Abstract
Anthocyanidin synthase (ANS) is one of the key enzymes in the biosynthesis of both anthocyanins and proanthocyanidins in grapevine. Although substantial researches have investigated ANS gene expression and regulation at the transcriptional level, little is yet known about protein expression and distribution in grapevine. Here, the expression and tissue and subcellular localization of ANS in different Cabernet sauvignon grapevine tissues were investigated by using the techniques of Western blotting, immunohistochemical localization, immuno-electron microscopy, and confocal microscopy. The results showed that the ANS was expressed in the grape berries, leaves, stems, petioles, and leaf buds. In grape berry skin and flesh, ANS expression is developmental dependent. Immunohistochemical analysis revealed that ANS is primarily distributed in the exocarp, mesocarp, and seed of the fruit; in palisade and spongy tissues of the leaves; in the primary phloem and pith ray in the stems; and in the growth point and leaf primordium of the leaf buds. Furthermore, at the subcellular level, the ANS was mainly localized in the cytoplasm regardless of cell types and some ANS were also found in the nucleus in the mesocarp vascular bundle and leaf bud cells. This research will give further insight for the biosynthesis and regulation of different flavonoid compounds in grapevine.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anthocyanins and proanthocyanidins (PAs) are the two major flavonoids involved in many plant functions. In grapevine, these compounds play important roles against UV irradiation and microbial and fungal infections in vegetative tissues (Chamkha et al. 2003; Harborne and Williams 2000) and help determine the color, astringency, and quality of grape berries and wine (Bogs et al. 2005; Chamkha et al. 2003). Furthermore, they can also act as antioxidants with beneficial effects for human health including protection against free radical-mediated injury and cardiovascular disease (Halliwell et al. 2005). So, the biosynthesis and regulation of anthocyanins and PAs in grapevine have attracted more and more attention over the past several years.
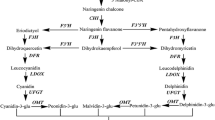
Common steps are shared in the biosynthesis of PAs and anthocyanins in the flavonoid pathway, in which anthocyanidin synthase (ANS; EC 1.14.11.19), one of the key enzymes at the branch point, converts the colorless leucoanthocyanidins to the colored anthocyanidins (Abrahams et al. 2003; Saito et al. 1999; Bogs et al. 2007). In the past years, the ANS gene has been cloned and characterized in many plants (Nakajima et al. 2001; Saito et al. 1999; Shimada et al. 2005; Turnbull et al. 2004). It is regulated developmentally in specific tissues or cell types and affected by stress (Bogs et al. 2005, 2007; Castellarin et al. 2007; Gollop et al. 2001; Deytieux et al. 2007). In Shiraz and Gamay grapevine, the transcript of ANS gene has been shown to be expressed in almost all the plant organs (Boss et al. 1996a, b; Gollop et al. 2001). However, little is yet known of the expression and regulation of ANS at the protein level in grapevine.
Evidence from the red form of Perilla frutescens has supported the fact that ANS is localized in the epidermal cells corresponding to the epidermis-specific accumulation pattern of anthocyanins (Saito and Yamazaki 2002). Previous studies from other vegetative tissues of plants have also suggested that the flavonoid biosynthetic enzymes are localized in the epidermal cells (Saslowsky and Winkel-Shirley 2001; Saslowsky et al. 2005). It is known that the accumulation of PAs and anthocyanins is tissue-specific in grapevine (Boss et al. 1996a, b). Anthocyanins are primarily accumulated in berries, and PAs are present in the leaves, tendrils, flowers, root, and berries of grapevine (Boss et al. 1996b). It is therefore necessary to further investigate the tissue distribution and localization of corresponding enzymes including ANS.
At the subcellular level, Pang et al. (2007) have found that Medicago ANS was localized in the cytosol. Substantial researches of vegetative tissues have demonstrated that flavonoids are biosynthesized in the cytoplasm (Hrazdina and Jensen 1992; Jorgensen et al. 2005; Saslowsky and Winkel-Shirley 2001; Winkel-Shirley 1999), which were transported to the vacuole for accumulation (Gomez et al. 2009; Grotewold and Davies 2008; Kitamura et al. 2004; Poustka et al. 2007; Zhao and Dixon 2010). Some flavonoids and flavonoid enzymes have also been found in the nucleus (Feucht et al. 2004; Polster et al. 2006; Saslowsky et al. 2005; Naoumkina et al. 2007; Yu et al. 2008). The subcellular localization of flavonoid biosynthesis in different grapevine tissues is unclear. Determining the subcellular localization of ANS is very important for understanding the sites of flavonoid metabolism in grapevine.
In this study, the accumulation of PAs and anthocyanins in different grapevine tissues was determined. The expression of ANS and its tissue and subcellular distribution patterns in different grapevine tissues were further studied with Western blotting, immunohistochemical localization, immuno-electron microscopy, and confocal microscopy techniques. The objectives of this study were to increase our understanding of flavonoid biosynthesis mechanism and provide additional insights into engineering this metabolic pathway in grapevine.
Materials and methods
Materials
Grape berries (Vitis vinifera L.cv. Cabernet Sauvignon) were collected from a commercial vineyard during the 2006 and 2007 seasons in the western suburbs of Beijing, People’s Republic of China. Sampling took place per 10 days from 20 days after full bloom to complete ripening (about 120 days after full bloom) according to the method described by Chen et al. (2006). The samples were transported to the laboratory, where the freshly harvested berries were randomly selected on the basis of similar size and the absence of physical injuries or infections. Once separated, the fresh berries were immediately peeled by hands to obtain the skin and flesh samples and then were either embedded for localization of ANS or quickly frozen in liquid nitrogen. These samples were stored at −80°C pending further analysis.
