; Cremer and Cremer 2010).
On the other hand, the chromosome locations in the nuclear space correlate with their sizes. The smallest chromosomes of the set are mainly localized to the center of the nucleus, while medium-sized and large chromosomes occupy medium and peripheral positions in the nucleus. Presumably, the mechanism providing for such pattern is determined by that the order established during anaphase chromosome movement is preserved in the interphase (Habermann et al. 2001; Stadler et al. 2004; Bolzer et al. 2005; Neusser et al. 2007).
The C. erythrocephala chromosome 6 is the smallest of the set and, in addition, it is a nucleolus organizer chromosome (Boyes and Shewell 1975), i.e., it is both transcriptionally active and enriched for rRNA genes. A central position of chromosome 6 material in the C. erythrocephala nurse cell nucleus at the stages of primary reticular nuclei, separated polytene chromosomes, and compact blob-like chromosomes is explainable by both a small size and a high transcription activity of chromosome 6 DNA at these stages.
The relocation of chromosome region taking place during chromatin polytenization and appearance of the signals at a considerable distance from it is likely to be connected with a functional rearrangement of the cells. Recent studies have demonstrated that expression activation entails intricate structural changes in the nucleus (Swedlow and Lamond 2001; Matarazzo 2007; Duan et al. 2010; Németh et al. 2010). This leads to the changes in relative positions of the adjacent chromosome regions. Presumably, this is the main reason of the changes in chromatin structure during the interphase, which leads to relocations of the chromosome territories in the nuclear space.
It is possible that the inactive regions of chromosome 6 (presumably, the dense pericentromeric heterochromatin blocks and the adjacent untranscribed regions) are moved to the periphery at the stage of decompaction of blob-like chromosomes and formation of secondary reticular nuclei due to an increase in the total expression activity of individual chromosomes and, possibly, the overall chromatin. The chromosome 6 regions that continue to actively transcribe are separated within chromatin loops from the centromeric regions moving to periphery, thereby becoming dispersed within the nuclear space. It has been shown that chromosome fibers are able to from giant loops with a length of up to 5 Mbp capable of carrying the genes located there to other chromosome territories (Volpi et al. 2000; Gondor and Ohlsson 2009; Mateos-Langerak et al. 2009; Amano et al. 2009).
Along with this, a single large nucleolus, observed in the nuclei with separate polytene chromosomes and later stages with compact blob-like chromosomes, is fragmented into micronucleoli dispersed all over the nucleus when the chromosomes commence dissociating into chromatids. Ribbert and Bier (1969) has demonstrated using an inbred C. erythrocephala strain (its chromatin is arranged in separate well-structured polytene chromosomes at all the stages of polytenization) that the number of nucleoli in the nucleus increases with the degree of polytenization of nurse cell nuclei, whereas only single nucleolus is observed at the initial stages of polytenization.
Our electron microscopy examination of the micronucleoli in highly polytenized nuclei with reticular chromatin structure has detected a large number of fibrillar centers within the micronucleoli. This demonstrates a high level of transcription activity, since it is known that the morphofunctional characteristics of the nucleoli to a certain degree reflect the total level of transcription activity in the nucleus. It is known that the integrated fibrillar center localized to the inactive nucleoli falls into several smaller fibrillar centers connected with one another via decompacted rDNA regions with an increase in the transcription of ribosomal genes (Zybina 1986; Thiry 1992; Zharskaya and Zatsepina 2007).
Thus, the obtained results suggest a large-scale chromatin relocation in the C. erythrocephala nurse cell nuclei, which is accompanied by a change in the chromosome territory of chromosome 6 associated with the change in expression activity of the nucleus and formation of reticular chromatin structure. Presumably, the chromosome 6 material during transition to a highly polytenized structure is redistributed in the nucleus so that the inactive pericentromeric regions are displaced to the nuclear periphery, while the chromosome regions carrying rDNA sequences loop out beyond the chromosome territory. Being dispersed over the entire nuclear space, rDNA sequences are likely to be amplified, thereby providing numerous small signals from the chromosome 6-specific DNA probe. Micronucleoli are formed around the actively transcribed nucleolar organizer regions.