The vegetative tissues used throughout the experiments were derived from 1-year-old potted grapevines of V. vinifera L. cv. Cabernet Sauvignon plants. The plants were raised in a greenhouse at 25°C with 16 h light/8 h dark photoperiod and 65% relative humidity. Plants with uniform vegetative growth were selected for the study, and the experiment was initiated when the plants were with 8∼10 functional leaves (Supplementary Fig. 1). The different plant parts were cut as shown in Supplementary Fig. 1 and were either embedded for localization of ANS or stored at −80°C after freezing in liquid nitrogen until further analysis.
All chemicals were purchased from Sigma unless otherwise noted. The polyclonal antibody against grape ANS was prepared in our laboratory (Wang et al. 2010). The purified anti-ANS immunoglobulin (IgG) was used in the following experiments.
Quantification of total anthocyanins and PAs
Total anthocyanins of the developing grape berry skins were extracted according to the method of Boss et al. (1996a). The content was determined using the pH-differential method described by Orak (2007). Total anthocyanin contents of the vegetative tissues of young grape plants were determined using the methods described by Sheoran et al. (2006) and Zhang et al. (2002). The anthocyanin contents were represented as color value (CV per gram fresh weight).
Total PAs were extracted according to Kennedy et al. (2000) with minor modifications. Tissue samples (0.5 g) were extracted twice with 20 ml 70% acetone solution (containing 0.1% Vc) for 6 h at 25°C in the dark with occasional shaking. After centrifugation at 8,000 rpm for 15 min, the combined supernatants were condensed by rotary evaporation, and then added ddH2O to a total volume of 50 ml. The concentration of PAs was determined by the method of Reddy et al. (2007) using procyanidins as standard.
RNA preparation and RT-PCR analysis
Total RNA was isolated from the different tissues with the method described by Wen et al. (2005). The complementary DNAs (cDNAs) were synthesized according to the manufacturer’s instruction of AMV reverse transcriptase (Promega A3500).
The messenger RNA (mRNA) expression patterns of ANS and Actin1 were examined by RT-PCR. The amplification of Actin1 cDNA was used as an internal control. According to the published sequences of grape ANS and Actin1 (GenBank accession no: EU156063 and AY680701), gene-specific primers for ANS (forward: 5′-AGGGCCTCAGGTTCCAACTAT-3′; reverse: 5′-CTCCCACTCAAGCTGTCCACT-3′) and Actin1 (forward: 5′-GATTCTGGTGATGGTGTGAGT-3′; reverse: 5′-GACAATTTCCCGTTCAGCAGT-3′) were selected for expression analysis. The expected sizes of the PCR products were 292 and 168 bp, respectively. For each primer set, PCR conditions were optimized for Tm using gradient PCR and optimized for the number of cycles by running the above PCR for 36 cycles and removing the reaction product every third cycle from cycles 15 to 36 to obtain the following cycling parameters: ANS, 26 cycles, Tm 50°C; Actin1, 29 cycles, Tm 47°C. The amplified products were separated on 1.5% agarose gel and analyzed with Gene analysis software package (Gene Company). For each sample, the RNA was extracted twice and RT-PCR was performed three times. The identity of all PCR products was confirmed by sequencing analysis at the Beijing Sunbiotech Company (Beijing, China).
Protein extraction and Western blotting analysis
Total proteins were extracted according to the method of Famiani et al. (2000). Sodium dodecyl sulfate polyacrylamide gel electrophoresis was carried out using 8% polyacrylamide gels according to Laemmli (1970). The total proteins (2 μg) were loaded per well. Here, it should be noted that these immunoblotting analyses were based on the same amount of loading proteins in each sample. After electrophoresis, the proteins were elector-transferred to nitrocellulose membranes (0.45 μm, Amersham Life Science) using a transfer apparatus (Bio-Rad) technique described by Isla et al. (1998). Immunological detection of proteins on the NC membrane was carried out using a primary polyclonal ANS antibody in a 1/1,000 dilution at 25°C with an alkaline phosphatase-conjugated anti-rabbit IgG antibody from a goat (1/800 dilution) as a secondary antibody (Sigma-Aldrich). The membrane was stained with 10 ml of 5-bromo-4-chloro-3-indolyl phosphate/nitro blue terazolium (Promega) in the dark, and the reaction was terminated by the addition of double-distilled water. The amount of ANS protein was quantified by scanning the nitrocellulose after immunoblotting with a densitometer using the ImageQuant software. For each sample, the total protein was extracted twice and Western blotting was performed three times.
Immunohistochemical localization of ANS
The process of ANS immunohistochemical localization was done essentially as described previously (Hou and Huang 2005) with some modifications. After the grape tissues were cut, they were fixed overnight in a solution containing 4% paraformaldehyde and 2.5% glutaraldehyde at 4°C, dehydrated with a graded ethanol series, embedded in paraffin, and sectioned into slices. The slides were spread with polylysine before the fixing of the sections. Dried sections were deparaffinized with xylene, hydrated in an ethanol–water series, and then washed by 10 mM phosphate-buffered saline (PBS, pH 7.0, containing 0.2 g/l KCl, 2.19 g/l Na2HPO4⋅12H2O, 0.482 g/l KH2PO4) for 5 min. Slides were blocked for 45 min at room temperature in a blocking solution [10 mM PBS, 0.1% (v/v) Tween-20, 1.5% (w/v) glycine, 5% (w/v) bovine serum albumin (BSA)], then rinsed in a regular salt rinse solution [RSR, 10 mM PBS, 0.1% (v/v) Tween-20, 0.8% (w/v) BSA, 0.88% (w/v) NaCl] for 5 min, and washed briefly with 10 mM PBS supplemented with 0.8% (w/v) BSA (PB) to remove Tween-20. 100 µl primary ANS antibody was added to each slide and incubated at 4°C overnight in a humidity chamber. They were washed vigorously twice in a high-salt rinse solution [10 mM PBS, 0.1% (v/v) Tween-20, 0.1% (w/v) BSA, 2.9% (w/v) NaCl] followed by a 10-min wash with RSR and a brief rinse with 10 mM PBS. Then, 100-µl secondary antibodies were added to the slides [1:100 (v/v)] with dilution of the anti-rabbit IgG–alkaline phosphatase conjugate (1 mg/l Promega) and incubated at room temperature in a humidity chamber. After rinsing twice in RSR and once in water, the slides were developed for approximately 15 min by adding 200 µl of Western Blue stabilized substrate for alkaline phosphatase. As the blue/green color was observed on the sections, they were rinsed with water, dehydrated, and mounted with a cover glass for photographing.
To verify the reliability of the immunolocalization technique and the specificity of the antibody, two types of negative controls were set up. In the first one, the antibody was omitted to test possible unspecific labeling of the goat anti-rabbit IgG–alkaline phosphatase conjugate. In the second one, rabbit preimmune serum was used instead of the rabbit antibody to test the specificity of the antibody. The other procedure of immunolocalization process was preceded as usual. The control experiments were performed more than three times for each sample.
Subcellular immunogold labeling of ANS
Tissue preparation was essentially conducted according to the methods as previously described (Chen et al. 2006). The immunostaining procedure followed that of Ruelland et al. (2003). The ultrathin sections were incubated with TBSTG buffer (100 mM Tris–HCl, pH 7.6, 150 mM NaCl, 0.1% Triton X-100, 3% (w/v) BSA) for 30 min and then crossed directly with affinity-purified IgG to ANS antibody in TBSTG. Following washing three times for 5 min each with TBSTG buffer, the sections were incubated for 1 h with a goat anti-rabbit IgG conjugated to 10-nm gold particles at 1:100 dilutions in the TBSTG buffer at room temperature. The sections were rinsed consecutively with TBSTG and double-distilled water, then stained with 2% uranyl acetate and 4% lead citrate. The ultrathin sections were examined with a JEM-100S electron microscope.
Two types of control were performed in order to test the specificity and reliability of the immunogold labeling. The first one was omission of the antiserum to test the possible unspecific labeling of goat anti-rabbit IgG antibody–gold conjugate, and the second one was the use of the rabbit preimmune serum instead of the rabbit antiserum (incubation of ultrathin sections in preimmune serum instead of antiserum before immunogold labeling) to test the specificity of the antibody. More than three repetitions of the control experiments were preformed for each sample.
Transient expression of ANS–GFP in Arabidopsis protoplasts
For observation of the subcellular localization of ANS in living cells, the full-length ORF of ANS cDNA was PCR-amplified by using primers 5′-CGCTCGAGATGGTGACTTCAGTGG-3′ (forward) and 5′-CCGGAATTCTAGATTTGGAGAGTAGAGC-3′ (reverse). The PCR product was then fused to the upstream of the enhanced green fluorescent protein (GFP; Cormack et al. 1996) at the XhoI (5′ end)/EcoRI (3′ end) sites in the cauliflower mosaic virus 35S-EGFP-Ocs 3′vector (pEZS-NL vector, Dr. Ehrhardt, http://deepgreen.stanford.edu). This vector does not express GFP well without adding coding sequence to the 5′ end of the ORF of GFP; thus, the control cells do not show fluorescence of GFP. Protoplasts were isolated from the leaves of 3- to 4-week-old plants of Arabidopsis (ecotype Columbia) and transiently transformed using PEG essentially according to Ueda et al. (2001). Fluorescence of GFP was observed by a confocal laser scanning microscope (Bio-Rad MRC 1024) after incubation at 23°C for 16 h.
Results
Total anthocyanin and PA contents in different grapevine tissues
In grape skin, anthocyanins were first detected in berries sampled at 70 days after full bloom (Fig. 1), and we considered that this sampling date most represented veraison stage (the onset of ripening) (see Wang et al. 2010; Fig. 3). Following veraison, a substantial increase was observed in anthocyanin levels up to harvest (120 days after full bloom). No anthocyanins were detected in grape berry flesh. The accumulation of PAs in grape skin (Fig. 1) occurred at early development stage, reaching a maximum at 60 days after full bloom, and then declining during ripening. A small amount of PAs was observed in grape berry flesh during grape berry development.
In order to investigate the distribution of anthocyanins and PAs in grapevine vegetative tissues, seven different tissues from young grape plants were selected including the leaves, stems (stem phloem and stem xylem), petioles, leaf buds, stem tips, and roots (Supplementary Fig. 1). As shown in Table 1, anthocyanins were accumulated in the stem tips, stem phloem, leaf buds, and petioles. The maximum concentration of anthocyanins was found in stem P (stem phloem), followed by leaf buds and petioles; the minimum concentration was found in stem tips. There were no anthocyanins detected in other sampled tissues. PAs were detected in all of the sampled tissues. The concentration of PAs in the various organs of young grape plants ranged from 6.5 mg/g FW (fresh weight) to 81.0 mg/g FW. Leaf buds contained the largest amount of PAs (81.0 mg/g FW); the stem tips (41.1 mg/g FW), leaves (36.6 mg/g FW), stem P (32.9 mg/g FW), roots (20.9 mg/g FW), and petioles (18.0 mg/g FW) showed lower concentration; the minimum was found in the stem xylem (6.5 mg/g FW).
The expression of ANS in developing grape berry skin and flesh
As shown in Fig. 2a, the changes of the ANS protein amounts and ANS gene transcript accumulation in developing grape berry skins were detected by Western blotting and RT-PCR. A single polypeptide with molecular mass of approximately 40 kDa was detected specifically from the protein fractions of the grape berry skins (Fig. 2a). The maximum accumulation of the ANS protein and ANS gene transcript occurred at early stage (20 days after full bloom) and then decreased gradually 30 to 60 days after full bloom. Following this period, the ANS amounts increased rapidly at veraison (70 days after full bloom), then decreased again during the later part of ripening stage.