There are certain general patterns in arrangement of chromosome territories within the nucleus. The disposition of chromosome territories in actively transcribed cells is dynamic, indicating a possible mechanism of gene regulation via an intranuclear positioning of individual chromosome loci. Our results suggest that the principle of spatial organization of the nucleus when genetically inactive chromatin is predominantly localized to the periphery, while the active chromatin is located in the center is true for the nurse cell nuclei. According to our opinion, this suggests the existence of general principles in spatial organization of the nuclei, which are implemented with certain variations in the cells with different transcription statuses.
References
Aizenshtadt TB, Baranov VS, Borovkov AYU (1977) Modern problems in ontogenesis. Nauka, Leningrad
Alexandrova O, Solovei I, Cremer T, David CN (2003) Replication labeling patterns and chromosome territories typical of mammalian nuclei are conserved in the early metazoan Hydra. Chromosoma 112:190–200
Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T (2009) Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell 16:47–57
Anan’ina TV, Kokhanenko AA, Khodzhanov AE, Stegniy VN (2007) Specific features in the structure of the egg tubes in the Calliphora erythrocephala (Mg.) (Diptera: Calliphoridae) ovaries. Vestn Tomsk Gos Univ 297:175–180
Bantignies F, Grimaud CH, Cavalli G (2007) Two-colour fluorescent in situ DNA hybridization on whole mount Drosophila embryos and larval imaginal discs. IOP Publishing epigenesys. http://mescaline.igh.cnrs.fr/EpiGeneSys/www/images/protopdf/p5.pdf. Accessed 24 November 2011
Bolzer A, Kreth G, Solovei I, Köhler D, Saracoglu K, Fauth C, Müller S, Eils R, Cremer C, Speicher MR, Cremer T (2005) Three-dimensional maps of all chromosome positions demonstrate aprobabilistic order in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3(5):e157
Boyes JW, Shewell GE (1975) Cytotaxonomy of Calliphoridae (Diptera). Genetica 45:435–488
Cremer T, Cremer C (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Genetics 2:292–301
Cremer T, Cremer C (2006) Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990’s, to the present. Eur J Histochem 50:223–272
Cremer T, Cremer M (2010) Chromosome territories. Cold Spring Harbor Laboratory Press. doi:10.1101/cshperspect.a003889
Cremer T, Dietzel S, Eils R, Lichter P, Cremer C (1995) Chromosome territories, nuclear matrix filaments and interchromatin channels: a topological view on nuclear architecture and function. Kew Chromosome Conference IV (P.E. Brandham, and M.D. Benett) Royal Botanic Gardens. Kew 63–81
Cremer M, Heintzmann R, Brero A (2000) Chromosome territories of small and large chromosomes are differently distributed in human lymphocyte nuclei. Medgen 12:95
Cremer M, Grasser F, Lanctot C, Muller S, Neusser M et al (2008) Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol 463:205–239
Croft JA, Bridger JM, Boyle S, Perry P, Teague P et al (1999) Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145:1119–1131
Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendur J, Fields S, Blau CA, Noble WS (2010) A three-dimensional model of the yeast genome. Nature 465:363–367
Gondor A, Ohlsson R (2009) Chromosome crosstalk in three dimensions. Nature 461:212–217
Habermann FA, Cremer M, Walter J, Kreth G, von Hase J, Bauer K, Wienberg J, Cremer C, Cremer T, Solovei I (2001) Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res 9:569–584
Karupu VY (1984) Electron microscopy. Vishcha Shkola, Kiev
Koehler D, Zakhartchenko V, Froenicke L, Stone G, Stanyo R, Wolf E, Cremer T, Brero A (2009) Changes of higher order chromatin arrangements during major genom activation in bovine preimplantation embryos. Exp Cell Res 315:2053–2063
Kokhanenko AA, Anan’ina TV, Stegniy VN (2010) Intranuclear dynamics of chromosome 6 in nurse cells of Calliphora erythrocephala Mg. (Diptera: Calliphoridae). Russ J Genet 46(9):1045–1047
Lichter P, Ledbetter SA, Ledbetter DH, Ward DC (1990) Fluorescence in situ hybridization with Alu and L1 polymerase chain reaction probes for rapid characterization of human chromosomes in hybrid cell lines. Proc Natl Acad Sci USA 87(17):6634–6638
Matarazzo MR (2007) Chromosome territory reorganization in a human disease with altered DNA methylation. PNAS 42:16546–16551