The expression of ANS protein and ANS gene transcript accumulation in the skin and flesh of developing grape berries. a Changes of expression of ANS protein and transcript accumulation of ANS gene in developing grape berry skins. Fragment of 292 bp (ANS) was amplified. As control, fragment of actin 1 (168 bp) was amplified simultaneously. Immunodetection of ANS in the soluble crude extracts throughout the berry development showed that only one band of polypeptide was immunodetected (about 40 kDa). The identical amount of protein (2 µg) was loaded per lane. b Changes of ANS protein and ANS gene transcript accumulation in berry flesh during grape berry development. The data of signal intensity in a and b are means from three analyses of one of three independent replicates. Bars are standard errors (n = 3). Different letters indicate a statistical difference at P ≤ 0.05 among samples according to Duncan’s multiple range tests
Changes in ANS accumulation in grape berry flesh samples showed similar pattern to that in skins (Fig. 2b). The ANS protein and ANS gene transcript showed maximum accumulation at 20 days after full bloom. There was then a reduction during the successive development stage. Following this period, a coordinate increase was observed in accumulation of the ANS protein and ANS gene transcript in the sample (70 days after full bloom) at approximately the time of veraison. The accumulation reached to a higher level at 80 days after full bloom, then decreased throughout the remainder of berry development.
The expression of ANS in grapevine vegetative tissues
As shown in Fig. 3, the maximum of transcript accumulation was found in the leaves. The lower level of ANS gene transcript accumulation was found in other tissues, and the minimum of that was investigated in stem tip and roots. The ANS protein was accumulated in leaves, stems, petioles, and leaf buds. The immune signal level in leaves and stems reached the maximum, and a significant accumulation of ANS protein was also detected in the petioles and leaf buds. There was no ANS detected in stem tip and roots.
The accumulation of ANS gene transcript and ANS protein in different grapevine vegetative tissues. The upper artwork shows the RT-PCR result of ANS. The bottom artwork shows Western blotting analysis of ANS in different tissues of young grape plants. The data of signal intensity are means from three analyses of one of two independent replicates. Bars are standard errors (n = 3). Different letters indicate a statistical difference at P ≤ 0.05 among samples according to Duncan’s multiple range tests
Immunohistochemical localization of ANS in different grapevine tissues
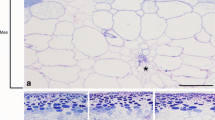
In order to observe the exact tissue localization of ANS in grapevine, five tissues were used including grape berry, leaves, stems, leaf buds, and roots. Endogenous ANS was immunohistochemically localized in grapevine tissues using the ANS polyclonal antibody (Fig. 4a–o). The blue-green color is the ANS immune signal, representing the natural distribution of endogenous ANS in the grapevine tissues. The results in Fig. 4a–c, d–f, g–i, j–l, and m–o showed the transverse sections of the grapevine tissues—grape berry, leaves, stems, leaf buds, and roots, respectively. Because maximum ANS protein was detected in grape berries at early stage, the grape berries at 20 days after full bloom were used. As shown in Fig. 4a, strong ANS signals were detected in the grape seed, mesocarp vascular bundles, and exocarp, while signals were also localized in mesocarp parenchyma cells. In grape seed (Fig. 4a), strong ANS signals were observed in epidermis, outer integument, inner integument, and the cells around the raphe. Slight ANS signals were distributed in medium integument and little ANS signals were investigated in the endosperm. In the grape leaves (Fig. 4d), intensive ANS signals were found in mesophyll and vein. In mesophyll (Fig. 4d), the ANS signals were mainly present in palisade and spongy tissue, while little ANS signals were investigated in the upper and lower epidermis. In leaf vein (Fig. 4d), the ANS signals were mainly localized in the vascular bundles, and obvious ANS signals were also found in the epidermis. In the stem (Fig. 4g), the ANS signals were mainly localized in the primary phloem and pith ray, and there were signals present in the epidermis of the stems, too. In leaf buds (Fig. 4j), the obvious ANS signals were detected in the growth point, leaf primordium, and young leaves. There was no obvious ANS signal in the grapevine roots (Fig. 4m).
Immunohistochemical localization of ANS in different organs of grapevine. a The distribution of ANS in grape berries; d tissue localization of ANS in grape leaves (mesophyll, vein); g immunohistochemical localization of ANS in grapevine stems; j immunohistochemical localization of ANS in grapevine leaf buds; m tissue localization of ANS in grapevine roots. b–c, e–f, h–i, n–o Negative controls of immunohistochemical localization of ANS in grapevine berries, leaves, stems, leaf buds, and roots, respectively. b, e, h, n Without the primary ANS antibody; c, f, i, o the primary ANS antibody was substituted with the preimmune serum of rabbit. No ANS signal was observed in these controls. EP exocarp, MP mesocarp, VB vascular bundle, Epi epidermis, OI outer integument, II inner integument, R raphe, Em endosperm, Sd seed, UE upper epidermis, LE lower epidermis, PT palisade tissue, ST spongy tissue, PPh primary phloem, PX protoxylem, PiR pith ray, CoP cortex parenchyma cells, GP growth point, LP leaf primordium, YL young leaf, Co cortex, En endoderm, Vc vascular cylinder. Bar = 10 µm
Additionally, little ANS signal was found in the controls without the antiserum (Fig. 4b, e, h, k, n), or in any of the preimmune serum controls (Fig. 4c, f, i, l, o), indicating that the antibody was specific and the unspecific signal was negligible.
Subcellular localization of ANS in grapevine
In order to determine the subcellular localization of ANS in grapevine, four different tissues were chosen to detect the subcellular distribution of ANS in grapevine, including the vascular tissue in the mesocarp of grape berry, the leaves, stems, and leaf buds. The subcellular localization of ANS in the mesocarp vascular tissue cells was shown in Fig. 5. In the sieve element (Fig. 5a, b), the gold particles representing ANS were mainly localized in the cytoplasm, and no other organelles were observed in the cells. For the companion cells (Fig. 5c, d), the cytoplasm was heavily labeled by gold particles; a few particles were also localized in the nucleus.
Immunogold electron microscope localization of ANS in mesocarp vascular tissue of grape berry. a-b Subcellular localization of ANS in sieve element. b The amplified portion of the boxed-in area of a, which shows that the ANS was mainly localized in the cytoplasm in sieve element. c-d Immunogold localization of ANS in companion cell. d Magnified image of the boxed-in area of c. The gold particles are mainly distributed in the cytoplasm and some also were found in the nucleus. e-f Negative controls of immunolocalization of ANS. e Antibody omission control; f preimmune serum control. No gold particles were detected. SE sieve element, Cyt cytoplasm, CW cell wall, V vacuole, CC companion cell, N nucleus. Bars = 0.5 µm
The subcellular localization of ANS in grapevine leaves was shown in Fig. 6. ANS visualized by gold particles were mainly localized in the cytoplasm. The cytoplasm localization of ANS in leaves was confirmed by the transient expression of ANS–GFP in the mesophyll protoplast of Arabidopsis (Supplementary Fig. 2). In the stem cells (Fig. 7a, b), gold particles representing ANS were also primarily found in the cytoplasm; few particles were localized in other organelles. For the leaf bud cells (Fig. 7c–e), the gold particles representing ANS were mainly localized in the cytoplasm and nucleus.
Immunogold electron microscope localization of ANS in the leaves of young grape plants. a Ultrastructure of grapevine leaf cells; b–c ANS visualized by gold particles mainly resides in the cytoplasm (×50,000); d-e negative controls of immunogold electron microscopy localization of ANS in grapevine leaf cells. d Without the primary ANS antibody; e the primary ANS antibody was substituted with the preimmune serum of rabbit. No gold particle was observed in these control cells. CW cell wall, Cyt cytoplasm, V vacuole, Chl chloroplast. Bar = 10 µm in a, 0.5 µm in b–e
Immunogold electron microscope localization of ANS in the stem (a–b), and leaf bud (c–e) of young grape plants. a Ultrastructure of grapevine stem cells; b ANS visualized by gold particles mainly resides in the cytoplasm (×60,000). Bar = 2 µm in a, 0.2 µm in b; c ultrastructure of grapevine leaf bud cells; c–e gold particles representing ANS mainly resides in the cytoplasm (d) and the nuclear (e). Bar = 10 µm in c, 0.5 µm in d–e; CW cell wall, Cyt cytoplasm, V vacuole, Chl chloroplast, N nuclear
Substantially, no gold particles were found in the controls without the antibody (Figs. 5e and 6d) and in any of the preimmune serum controls (Figs. 5f and 6e), indicating that the antibodies were specific and unspecific labeling was negligible.
Discussion
The ANS gene has been detected to be expressed in almost all the organs of Shiraz grapevine: berry skin, berry flesh, seed, leaves, tendril, and green cane (Boss et al. 1996a, b). Gollop et al. (2001) have also found that the ANS promoter fusion was expressed in the leaves, meristem tissues, stems, and root of Gamay grapevine plants. Our present results from Cabernet Sauvignon grapevine revealed that the ANS protein was accumulated in the berry skin, flesh, seed, leaves, stems (stem P and stem X), petioles, and leaf buds, but not in the stem tip and root. Boss et al. (1996b) have suggested that the expression of flavonoid biosynthesis genes varied according to the cultivar. It indicates that the differences in the patterns of ANS expression in Cabernet Sauvignon, Shiraz, and Gamay may be due to varietal influences. Charrier et al. (1998) have also observed the difference of flavanone 3-hydroxylase expression and flavonoid localization in the nodules of three legume plants. In berry skin and flesh, it was noteworthy that the expression of the ANS gene and protein is developmental-dependent (Fig. 2), which is consistent with previous studies (Boss et al. 1996a, 2005, 2007; Deytieux et al. 2007), and the ANS was subjected to both transcriptional and translational regulation. In grape skin, the expression of ANS corresponds well to the accumulation of PAs and anthocyanins. However, no anthocyanins and a small amount of PAs were detected in berry flesh. It might be that the PAs synthesized in fruit flesh were transported to the grape berries skin or some other tissues to accumulate (Braidot et al. 2008a).
As to ANS expression in grapevine vegetative tissues, a large amount of ANS protein and ANS gene transcript accumulation was observed in the leaves and stem phloem (Fig. 3). In grapevine leaves, ANS was mainly involved in the PAs biosynthesis; in stem phloem, ANS responded to both anthocyanins and PAs synthesis (Table 1). In leaf buds and petiole, the accumulation of ANS protein did not correspond to the levels of mRNA. A possible post-transcriptional regulatory mechanism may be involved, whereas in the stem xylem, a great quantity of ANS was investigated, but little PAs and anthocyanins were detected. Several authors have demonstrated the existence of a transport mechanism for several polyphenolic compounds, including flavonoids in grapevine (Fischer 1968; Berhow and Vandercook 1989). The flavonoids synthesized in stem xylem might be transported to other tissues such as the root and stem tip, where no ANS protein was detected, but numerous of PAs and anthocyanins were accumulated (Table 1). The accumulation of flavonoids in different grapevine tissues is the result of in situ biosynthesis and a translocation process from the synthesis sites such as the fruit and leaves to the target organs (stem tip, root, and so on). However, the mechanism of flavonoid transport between different grapevine tissues needs to be made clear further.