Mateos-Langerak J, Bohn M, de Leeuw W, Giromus O, Manders EM, Verschure PJ et al (2009) Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci USA 106:3812–3817
Mayr C, Jasencakova Z, Meister A, Schubert I, Zink D (2003) Comparative analysis of the functional genome architecture of animal and plant cell nuclei. Chromosome Res 11(5):471–484
Németh A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Péterfia B, Solovei I, Cremer T, Dopazo J, Längst G (2010) Initial genomics of the human nucleolus. PLoS Genet. doi:10.1371/journal.pgen.1000889
Neusser M, Schubel V, Koch A, Cremer T, Muller S (2007) Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma 116:307–320
Parada L, Misteli T (2002) Chromosome positioning in the interphase nucleus. Trends Cell Biol 12:425–432
Postberg J, Alexandrova O, Cremer T, Lipps HJ (2005) Exploiting nuclear duality of ciliates to analyse topological requirements for DNA replication and transcription. J Cell Sci 118:3973–3983
Ribbert D (1979) Chromosomes and puffing in experimentally induced polytene chromosomes of Calliphora erythrocephala. Chromosoma (Berl) 74:269–298
Ribbert D, Bier K (1969) Multiple nucleoli and enhanced nucleolar activity in the nurse cells of the insect ovary. Chromosoma (Berl) 27:178–197
Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, Fakan S (2009) Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res 17:801–810
Rubtsov NB (2006) Methods for work with mammalian chromosomes. Novosibirsk Gos. Universitet, Novosibirsk
Rubtsov NB, Alekseenko AA, Belyaeva ES (1999) Microcloning and characteristics of DNA from regions of the centromeric heterochromatin of Drosophila melanogaster polytene chromosomes. Russ J Genet 1:55–61
Stadler S, Schnapp V, Mayer R, Stein S, Cremer C, Bonifer C, Cremer T, Dietzel S (2004) The architecture of chicken chromosome territories changes during differentiation. BMC Cell Biol 5(1):44
Stegniy VN, Vasserlauf IE, Anan’ina TV (1999) Identification, mutual arrangement, and development of primary polytene chromosomes in the Calliphora erythrocephala (Diptera: Calliphoridae) nurse cell nuclei. Genetika 7:912–918
Swedlow JR, Lamond A (2001) Nuclear dynamics: where genes are how they got there. Genome Biol 3:1–7
Thiry M (1992) New data concerning the functional organization of the mammalian cell nucleolus: detection of RNA and rRNA by in situ molecular immunocytochemistry. Nucleic Acids Res 20(23):6195–6200
van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI (2010) High resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 21:3735–3748
Vasserlauf IE, Anan’ina TV, Unger MF, Karamysheva TV, Mel’nikova NN, Rubtsov NB, Stegniĭ VN (2003) Organization and differential staining of the chromosomes of Calliphora erythrocephala (Diptera: Calliphoridae) nurse cell endomitotic nuclei. Genetika 39(9):1193–1202
Vinogradova LB (1984) Bluebottle Blowfly (Calliphora vicina): a model object of ecological and physiological studies. Nauka, Leningrad
Visser AE, Jaunin F, Fakan S, Aten JA (2000) High resolution analysis of interphase chromosome domains. J Cell Sci 113:2585–2593
Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J et al (2000) Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci 113:1565–1576
Wickly B, Polyakov YuV (1975) Electron microscopy for the beginners. Mir, Moscow
Zharskaya OO, Zatsepina OV (2007) The dynamics and mechanisms of nucleolus reorganization in mitosis. Tsitologiya 5:355–369
Zorn C, Cremer C, Cremer T, Zimmer J (1979) Unscheduled DNA synthesis after partial UV irradiation of the cell nucleus. Distribution in interphase and metaphase. Exp Cell Res 124:111–119
Zybina EV (1986) Cytology of the nurse cell. Nauka, Leningrad
Acknowledgments
We thank Dr. A.A. Miller for the helpful in electron microscopical study and Dr. S.I. Baiborodin for the helpful in confocal microscopical analysis. The work was supported by the Russian Foundation for Basic Research (grant no. 10-04-01059a).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Peter Nick
Rights and permissions
About this article
Cite this article
Kokhanenko, A.A., Anan’ina, T.V. & Stegniy, V.N. The changes in chromosome 6 spatial organization during chromatin polytenization in the Calliphora erythrocephala Mg. (Diptera: Calliphoridae) nurse cells. Protoplasma 250, 141–149 (2013). https://doi.org/10.1007/s00709-012-0385-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-012-0385-7