Western blotting analyses have revealed ANS protein mainly in the fruit, leaves, stems, and leaf buds (Fig. 3). To determine the exact localization of the ANS protein in these organs, an immunohistochemical analysis of fixed samples was performed with the anti-ANS IgG, which has high specificity and sensitivity (Wang et al. 2010). In grape berries, ANS was found to be distributed ubiquitously. The distribution of ANS in the excarp and mesocarp has been discussed in our previous study (Wang et al. 2010). Here, we focused on the location of ANS in seed. Previous studies have suggested that flavonoids are localized not only in surface tissues of the seed coat, but also in the inner layers (Adams 2006; Pinelo et al. 2006; Cadot et al. 2006). Our present result showed that ANS was localized in the epidermis, outer integument, inner integument, and the cells around the raphe, which corresponded well to the distribution of PAs in grape seed (Cadot et al. 2006). It indicates that PAs accumulated in the different structures of grape seed are biosynthesized in situ. For vegetative organs, tissue-specific distribution of ANS in different organs was observed. Previous studies have detected that flavonoid biosynthetic enzymes are specifically localized in the epidermal cells in Arabidopsis and Perilla (Saito and Yamazaki 2002; Saslowsky et al. 2005; Saslowsky and Winkel-Shirley 2001). However, we found different distributed patterns of ANS in grapevine leaves and stems (Fig. 4d, g). These discrepancies imply that there exist different mechanisms of flavonoid biosynthesis and accumulation in different species (Braidot et al. 2008a). Although the physiological functions of anthocyanins and PAs biosynthesized in these tissues are not known, we cannot exclude the proposal that the sites of accumulation are not necessarily the sites of biosynthesis and that the transport of flavonoid end products between cells is involved (Hrazdina and Jensen 1992; Kaltenbach et al. 1999; Braidot et al. 2008a). The anthocyanins and PAs are transported to the epidermis to play an important role in protecting the tissues from unavoidable damages in grapevine. This is the first time to investigate the distribution of the ANS enzyme in leaf buds, but the roles the enzyme plays in these tissues need to be made clear. Meanwhile, there was little ANS signal observed in the root, which corresponded well to the result of Western blotting analysis.
Interestingly, the ANS signals were obviously distributed in vascular bundles (Fig. 4). Flavonoid 3′5′-hydroxylase has been only detected in the phloem in cotyledons and flower buds from Catharanthus roseus (Kaltenbach et al. 1999). It is suggested that there are some other flavonoid pathway enzymes distributed in the vascular bundles of grapevine, which combine with ANS to biosynthesize different flavonoid end products in situ. Many studies have confirmed the distribution of flavonoids in transport tissues (Gould et al. 2000; Gholami 2004). In addition, the vascular bundle was considered to function in the long distance transport of water, signaling molecules, and for nutrient reallocation (Oparka and Turgeon 1999; Bondada et al. 2005), and it is suggested that the flavonoids are transported from the vascular bundle to the different tissues/organs in grape plants subjected to biotic and abiotic stresses. Recent findings have confirmed flavonoid occurrence in vascular bundles, where they move towards the root tip (Buer et al. 2007). The identification of the flavonoid translocator BTL homolog associated to vascular bundles further indicated the existence of flavonoid transport in grapevine (Braidot et al. 2008b).
At the subcellular level, distributed patterns of ANS in the cytoplasm/plastid/nucleus have been found in the exocarp of developing grape berries in our previous study (Wang et al. 2010). However, in mesocarp vascular tissues, ANS was mainly localized in the cytoplasm, and the nuclear distributed pattern of ANS was only observed in the companion cells (Fig. 5). The data show that ANS exhibits differences in subcellular localization between fruit skin and flesh, but its functions in different sites of grape berry need further study. Substantial studies have confirmed the cytoplasm ER-localized pattern of flavonoid pathway in vegetative tissues and suggested that flavonoid metabolism is catalyzed, not by free-floating “soluble” enzymes, but via one or more membrane-associated multienzyme complexes (Hrazdina and Jensen 1992; Jorgensen et al. 2005; Saslowsky and Winkel-Shirley 2001; Winkel-Shirley 1999). Here in the sampled grapevine vegetative tissues, the ANS enzyme was also mainly localized in the cytoplasm. It appeared that ANS was with other flavonoid enzymes organized into a complex to facilitate the synthesis of anthocyanins and PAs in the cytoplasm of different grapevine tissues. Pang et al. (2007) have found that Medicago ANS was localized in the cytosol. But the experiment was performed by particle bombardment of single-enzyme construct into tobacco leaf epidermal cells, not into the natural milieu of the endothelial layer of the Medicago seed coat. It cannot rule out possible complex formation when all the enzymes are present together in the same cell. The nuclear distributed pattern of ANS was also detected in grape berry vascular bundle cells and leaf bud cells. It was not surprising. Several other recent reports have described the localization of some flavonoid enzymes in the nucleus and suggested that these specially localized enzymes are responsible for in situ synthesis of flavonoids in the nucleus (Chen et al. 2006; Naoumkina et al. 2007; Saslowsky et al. 2005; Tian et al. 2008; Yu et al. 2008). Much evidence has described the accumulation of flavonoids in the nucleus, which serves to protect DNA from UV and oxidative damage (Feucht et al. 2004; Polster et al. 2006; Saslowsky et al. 2005) and so on.
In conclusion, ANS was mainly distributed in grape berries, leaves, stems, petioles, and leaf buds in Cabernet Sauvignon grapevine. The ANS exhibits tissue-specific localization in different organs. At the subcellular level, the cytoplasm and nucleus distributed patterns of ANS were observed in different grapevine tissue cells. No matter what mechanisms were involved, the elucidation of the compartmentations of ANS in different grapevine tissues was of particular interest in the future.
Abbreviations
- ANR:
-
Anthocyanidin reductase
- ANS:
-
Anthocyanidin synthase
- BSA:
-
Bovine serum albumin
- F3′5′H:
-
Flavonoid 3′, 5′-hydroxylase
- IgG:
-
Immunoglobulin fractions
- PAs:
-
Proanthocyanidins
- PBS:
-
Phosphate-buffered saline
- RSR:
-
Regular salt rinse solution
References
Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35:624–636
Adams DO (2006) Phenolics and ripening in grape berries. Am J Enol Vitic 57:249–256
Berhow MA, Vandercook CE (1989) Biosynthesis of naringin and prunin in detached grapefruit. Phytochemistry 28:1627–1630
Bogs J, Downey MO, Harvey JS, Ashton AR, Tanner GJ, Robinson SP (2005) Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol 139:652–663
Bogs J, Jaffe FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143:1347–1361
Bondada BR, Matthews MA, Shackel KA (2005) Functional xylem in the post-veraison grape berry. J Exp Bot 56:2949–2957
Boss PK, Davies C, Robinson SP (1996a) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111:1059–1066
Boss PK, Davies C, Robinson SP (1996b) Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol Biol 32:565–569
Braidot E, Zancani M, Petrussa E, Peresson C, Bertolini A, Patui S, Macrì F, Vianello A (2008a) Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.). Plant Signal Behav 3:626–632
Braidot E, Petrussa E, Bertolini A, Peresson C, Ermacora P, Loi N, Passamonti S, Terdoslavich M, Macri F, Vianello A (2008b) Evidence for a putative flavonoid translocator similar to mammalian bilitranslocase in grape berries (Vitis vinifera. L.) during ripening. Planta 228:203–213
Buer CS, Muday GK, Djordjevic MA (2007) Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol 145:478–490
Cadot Y, Minana-castelloa MT, Chevalier M (2006) Anatomical, histological, and histochemical changes in grape seeds from Vitis vinifera L. cv Cabernet Franc during fruit development. J Agric Food Chem 54:9206–9215
Castellarin SD, PfeiVer A, Sivilotti P, Degan M, Peterlunger E, Di Gaspero G (2007) Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ 30:1381–1399
Chamkha M, Cathala B, Cheynier V, Douillard R (2003) Phenolic composition of champagnes from Chardonnay and Pinot Noir vintages. J Agric Food Chem 51:3179–3184
Charrier B, Trinh H, Poirier S, Kondorosi A, Ratet P (1998) Flavanone 3-hydroxylase (F3H) expression and flavonoid localization in nodules of three legume plants reveal distinct tissue specificities. Mol Plant Microbe In 11:924–932
Chen JY, Wen PF, Kong WF, Pan QH, Wan SB, Huang WD (2006) Changes and subcellular localizations of the enzymes that involved in phenylpropanoid metabolism during grape berry development. J Plant Physiol 163:115–127
Cormack BP, Valdivia RH, Falkow S (1996) FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38
Deytieux C, Geny L, Lapaillerie D, Claverol S, Bonneu M, Donèche B (2007) Proteome analysis of grape skins during ripening. J Exp Bot 58:1851–1862
Famiani F, Walker RP, Tecsi L, Chen ZH, Proietti P, Leegood RC (2000) An immunohistochemical study of the compartmentation of metabolism during the development of grape berries. J Exp Bot 51:675–683
Feucht W, Dithmar H, Polster J (2004) Nuclei of tea flowers as targets for flavanols. Plant Biol 6:696–701
Fischer JFA (1968) Procedure for obtaining radioactive naringin from grapefruit leaves fed L-phenylalanine-14C. Phytochemistry 7:769–771
Gholami M (2004) Biosynthesis of anthocyanins in Shiraz grape berries. Acta Hortic (ISHS) 640:353–359
Gollop R, Farhi S, Perl A (2001) Regulation of the leucoanthocyanidin dioxygenase gene expression in Vitis vinifera. Plant Sci 161:579–588
Gomez C, Terrier N, Torregrosa L, Vialet S, Fournier-Level A, Clotilde Verriès C, Souquet JM, Mazauric JP, Klein M, Cheynier V, Ageorges A (2009) Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol 150:402–415
Gould KS, Markham KR, Smith RH, Goris JJ (2000) Functional role of anthocyanins in the leaves of Quintinia serrata A Cunn. J Exp Bot 51:1107–1115
Grotewold E, Davies K (2008) Trafficking and sequestration of anthocyanins. Nat Prod Commun 3:1251–1258
Halliwell B, Rafter J, Jenner A (2005) Health promotion by flavonoids, tocopherols, tocotrienols, and other phenols: direct or indirect effects? Antioxidant or not? Am J Clin Nutr 81:268s–276s
Harborne JB, Williams CA (2000) Advances in flavonoid research since 1992. Phytochemistry 55:481–504
Hou ZX, Huang WD (2005) Immunohistochemical localization of IAA and ABP1 in strawberry shoot apexes during floral induction. Planta 222:678–687
Hrazdina G, Jensen RA (1992) Spatial organization of enzymes in plant metabolic pathways. Ann Rev Plant Physiol Plant Mol Biol 43:241–267
Isla MI, Vattuone MA, Sampietro AR (1998) Essential group at the active site of Frapaeolum invertase. Phytochemistry 47:1189–1193
Jorgensen K, Rasmussen AV, Morant M, Nielsen AH, Bjarnholt N, Zagrobelny M, Bak S, Møller BL (2005) Metabolon formation and metabolic channeling in the biosynthesis of plant natural products. Curr Opin Plant Biol 8:280–291
Kaltenbach M, Schroder G, Schmelzer E, Lutzl V, Schroder J (1999) Flavonoid hydroxylase from Catharanthus roseus: cDNA, heterologous expression, enzyme properties and cell-type specific expression in plants. Plant J 19:183–193
Kennedy JA, Troup GJ, Pilbrow JR, Hutton DR, Hewitt D, Hunter CR, Ristic R, Iland PG, Jones GP (2000) Development of seed polyphenols in berries from Vitis vinifera L.cv. Shiraz. Aust J Grape Wine Res 6:244–254
Kitamura S, Shikazono N, Tanaka A (2004) TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 37:104–114
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Nakajima J, Tanaka Y, Yamazaki M, Saito K (2001) Reaction mechanism from leucoanthocyanidin to anthocyanidin 3-glucoside, a key reaction for coloring in anthocyanin biosynthesis. J Biol Chem 276:25797–25803
Naoumkina M, Farag MA, Sumner LW, Tang Y, Liu CJ, Dixon RA (2007) Different mechanisms for phytoalexin induction by pathogen and wound signals in Medicago truncatula. Proc Natl Acad Sci U S A 104:17909–17915
Oparka KJ, Turgeon R (1999) Sieve elements and companion cells—traffic control centers of the phloem. Plant Cell 11:739–750
Orak HH (2007) Total antioxidant activities, phenolics, anthocyanins, polyphenoloxidase activities of selected red grape cultivars and their correlations. Sci Hortic 111:235–241
Pang YZ, Peel GJ, Wright E, Wang ZY, Dixon RA (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145:601–615
Pinelo M, Arnous A, Meyer AS (2006) Upgrading of grape skins: significance of plant cell-wall structural components and extraction techniques for phenol release. Trends Food Sci Tech 17:579–590
Polster J, Dithmar H, Burgemeister R, Friedemann G, Feucht W (2006) Flavonoids in plant nuclei: detection by laser microdissection and pressure catapulting (LMPC), in vivo staining, and uv–visible spectroscopic titration. Physiol Plantarum 128:163–174
Poustka F, Irani NG, Feller A, Lu Y, Pourcel L, Frame K, Grotewold E (2007) A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol 145:1323–1335
Reddy AM, Reddy VS, Scheffler BE, Wienand U, Reddy AR (2007) Novel transgenic rice overexpressing anthocyanidin synthase accumulates a mixture of flavonoids leading to an increased antioxidant potential. Metab Eng 9:95–111
Ruelland E, Campalans A, Selman-Housein G, Puigdomenech P, Rigau J (2003) Cellular and subcellular localization of the lignin biosynthetic enzymes caffeic acid-O-methyltransferase, cinnamyl alcohol dehydrogenase and cinnamoyl-coenzyme A reductase in two monocots, sugarcane and maize. Plant Physiol 117:93–98
Saito K, Yamazaki M (2002) Biochemistry and molecular biology of the late-stage of biosynthesis of anthocyanin: lessons from Perilla frutescens as a model plant. New Phytol 155:9–23
Saito K, Kobayshi M, Gong ZZ, Tanaka Y, Yamazaki M (1999) Direct evidence for anthocyanidin synthase as a 2-Oxoglutarate-dependent oxygenase: molecular cloning and functional expression of cDNA from a red forma of perilla frutescens. Plant J 17:181–189
Saslowsky D, Winkel-Shirley B (2001) Localization of flavonoid enzymes in Arabidopsis roots. Plant J 27:37–48
Saslowsky DE, Warek U, Winkel BS (2005) Nuclear localization of flavonoid enzymes in Arabidopsis. J Biol Chem 280:23735–23740
Sheoran IS, Dumonceaux T, Datla R, Vipen K, Sawhney VK (2006) Anthocyanin accumulation in the hypocotyl of an ABA-over producing male-sterile tomato (Lycopersicon esculentum) mutant. Physiol Plantarum 127:681–689
Shimada S, Inoue YT, Sakuta M (2005) Anthocyanidin synthase in non-anthocyanin-producing Caryophyllales species. Plant J 44:950–959
Tian L, Wan SB, Pan QH, Zheng JC, Huang WD (2008) A novel plastid localization of chalcone synthase in developing grape berry. Plant Sci 175:431–436
Turnbull JJ, Nakajima J, Welford RW, Yamazaki M, Saito K, Schofield CJ (2004) Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3′-hydroxylase. J Biol Chem 279:1206–1216
Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. J Exp Bot 20:4730–4741
Wang HL, Wang W, Zhang P, Pan QH, Zhan JC, Huang WD (2010) Gene transcript accumulation, tissue and subcellular localization of anthocyanidin synthase (ANS) in developing grape berries. Plant Sci. 10.1016/j.plantsci.2010.04.002
Wen PF, Chen JY, Kong WF, Pan QH, Wan SB, Huang WD (2005) Salicylic acid induced the expression of phenylalanine ammonia-lyase gene in grape berry. Plant Sci 169:928–934
Winkel-Shirley B (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Plant Physiol 107:142–149
Yu XH, Chen MH, Liu CJ (2008) Nucleocytoplasmic-localized acyltransferases catalyze the malonylation of 7-O-glycosidic (iso)flavones in Medicago truncatula. Plant J 55:382–396
Zhang W, Curtin C, Kikuchi M, Franco C (2002) Integration of jasmonic acid and light irradiation for enhancement of anthocyanin biosynthesis in Vitis vinifera suspension cultures. Plant Sci 162:459–468
Zhao J, Dixon RA (2010) The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci 15:72–80
Acknowledgments
This research was supported by the National Natural Science Foundation of China (grant numbers 30871747) and major program of Beijing Municipal Science and Technology Commission (no. D07060500160701).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Hanns H. Kassemeyer
Huiling Wang and Wei Wang contribute equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Scheme of the grapevine seedling to show the experimental sampling sites, including: leaf bud, stem tip, leaf, root, stem (stem phloem and stem xylem). (GIF 345 kb)
Supplementary Fig. 2
Subcellular localization of ANS–GFP fusion protein in the Arabidopsis protoplasts. ANS–GFP fusion protein was transiently expressed from its chimeric gene, and the localization of the fusion proteins was detected by confocal laser scanning microscopical imaging of GFP fluorescence. a–c Transient expression of the green fluorescent protein (GFP)–ANS fusion protein showing that the fusion protein was localized to the cytoplasm. d–f GFP of PEZS-NL empty vector expressed in the Arabidopsis protoplast, and no fluorescences were observed in the control protoplasts. The laser-scanning confocal microscopy images are the ANS–GFP fluorescence (a, d), merged images (b, e), and bright-field image (c, f) (GIF 128 kb)
Rights and permissions
About this article
Cite this article
Wang, H., Wang, W., Li, H. et al. Expression and tissue and subcellular localization of anthocyanidin synthase (ANS) in grapevine. Protoplasma 248, 267–279 (2011). https://doi.org/10.1007/s00709-010-0160-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-010-0160-6